Molecular Dynamics Simulations of a Catalytic Multivalent Peptide–Nanoparticle Complex
Abstract
:1. Introduction
2. Results
2.1. MD of Free Peptide and Free Au-MPC in Water
2.1.1. MD of Peptide in Water
2.1.2. MD of Au-MPC in Water
2.2. Binding Poses and Binding Energies of Peptide-Au-MPC Complexes
2.3. MD Simulations of Peptide-Au-MPC Complexes
3. Discussion
4. Materials and Methods
4.1. Set-Up of the H1 and H3 Peptide Systems
4.2. Set-Up of the Nanocluster Model Au144(L)60 (L = S(CH2)8NH2+)
4.3. Set-Up of the Flexible Docking Simulations
4.4. Set-Up of the MD Refinement of Supramolecular Complexes
4.5. Analysis of MD Trajectories
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pasquato, L.; Pengo, P.; Scrimin, P. Nanozymes: Functional Nanoparticle-based Catalysts. Supramol. Chem. 2005, 17, 163–171. [Google Scholar] [CrossRef]
- Manea, F.; Houillon, F.B.; Pasquato, L.; Scrimin, P. Nanozymes: Gold-nanoparticle-based transphosphorylation catalysts. Angew. Chem. 2004, 43, 6165–6169. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, H.; Zhang, Z.; Wang, E.; Dong, S. Nanozyme: An emerging alternative to natural enzyme for biosensing and immunoassay. Trends Anal. Chem. 2018, 105, 218–224. [Google Scholar] [CrossRef]
- Huang, Y.; Ren, J.; Qu, X. Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications. Chem. Rev. 2019, 119, 4357–4412. [Google Scholar] [CrossRef]
- Jiang, D.; Ni, D.; Rosenkrans, Z.T.; Huang, P.; Yan, X.; Cai, W. Nanozyme: New horizons for responsive biomedical applications. Chem. Soc. Rev. 2019, 48, 3683–3704. [Google Scholar] [CrossRef]
- Wang, P.; Wang, T.; Hong, J.; Yan, X.; Liang, M. Nanozymes: A New Disease Imaging Strategy. Front. Bioeng. Biotechnol. 2020, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Stasyuk, N.; Smutok, O.; Demkiv, O.; Prokopiv, T.; Gayda, G.; Nisnevitch, M.; Gonchar, M. Synthesis, Catalytic Proper-ties and Application in Biosensorics of Nanozymes and Electronanocatalysts: A Review. Sensors 2020, 20, 4509. [Google Scholar] [CrossRef]
- Gabrielli, L.; Prins, L.J.; Rastrelli, F.; Mancin, F.; Scrimin, P. Hydrolytic Nanozymes. Eur. J. Org. Chem. 2020, 32, 5044–5055. [Google Scholar] [CrossRef]
- Das, R.; Landis, R.F.; Tonga, G.Y.; Cao-Milán, R.; Luther, D.C.; Rotello, V.M. Control of Intra-versus Extracellular Bioorthogonal Catalysis Using Surface-Engineered Nanozymes. ACS Nano 2019, 13, 229–235. [Google Scholar] [CrossRef]
- Cao-Milán, R.; He, L.D.; Shorkey, S.; Tonga, G.Y.; Wang, L.-S.; Zhang, X.; Uddin, I.; Das, R.; Sulak, M.; Rotello, V.M. Modulating the catalytic activity of enzyme-like nanoparticles through their surface functionalization. Mol. Syst. Des. Eng. 2017, 2, 624–628. [Google Scholar] [CrossRef]
- Cao-Milán, R.; Gopalakrishnan, S.; He, L.D.; Huang, R.; Wang, L.-S.; Castellanos, L.; Luther, D.C.; Landis, R.F.; Makabenta, J.M.V.; Li, C.-H.; et al. Thermally Gated Bio-orthogonal Nanozymes with Supramolecularly Confined Porphyrin Catalysts for Antimicrobial Uses. Chem 2020, 6, 1113–1124. [Google Scholar] [CrossRef]
- Lou-Franco, J.; Das, B.; Elliott, C.; Cao, C. Gold Nanozymes: From Concept to Biomedical Applications. Nano-Micro Lett. 2021, 13, 1–36. [Google Scholar] [CrossRef]
- Pieters, G.; Prins, L.J. Catalytic self-assembled monolayers on gold nanoparticles. New J. Chem. 2012, 36, 1931–1939. [Google Scholar] [CrossRef]
- Taguchi, T.; Isozaki, K.; Miki, K. Enhanced Catalytic Activity of Self-Assembled-Monolayer-Capped Gold Nanoparticles. Adv. Mater. 2012, 24, 6462–6467. [Google Scholar] [CrossRef]
- Higaki, T.; Li, Y.; Zhao, S.; Li, Q.; Li, S.; Du, X.S.; Yang, S.; Chai, J.; Jin, R. Atomically Tailored Gold Nanoclusters for Catalytic Application. Angew. Chem. 2019, 58, 8291–8302. [Google Scholar] [CrossRef]
- Duncan, K.L.; Ulijn, R.V. Short Peptides in Minimalistic Biocatalyst Design. Biocatalysis 2015, 1, 67–81. [Google Scholar] [CrossRef] [Green Version]
- Pengo, P.; Baltzer, L.; Pasquato, L.; Scrimin, P. Substrate modulation of the activity of an artificial nanoesterase made of peptide-functionalized gold nanoparticles. Angew. Chem. 2007, 46, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Levy, R. Peptide-capped gold nanoparticles: Towards artificial proteins. ChemBioChem Eur. J. Chem. Biol. 2006, 7, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Mikolajczak, D.J.; Koksch, B. Peptide-Gold Nanoparticle Conjugates as Sequential Cascade Catalysts. ChemCatChem 2018, 10, 4324–4328. [Google Scholar] [CrossRef]
- Duchesne, L.; Wells, G.; Fernig, D.G.; Harris, S.A.; Lévy, R.; Wells, G. Supramolecular Domains in Mixed Peptide Self-Assembled Monolayers on Gold Nanoparticles. ChemBioChem 2008, 9, 2127–2134. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Wang, Y.; Zhao, L.; Liu, R.; Gao, F.-P.; Gao, L.; Gao, X. Peptide protected gold clusters: Chemical synthesis and biomedical applications. Nanoscale 2016, 8, 12095–12104. [Google Scholar] [CrossRef] [PubMed]
- Kyrychenko, A. NANOGOLD decorated by pHLIP peptide: Comparative force field study. Phys. Chem. Chem. Phys. 2015, 17, 12648–12660. [Google Scholar] [CrossRef] [PubMed]
- Colangelo, E.; Chen, Q.; Davidson, A.M.; Paramelle, D.; Sullivan, M.B.; Volk, M.; Levy, R. Computational and Experi-mental Investigation of the Structure of Peptide Monolayers on Gold Nanoparticles. Langmuir ACS J. Surf. Colloids 2017, 33, 438–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samieegohar, M.; Sha, F.; Clayborne, A.Z.; Wei, T. ReaxFF MD Simulations of Peptide-Grafted Gold Nanoparticles. Langmuir 2019, 35, 5029–5036. [Google Scholar] [CrossRef]
- Monti, S.; Barcaro, G.; Sementa, L.; Carravetta, V.; Ågren, H. Dynamics and self-assembly of bio-functionalized gold nanoparticles in solution: Reactive molecular dynamics simulations. Nano Res. 2018, 11, 1757–1767. [Google Scholar] [CrossRef]
- Chew, A.K.; Dallin, B.C.; Van Lehn, R.C. The Interplay of Ligand Properties and Core Size Dictates the Hydrophobi-city of Monolayer-Protected Gold Nanoparticles. ACS Nano 2021, 15, 4534–4545. [Google Scholar] [CrossRef]
- Chew, A.K.; Van Lehn, R.C. Effect of Core Morphology on the Structural Asymmetry of Alkanethiol Monolayer Pro-tected Gold Nanoparticles. J. Phys. Chem. C 2018, 122, 26288–26297. [Google Scholar] [CrossRef]
- Van Lehn, R.C.; Alexander-Katz, A. Structure of Mixed-Monolayer-Protected Nanoparticles in Aqueous Salt Solution from Atomistic Molecular Dynamics Simulations. J. Phys. Chem. C 2013, 117, 20104–20115. [Google Scholar] [CrossRef]
- Kyrychenko, A.; Korsun, O.M.; Gubin, I.I.; Kovalenko, S.M.; Kalugin, O.N. Atomistic Simulations of Coating of Silver Nanoparticles with Poly(vinylpyrrolidone) Oligomers: Effect of Oligomer Chain Length. J. Phys. Chem. C 2015, 119, 7888–7899. [Google Scholar] [CrossRef]
- Kyrychenko, A.; Pasko, D.A.; Kalugin, O.N. Poly(vinyl alcohol) as a water protecting agent for silver nanoparticles: The role of polymer size and structure. Phys. Chem. Chem. Phys. 2017, 19, 8742–8756. [Google Scholar] [CrossRef] [PubMed]
- Kyrychenko, A.; Blazhynska, M.M.; Kalugin, O.N. Protonation-dependent adsorption of polyarginine onto silver na-noparticles. J. Appl. Phys. 2020, 127, 075502. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C.; Alexander-Katz, A. Computational Insights into the Binding of Monolayer-Capped Gold Nanoparticles onto Amyloid-beta Fibrils. ACS Chem. Neurosci. 2020, 11, 3153–3160. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Vukovic, L.; Kral, P. Computational screening of nanoparticles coupling to Abeta40 peptides and fibrils. Sci. Rep. 2019, 9, 17804. [Google Scholar] [CrossRef] [Green Version]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Disclosing the Interaction of Gold Nanoparticles with Abeta(1-40) Monomers through Replica Exchange Molecular Dynamics Simulations. Int. J. Mol. Sci. 2020, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Multiscale Molecular Dynamics Simulation of Multiple Protein Adsorption on Gold Nanoparticles. Int. J. Mol. Sci. 2019, 20, 3539. [Google Scholar] [CrossRef] [Green Version]
- Power, D.; Rouse, I.; Poggio, S.; Brandt, E.; Lopez, H.; Lyubartsev, A.; Lobaskin, V. A multiscale model of protein ad-sorption on a nanoparticle surface. Model. Simul. Mater. Sci. Eng. 2019, 27, 084003. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. A closer look into the ubiquitin corona on gold nanoparticles by computation-al studies. New J. Chem. 2015, 39, 2474–2482. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Competitive Binding of Proteins to Gold Nanoparticles Disclosed by Molecular Dynamics Simulations. J. Phys. Chem. C 2015, 119, 22172–22180. [Google Scholar] [CrossRef]
- Lopez, H.; Lobaskin, V. Coarse-grained model of adsorption of blood plasma proteins onto nanoparticles. J. Chem. Phys. 2015, 143, 243138. [Google Scholar] [CrossRef]
- Mikolajczak, D.J.; Heier, J.L.; Schade, B.; Koksch, B. Catalytic Activity of Peptide–Nanoparticle Conjugates Regulated by a Conformational Change. Biomacromolecules 2017, 18, 3557–3562. [Google Scholar] [CrossRef] [PubMed]
- Zaramella, D.; Scrimin, P.; Prins, L.J. Self-Assembly of a Catalytic Multivalent Peptide–Nanoparticle Complex. J. Am. Chem. Soc. 2012, 134, 8396–8399. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Bruce, N.J.; Romanowska, J.; Kokh, D.B.; Ozboyaci, M.; Yu, X.; Ozturk, M.A.; Richter, S.; Wade, R.C. SDA 7: A modular and parallel implementation of the simulation of diffusional association software. J. Comput. Chem. 2015, 36, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, L.; Corni, S. Interaction with a Gold Surface Reshapes the Free Energy Landscape of Alanine Dipeptide. J. Phys. Chem. C 2014, 118, 11357–11364. [Google Scholar] [CrossRef]
- Penna, M.J.; Mijajlovic, M.; Biggs, M.J. Molecular-Level Understanding of Protein Adsorption at the Interface between Water and a Strongly Interacting Uncharged Solid Surface. J. Am. Chem. Soc. 2014, 136, 5323–5331. [Google Scholar] [CrossRef]
- Frenkel, D.; Smit, B. Understanding Molecular Simulation from Alogorithms to Applications; Academic Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids, 2nd ed.; Oxford Science Publications: Oxford, UK, 1989. [Google Scholar]
- Banks, D.S.; Fradin, C. Anomalous Diffusion of Proteins Due to Molecular Crowding. Biophys. J. 2005, 89, 2960–2971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccardi, L.; Gabrielli, L.; Sun, X.; De Biasi, F.; Rastrelli, F.; Mancin, F.; De Vivo, M. Nanoparticle-Based Receptors Mimic Protein-Ligand Recognition. Chem 2017, 3, 92–109. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Riccardi, L.; De Biasi, F.; Rastrelli, F.; De Vivo, M.; Mancin, F. Molecular-Dynamics-Simulation-Directed Rational Design of Nanoreceptors with Targeted Affinity. Angew. Chem. 2019, 58, 7702–7707. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Poma, A.B.; Guzman, H.V.; Li, M.S.; Theodorakis, P.E. Mechanical and thermodynamic properties of Abeta42, Abeta40, and alpha-synuclein fibrils: A coarse-grained method to complement experimental studies. Beilstein J. Nanotechnol. 2019, 10, 500–513. [Google Scholar] [CrossRef] [Green Version]
- Poma, A.B.; Cieplak, M.; Theodorakis, P.E. Combining the MARTINI and Structure-Based Coarse-Grained Approaches for the Molecular Dynamics Studies of Conformational Transitions in Proteins. J. Chem. Theory Comput. 2017, 13, 1366–1374. [Google Scholar] [CrossRef]
- Mahmood, M.I.; Poma, A.B.; Okazaki, K.I. Optimizing Go-MARTINI Coarse-Grained Model for F-BAR Protein on Lipid Membrane. Front. Mol. Biosci. 2021, 8, 619381. [Google Scholar] [CrossRef]
- Byler, K.G.; Li, Y.; Houghten, R.A.; Martinez-Mayorga, K. The role of imidazole in peptide cyclization by transesterifi-cation: Parallels to the catalytic triads of serine proteases. Org. Biomol. Chem. 2013, 11, 2979–2987. [Google Scholar] [CrossRef] [Green Version]
- Brancolini, G.; Bellucci, L.; Maschio, M.C.; Di Felice, R.; Corni, S. The interaction of peptides and proteins with nanostructures surfaces: A challenge for nanoscience. Curr. Opin. Colloid Interface Sci. 2019, 41, 86–94. [Google Scholar] [CrossRef]
- Brancolini, G.; Tozzini, V. Multiscale modeling of proteins Interaction with functionalized nanoparticles. Curr. Opin. Colloid Interface Sci. 2019, 41, 66–73. [Google Scholar] [CrossRef]
- Brancolini, G.; Lopez, H.; Corni, S.; Tozzini, V. Low-Resolution Models for the Interaction Dynamics of Coated Gold Nanoparticles with beta2-microglobulin. Int. J. Mol. Sci. 2019, 20, 3866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, S.; Bellucci, L.; Agostini, M.; Gagliardi, M.; Corni, S.; Cecchini, M.; Brancolini, G. Atomistic simulations of gold surface functionalization for nanoscale biosensors applications. Nanotechnology 2020, 32, 095702. [Google Scholar] [CrossRef] [PubMed]
- Iori, F.; Di Felice, R.; Molinari, E.; Corni, S. GolP: An atomistic force-field to describe the interaction of proteins with Au(111) surfaces in water. J. Comput. Chem. 2009, 30, 1465–1476. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conforma-tional energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Brancolini, G.; Toroz, D.; Corni, S. Can small hydrophobic gold nanoparticles inhibit β2-microglobulin fibrillation? Nanoscale 2014, 6, 7903–7911. [Google Scholar] [CrossRef]
- Cantarutti, C.; Raimondi, S.; Brancolini, G.; Corazza, A.; Giorgetti, S.; Ballico, M.; Zanini, S.; Palmisano, G.; Bertoncin, P.; Marchese, L.; et al. Citrate-stabilized gold nanoparti-cles hinder fibrillogenesis of a pathological variant of β2-microglobulin. Nanoscale 2017, 9, 3941–3951. [Google Scholar] [CrossRef]
- Brancolini, G.; Maschio, M.C.; Cantarutti, C.; Corazza, A.; Fogolari, F.; Bellotti, V.; Corni, S.; Esposito, G. Citrate stabilized gold nanoparticles interfere with amyloid fibril formation: D76N and DeltaN6 β2-microglobulin variants. Nanoscale 2018, 10, 4793–4806. [Google Scholar] [CrossRef] [Green Version]
- Brancolini, G.; Kokh, D.B.; Calzolai, L.; Wade, R.C.; Corni, S. Docking of ubiquitin to gold nanoparticles. ACS Nano 2012, 6, 9863–9878. [Google Scholar] [CrossRef] [PubMed]
- Pronk, S.; Pall, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. B 2001, 105, 24a. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Mcsherry, T.M. General Steepest Descent Algorithm. IEEE Trans. Aerosp. Electron. Syst. 1976, 12, 12–22. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Franco-Ulloa, S.; Riccardi, L.; Rimembrana, F.; Pini, M.; De Vivo, M. NanoModeler: A Webserver for Molecular Simulations and Engineering of Nanoparticles. J. Chem. Theory Comput. 2019, 15, 2022–2032. [Google Scholar] [CrossRef] [PubMed]
- Pohjolainen, E.; Chen, X.; Malola, S.; Groenhof, G.; Häkkinen, H. A Unified AMBER-Compatible Molecular Mechanics Force Field for Thiolate-Protected Gold Nanoclusters. J. Chem. Theory Comput. 2016, 12, 1342–1350. [Google Scholar] [CrossRef]
- Heinz, H.; Vaia, R.A.; Farmer, B.L.; Naik, R.R. Accurate Simulation of Surfaces and Interfaces of Face-Centered Cubic Metals Using 12−6 and 9−6 Lennard-Jones Potentials. J. Phys. Chem. C 2008, 112, 17281–17290. [Google Scholar] [CrossRef]
- Lundborg, M.; Lindahl, E. Automatic GROMACS Topology Generation and Comparisons of Force Fields for Solvation Free Energy Calculations. J. Phys. Chem. B 2015, 119, 810–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y. R.E.D. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanzan, M.; Rosa, M.; Corni, S. Atomistic insight into the aggregation of [Au25(SR)18]q nanoclusters. Nanoscale Adv. 2020, 2, 2842–2852. [Google Scholar] [CrossRef]
- Evans, D.J.; Holian, B.L. The Nose–Hoover thermostat. J. Chem. Phys. 1985, 83, 4069. [Google Scholar] [CrossRef]
- Heikkilä, E.; Gurtovenko, A.A.; Martinez-Seara, H.; Häkkinen, H.; Vattulainen, I.; Akola, J. Atomistic Simulations of Functional Au144(SR)60 Gold Nanoparticles in Aqueous Environment. J. Phys. Chem. C 2012, 116, 9805–9815. [Google Scholar] [CrossRef] [Green Version]
- Gabdoulline, R.; Wade, R. Simulation of the diffusional association of barnase and barstar. Biophys. J. 1997, 72, 1917–1929. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtu-bules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, A.; Amoros, D.; Garcia de la Torre, J. Prediction of hydrodynamic and other solution properties of rigid pro-teins from atomic- and residue-level models. Biophys. J. 2011, 101, 892–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grell, L.; Parkin, C.; Slatest, L.; Craig, P.A. EZ-Viz, a tool for simplifying molecular viewing in PyMOL. Biochem. Mol. Biol. Educ. 2006, 34, 402–407. [Google Scholar] [CrossRef] [PubMed]
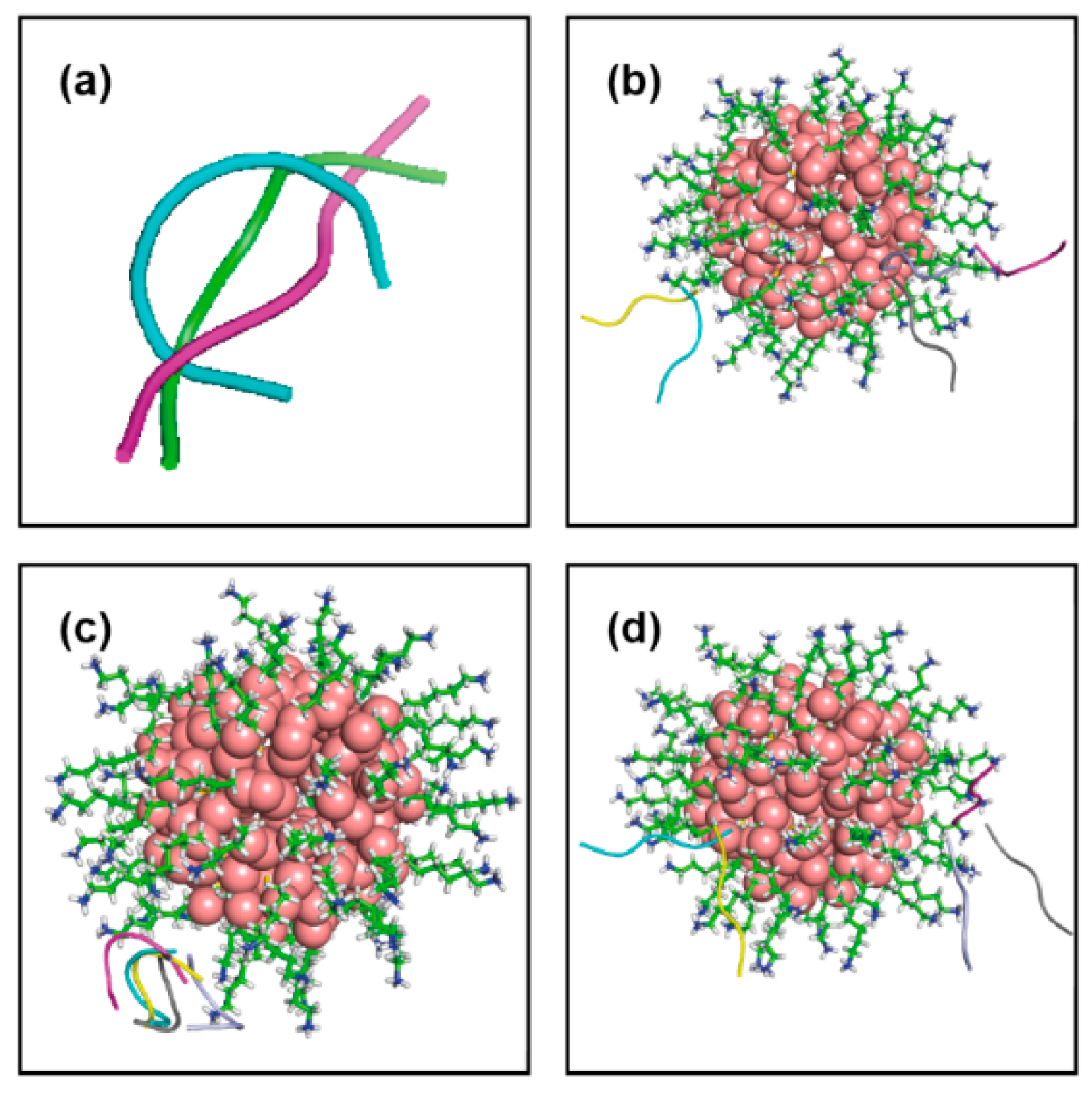
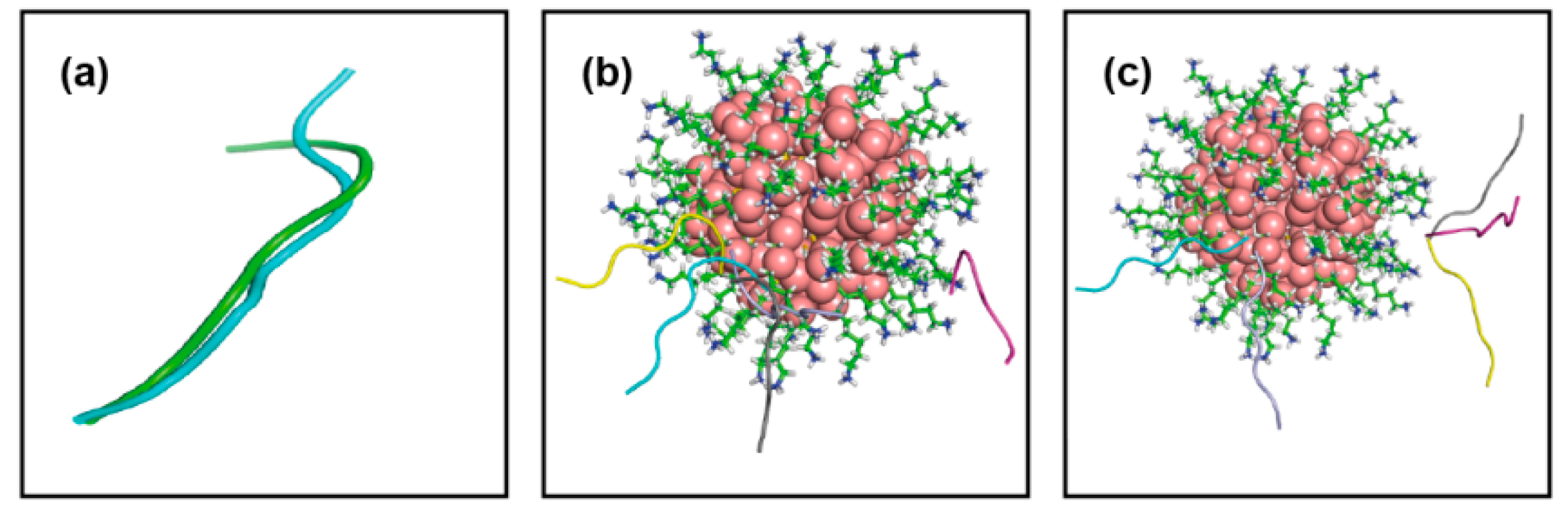



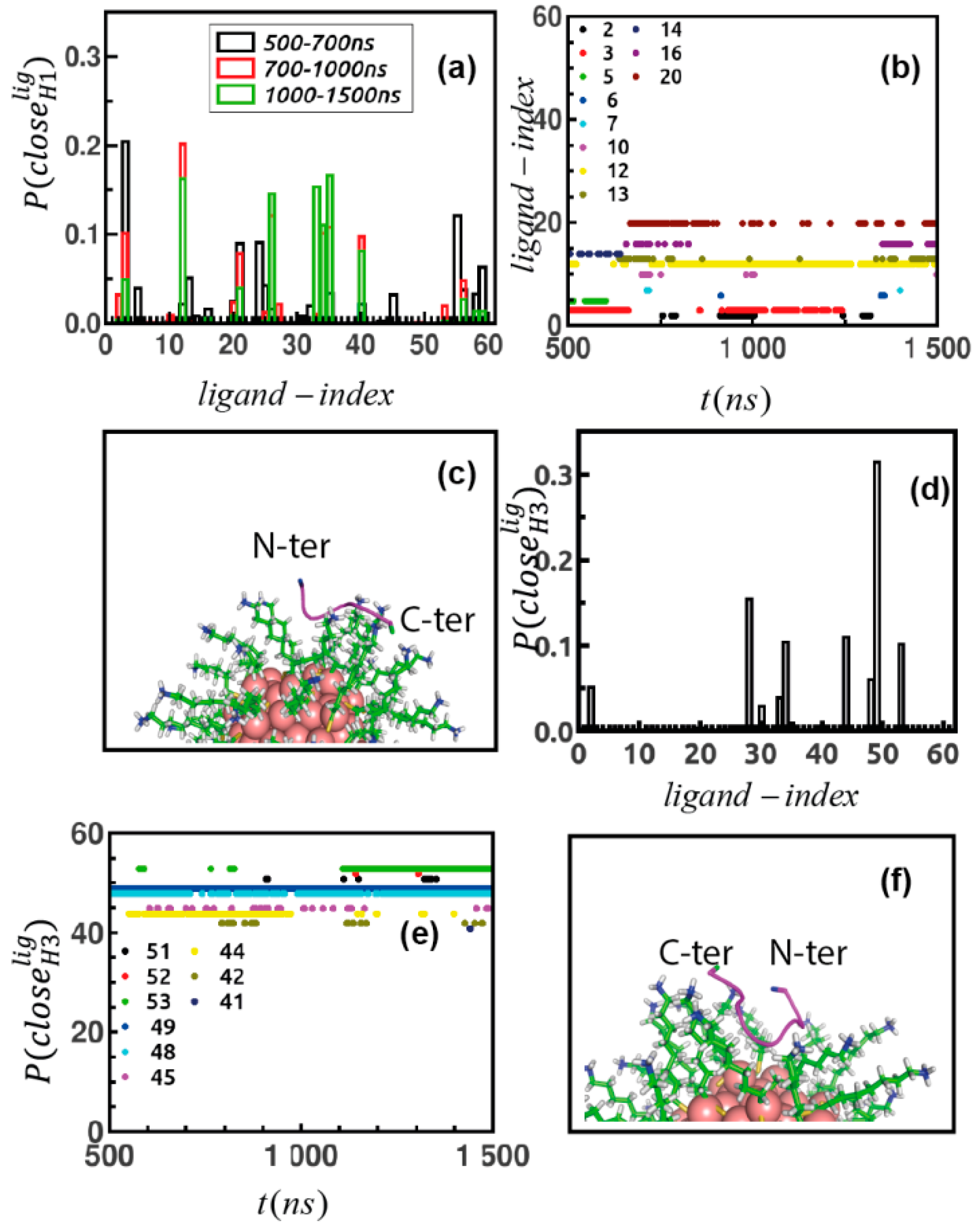
| Conf Index | Clust Index | RelPop (%) (a) | kT (b) | kT (c) | kT (d) | kT (e) | Spread (f) |
|---|---|---|---|---|---|---|---|
| 1 | A1 | 70.8 | 52.815 | 50.607 | 8.886 | 11.094 | 1.203 |
| 1 | A2 | 12.0 | 51.375 | 51.288 | 9.046 | 9.132 | 1.172 |
| 1 | A3 | 7.0 | 50.788 | 50.638 | 9.936 | 10.085 | 0.571 |
| 1 | A4 | 8.4 | 51.005 | 51.168 | 9.391 | 9.227 | 0.343 |
| 1 | A5 | 1.8 | 51.006 | 49.864 | 9.771 | 10.913 | 0.495 |
| 1 | B1 | 63.5 | 50.890 | 47.567 | 8.618 | 11.841 | 0.342 |
| 1 | B2 | 18.9 | 50.631 | 45.628 | 7.283 | 12.287 | 0.500 |
| 1 | B3 | 11.4 | 50.676 | 45.877 | 6.823 | 11.622 | 0.464 |
| 1 | B4 | 3.8 | 50.268 | 47.143 | 8.347 | 11.472 | 0.254 |
| 1 | B5 | 2.4 | 50.361 | 48.345 | 8.667 | 10.683 | 0.273 |
| 1 | C1 | 37.3 | 51.075 | 50.940 | 10.970 | 11.105 | 0.626 |
| 1 | C2 | 21.7 | 51.325 | 49.756 | 9.965 | 11.534 | 0.518 |
| 1 | C3 | 33.9 | 50.266 | 49.160 | 10.833 | 11.940 | 0.601 |
| 1 | C4 | 5.6 | 50.346 | 50.611 | 10.391 | 10.126 | 0.387 |
| 1 | C5 | 1.5 | 50.230 | 50.292 | 10.715 | 9.654 | 0.001 |
| Conf Index | Clust Index | RelPop (%) (a) | kT (b) | kT (c) | kT (d) | kT (e) | Spread (f) |
|---|---|---|---|---|---|---|---|
| 1 | D1 | 78.5 | 55.104 | 51.538 | 9.048 | 12.613 | 1.271 |
| 1 | D2 | 10.7 | 53.506 | 52.317 | 10.716 | 11.905 | 0.821 |
| 1 | D3 | 3.6 | 53.496 | 52.045 | 9.897 | 11.347 | 0.369 |
| 1 | D4 | 3.7 | 53.642 | 50.904 | 10.819 | 13.557 | 0.292 |
| 1 | D5 | 3.5 | 53.541 | 49.327 | 9.270 | 13.484 | 0.267 |
| 1 | E1 | 44.4 | 54.087 | 52.973 | 10.398 | 11.511 | 2.127 |
| 1 | E2 | 26.2 | 54.218 | 53.461 | 9.214 | 9.971 | 1.573 |
| 1 | E3 | 15.8 | 53.681 | 54.362 | 10.045 | 9.364 | 4.559 |
| 1 | E4 | 10.8 | 54.442 | 51.790 | 8.913 | 11.565 | 0.578 |
| 1 | E5 | 2.8 | 53.662 | 53.151 | 9.899 | 10.410 | 1.772 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutta, S.; Corni, S.; Brancolini, G. Molecular Dynamics Simulations of a Catalytic Multivalent Peptide–Nanoparticle Complex. Int. J. Mol. Sci. 2021, 22, 3624. https://doi.org/10.3390/ijms22073624
Dutta S, Corni S, Brancolini G. Molecular Dynamics Simulations of a Catalytic Multivalent Peptide–Nanoparticle Complex. International Journal of Molecular Sciences. 2021; 22(7):3624. https://doi.org/10.3390/ijms22073624
Chicago/Turabian StyleDutta, Sutapa, Stefano Corni, and Giorgia Brancolini. 2021. "Molecular Dynamics Simulations of a Catalytic Multivalent Peptide–Nanoparticle Complex" International Journal of Molecular Sciences 22, no. 7: 3624. https://doi.org/10.3390/ijms22073624
APA StyleDutta, S., Corni, S., & Brancolini, G. (2021). Molecular Dynamics Simulations of a Catalytic Multivalent Peptide–Nanoparticle Complex. International Journal of Molecular Sciences, 22(7), 3624. https://doi.org/10.3390/ijms22073624







