Structural Mutations in the Organellar Genomes of Valeriana sambucifolia f. dageletiana (Nakai. ex Maekawa) Hara Show Dynamic Gene Transfer
Abstract
1. Introduction
2. Results
2.1. Variation in the V. sambucifolia f. dageletiana Plastome
2.2. Features of the V. sambucifolia f. dageletiana Mitochondrial Genome
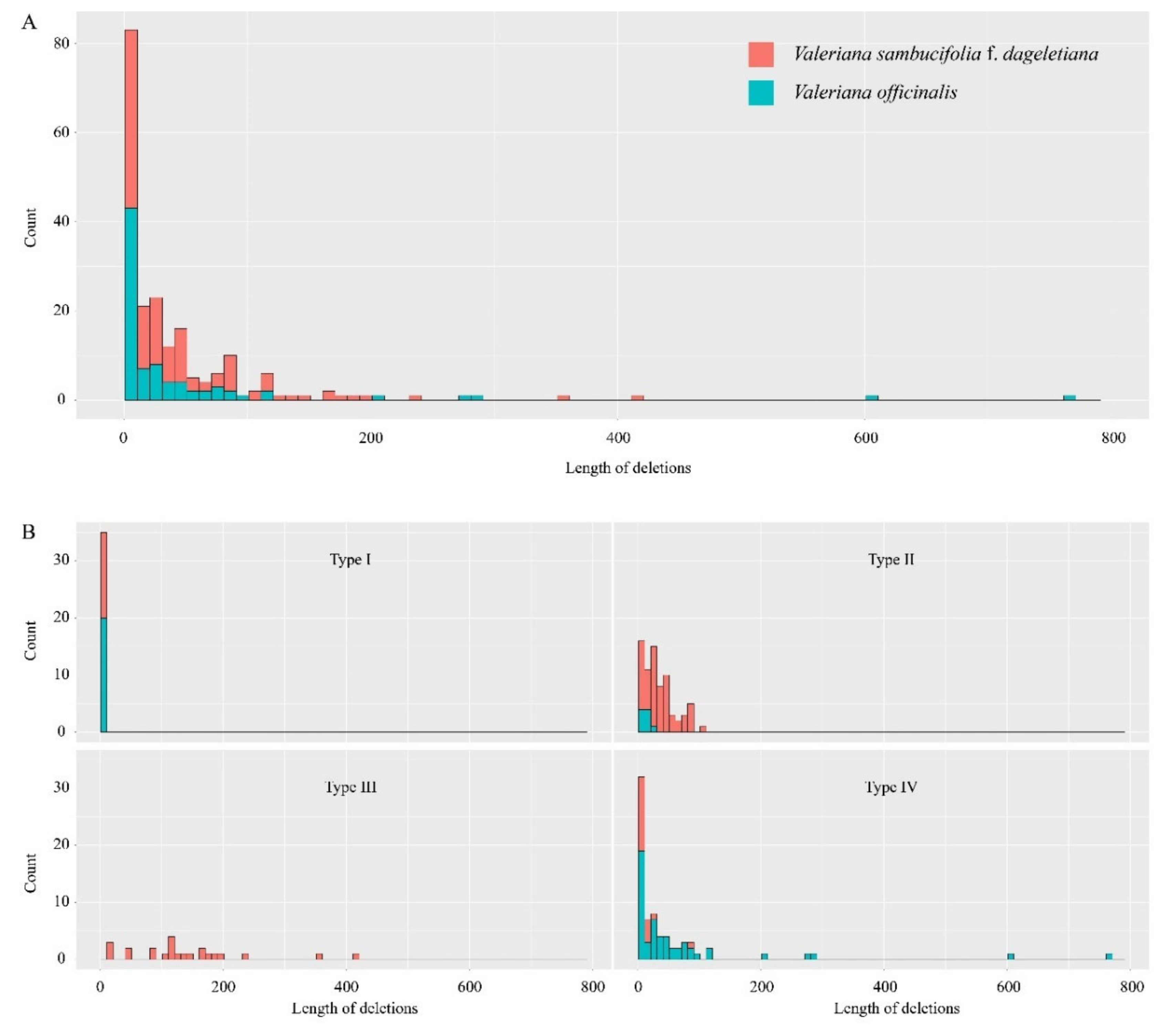
2.3. Intercompartmental Gene Transfer in the Mitogenome of V. sambucifolia f. dageletiana
3. Discussion
3.1. Frequent Gene Transfers from the V. sambucifolia f. dageletiana mitogenome
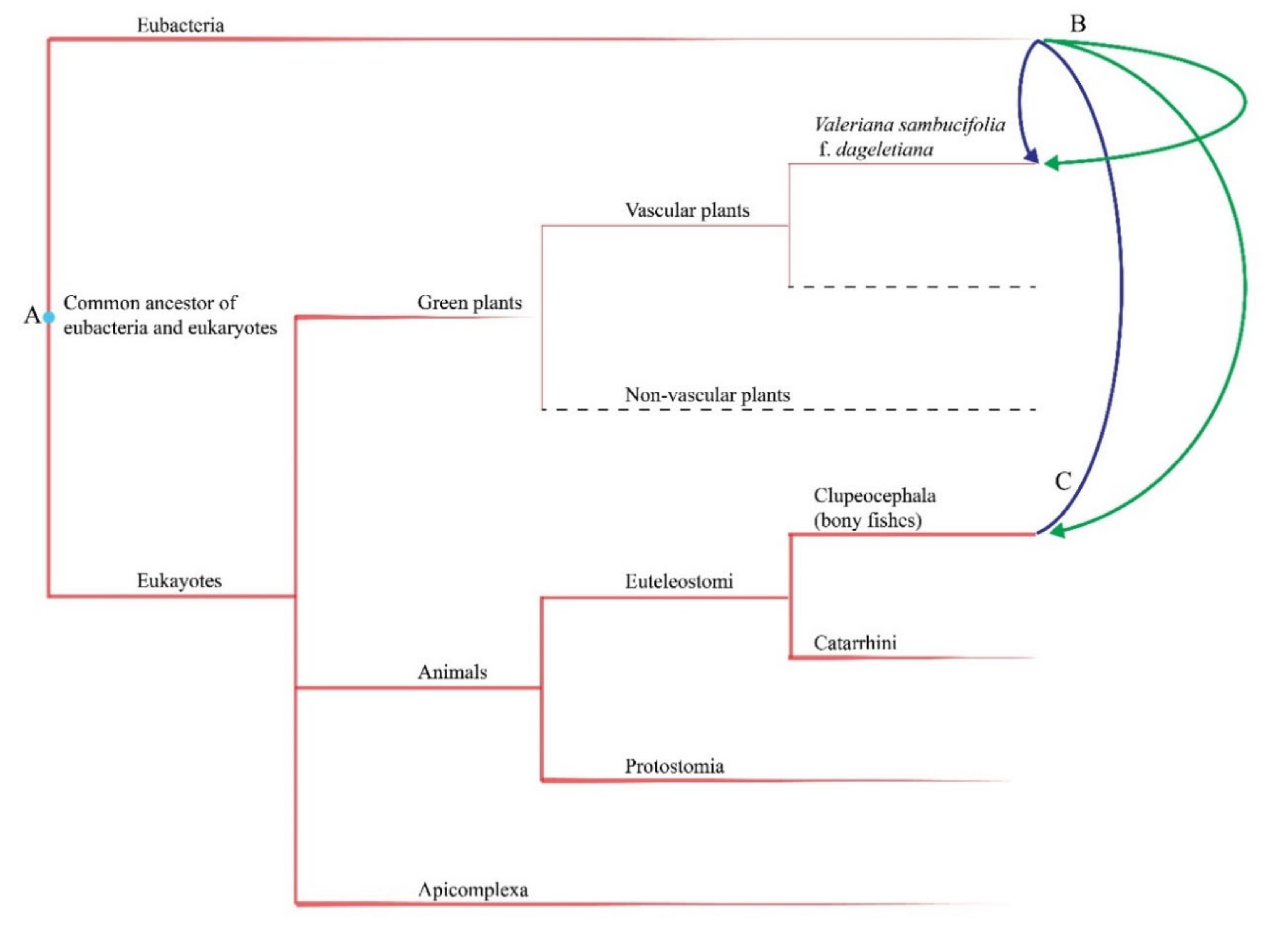
3.2. Origins of Non-Plastomic Regions Found in the V. sambucifolia f. dageletiana Plastome
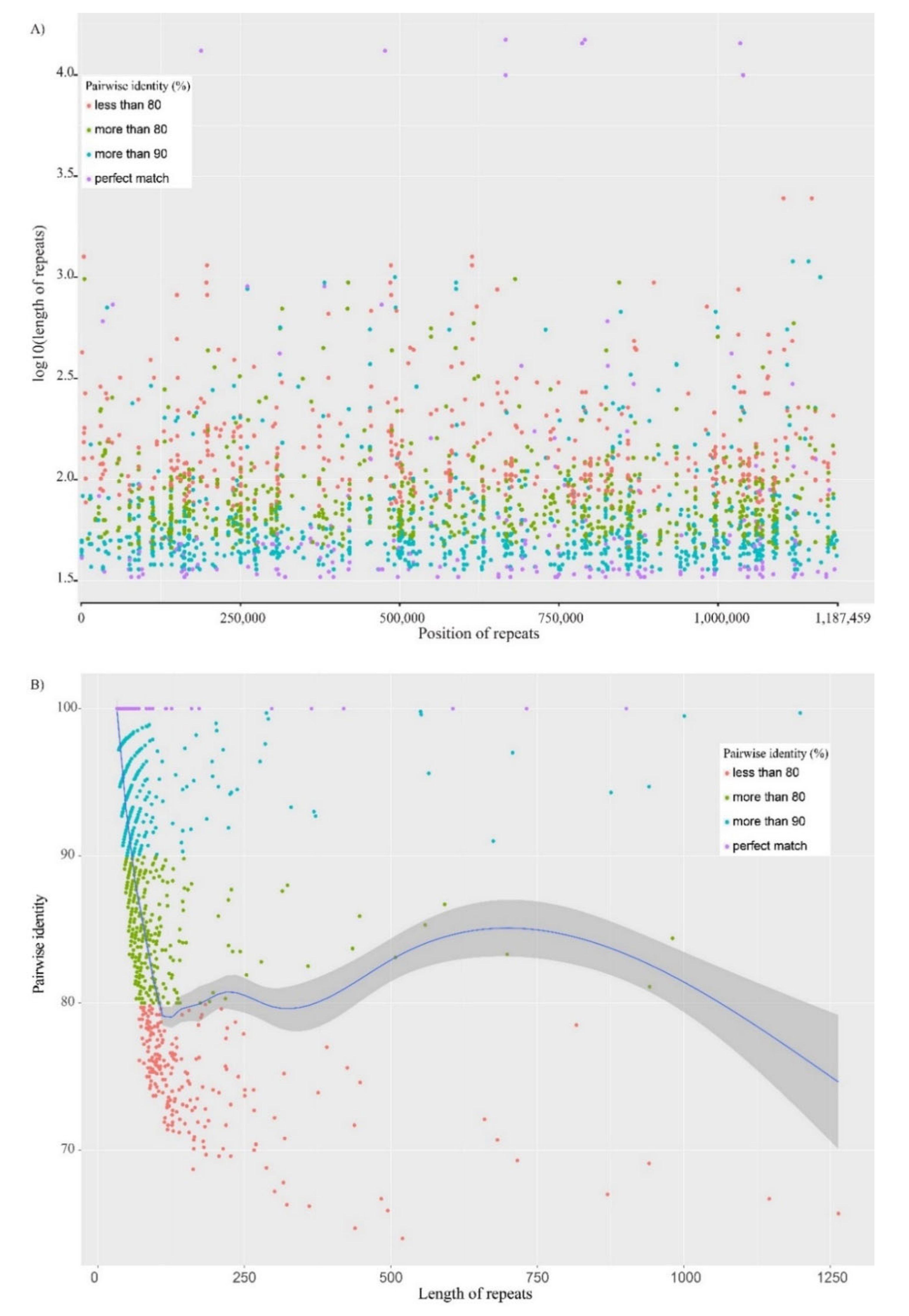
3.3. Rapid Structural Mutation of Plastomes in the Genus Valeriana
3.4. Genetic Consequence of Anagenetic Speciation on Ulleung Island
4. Materials and Methods
4.1. Sampling, DNA Extraction, and Next-Generation Sequencing
4.2. Assembly and Annotation of the Plastome and Mitogenome
4.3. Analysis of Repetitive Sequences in Organellar Genomes
4.4. Identification of Mitochondrial Plastid DNAs in the Mitogenome of V. sambucifolia f. dageletiana
4.5. Identification of non-plastome-originated DNAs in the plastome of V. sambucifolia f. dageletiana
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiang, C.L.; Dong, H.J.; Landrein, S.; Zhao, F.; Yu, W.B.; Soltis, D.E.; Soltis, P.S.; Backlund, A.; Wang, H.F.; Li, D.Z.; et al. Revisiting the phylogeny of Dipsacales: New insights from phylogenomic analyses of complete plastomic sequences. J. Syst. Evol. 2020, 58, 103–117. [Google Scholar] [CrossRef]
- Jacobs, B.; Bell, C.; Smets, E. Fruits and seeds of the valeriana clade (Dipsacales): Diversity and evolution. Int. J. Plant Sci. 2010, 171, 421–434. [Google Scholar] [CrossRef]
- Bell, C.D.; Donoghue, M.J. Phylogeny and biogeography of Valerianaceae (Dipsacales) with special reference to the South American valerians. Org. Divers. Evol. 2005, 5, 147–159. [Google Scholar] [CrossRef]
- Backlund, A.; Moritz, T. Phylogenetic implications of an expanded valepotriate distribution in the Valerianaceae. Biochem. Syst. Ecol. 1998, 26, 309–335. [Google Scholar] [CrossRef]
- eFloras. 2008. Available online: http://www.efloras.org (accessed on 10 April 2020).
- Bjørn, G.K.; Olsson, K.; Bladh, K.W. Valeriana officinalis L.(Common valerian). Spice-and Medicinal Plants in the Nordic and Baltic Countries. 2006. Available online: https://www.nordgen.org/ngdoc/plants/publications/SPIMED_report_maj_2006.pdf (accessed on 5 April 2020).
- Flora of Korea Editorial Committe. The Genera of Vascular Plants of Korea; Academy Publishing, Co.: Seoul, Korea, 2007. [Google Scholar]
- Wang, Z.; Hwang, S.H.; Kim, J.H.; Lim, S.S. Anti-obesity effect of the above-ground part of valeriana dageletiana nakai ex f. maek extract in high-fat diet-induced obese C57BL/6N Mice. Nutrients 2017, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.-Y.; Suh, H.-W.; Choi, E.-J.; Chung, S.-Y.; Kim, J.-W. Complementary and alternative medicine in clinical practice guideline for insomnia. Korean Soc. Orient. Neuropsychiatry 2016, 27, 235–248. [Google Scholar] [CrossRef][Green Version]
- Ryu, K.-S. Monoterpenoid of Korean valerian roots. Korean J. Pharmacogn. 1974, 5, 1–6. [Google Scholar]
- Federov, A.A.; Bobrov, A. Flora of Russia, the European Part and Bordering Regions; AA Balkema Publishers: Rotterdam, The Netherlands, 2001. [Google Scholar]
- Skalińska, M. Meiosis in a polyhaploid twin plant and a hexaploid hybrid of Valeriana sambucifolia Mikan. Acta Soc. Bot. Poloniae 1954, 23, 359–374. [Google Scholar] [CrossRef]
- Lövkvist, B.R. Chromosome numbers in south Swedish vascular plants. Opera Bot. 1999, 137, 1–42. [Google Scholar]
- Lee, Y. Chromosome numbers of flowering plants in Korea (2). J. Korean Res. Inst. Better Living 1969, 2, 141–145. [Google Scholar]
- Bock, R.; Knoop, V. Genomics of Chloroplasts and Mitochondria; Springer Science & Business Media: Berlin/Heidelberg, Germany; Amsterdam, The Netherlands, 2012; Volume 35. [Google Scholar]
- Kim, H.T.; Lee, J.M. Organellar genome analysis reveals endosymbiotic gene transfers in tomato. PLoS ONE 2018, 13, e0202279. [Google Scholar] [CrossRef]
- Ruhlman, T.A.; Jansen, R.K. The plastid genomes of flowering plants. Methods Mol. Biol. 2014, 1132, 3–38. [Google Scholar] [CrossRef]
- Wolf, P.G.; Roper, J.M.; Duffy, A.M. The evolution of chloroplast genome structure in ferns. Genome 2010, 53, 731–738. [Google Scholar] [CrossRef]
- Wolf, P.G.; Karol, K.G.; Mandoli, D.F.; Kuehl, J.; Arumuganathan, K.; Ellis, M.W.; Mishler, B.D.; Kelch, D.G.; Olmstead, R.G.; Boore, J.L. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene 2005, 350, 117–128. [Google Scholar] [CrossRef] [PubMed]
- De Pamphilis, C.W.; Palmer, J.D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 1990, 348, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Freudenstein, J.V.; Li, J.; Mayfield-Jones, D.R.; Perez, L.; Pires, J.C.; Santos, C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 2014, 31, 3095–3112. [Google Scholar] [CrossRef] [PubMed]
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.; Clements, M.A.; Arroyo, M.T.; Leebens-Mack, J.; et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. Biol. Sci. 2015, 282, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Kim, H.T.; Shin, C.-H.; Sun, H.; Kim, J.-H. Sequencing of the plastome in the leafless green mycoheterotroph Cymbidium macrorhizon helps us to understand an early stage of fully mycoheterotrophic plastome structure. Plant Syst. Evol. 2017, 304, 1–14. [Google Scholar] [CrossRef]
- Barrett, C.F.; Davis, J.I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 2012, 99, 1513–1523. [Google Scholar] [CrossRef]
- Wicke, S.; Muller, K.F.; de Pamphilis, C.W.; Quandt, D.; Wickett, N.J.; Zhang, Y.; Renner, S.S.; Schneeweiss, G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 2013, 25, 3711–3725. [Google Scholar] [CrossRef] [PubMed]
- Karol, K.G.; Arumuganathan, K.; Boore, J.L.; Duffy, A.M.; Everett, K.D.; Hall, J.D.; Hansen, S.K.; Kuehl, J.V.; Mandoli, D.F.; Mishler, B.D.; et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol. Biol. 2010, 10, 321. [Google Scholar] [CrossRef]
- Raubeson, L.A.; Jansen, R.K. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 1992, 255, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Herbon, L.A. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988, 28, 87–97. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Wang, D.; Wu, Y.W.; Shih, A.C.; Wu, C.S.; Wang, Y.N.; Chaw, S.M. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 2007, 24, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Moreno, L.; Gonzalez, V.M.; Benjak, A.; Marti, M.C.; Puigdomenech, P.; Aranda, M.A.; Garcia-Mas, J. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genom. 2011, 12, 424. [Google Scholar] [CrossRef] [PubMed]
- Naito, K.; Kaga, A.; Tomooka, N.; Kawase, M. De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed. Sci. 2013, 63, 176–182. [Google Scholar] [CrossRef]
- Jo, Y.D.; Choi, Y.; Kim, D.H.; Kim, B.D.; Kang, B.C. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genom. 2014, 15, 561. [Google Scholar] [CrossRef]
- Chang, S.; Wang, Y.; Lu, J.; Gai, J.; Li, J.; Chu, P.; Guan, R.; Zhao, T. The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS ONE 2013, 8, e56502. [Google Scholar] [CrossRef]
- Ellis, J. Promiscuous DNA-chloroplast genes inside plant mitochondria. Nature 1982, 299, 678–679. [Google Scholar] [CrossRef]
- Stern, D.B.; Lonsdale, D.M. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature 1982, 299, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Chen, H.; Yang, D.; Liu, C. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2018, 29, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Goremykin, V.V.; Salamini, F.; Velasco, R.; Viola, R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2009, 26, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Spooner, D.M.; Ruess, H.; Iorizzo, M.; Senalik, D.; Simon, P. Entire plastid phylogeny of the carrot genus (Daucus, Apiaceae): Concordance with nuclear data and mitochondrial and nuclear DNA insertions to the plastid. Am. J. Bot. 2017, 104, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. Mitochondrion-to-plastid DNA transfer: It happens. New Phytol. 2014, 202, 736–738. [Google Scholar] [CrossRef] [PubMed]
- Rabah, S.O.; Lee, C.; Hajrah, N.H.; Makki, R.M.; Alharby, H.F.; Alhebshi, A.M.; Sabir, J.S.M.; Jansen, R.K.; Ruhlman, T.A. Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Jung, E.-H.; Lim, C.E.; Lee, B.Y.; Hong, S.-P. Complete chloroplast genome sequence of Patrinia saniculifolia hemsl.(Disacales: Caprifoliaceae), an endemic plant in Korea. Mitochondrial DNA Part B 2018, 3, 60–61. [Google Scholar] [CrossRef]
- Wang, H.-X.; Liu, H.; Moore, M.J.; Landrein, S.; Liu, B.; Zhu, Z.-X.; Wang, H.-F. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. (Dipsacales). Mol. Phylogenet. Evol. 2020, 142, 106641. [Google Scholar] [CrossRef]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T.; Maier, U.G.; Leister, D. DNA transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 2009, 60, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Medina, R.; Goffinet, B. 350 my of mitochondrial genome stasis in mosses, an early land plant lineage. Mol. Biol. Evol. 2014, 31, 2586–2591. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Mower, J.P. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017, 213, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.M.; Shih, A.C.; Wang, D.; Wu, Y.W.; Liu, S.M.; Chou, T.Y. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 2008, 25, 603–615. [Google Scholar] [CrossRef]
- Alverson, A.J.; Wei, X.X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 2005, 21, 655–663. [Google Scholar] [CrossRef]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; de Pamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Dong, S.; Chen, L.; Liu, Y.; Wang, Y.; Zhang, S.; Yang, L.; Lang, X.; Zhang, S. The draft mitochondrial genome of Magnolia biondii and mitochondrial phylogenomics of angiosperms. PLoS ONE 2020, 15, e0231020. [Google Scholar] [CrossRef]
- Satoh, M.; Kubo, T.; Nishizawa, S.; Estiati, A.; Itchoda, N.; Mikami, T. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol. Genet. Genom. 2004, 272, 247–256. [Google Scholar] [CrossRef]
- Michalovova, M.; Vyskot, B.; Kejnovsky, E. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: Size, relative age and chromosomal localization. Heredity 2013, 111, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Richly, E.; Leister, D. NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol. Biol. Evol. 2004, 21, 1972–1980. [Google Scholar] [CrossRef]
- Downie, S.R.; Jansen, R.K. A comparative analysis of whole plastid genomes from the apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 2015, 40, 336–351. [Google Scholar] [CrossRef]
- Ma, P.F.; Zhang, Y.X.; Guo, Z.H.; Li, D.Z. Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci. Rep. 2015, 5, 11608. [Google Scholar] [CrossRef]
- Straub, S.C.; Cronn, R.C.; Edwards, C.; Fishbein, M.; Liston, A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol. Evol. 2013, 5, 1872–1885. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.W.; Palmer, J.D. An exceptional horizontal gene transfer in plastids: Gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006, 4, 31. [Google Scholar] [CrossRef]
- Sheveleva, E.V.; Hallick, R.B. Recent horizontal intron transfer to a chloroplast genome. Nucleic Acids Res. 2004, 32, 803–810. [Google Scholar] [CrossRef]
- Brouard, J.S.; Otis, C.; Lemieux, C.; Turmel, M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): Unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genom. 2008, 9, 290. [Google Scholar] [CrossRef]
- Delwiche, C.F.; Palmer, J.D. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 1996, 13, 873–882. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, K.J. Evolution of six novel ORFs in the plastome of Mankyua chejuense and phylogeny of eusporangiate ferns. Sci. Rep. 2018, 8, 16466. [Google Scholar] [CrossRef]
- Robison, T.A.; Grusz, A.L.; Wolf, P.G.; Mower, J.P.; Fauskee, B.D.; Sosa, K.; Schuettpelz, E. Mobile elements shape plastome evolution in ferns. Genome Biol. Evol. 2018, 10, 2558–2571. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, J.S. The dynamic evolution of mobile open reading frames in plastomes of Hymenophyllum Sm. and new insight on Hymenophyllum coreanum Nakai. Sci. Rep. 2020, 10, 11059. [Google Scholar] [CrossRef]
- Yue, J.; Hu, X.; Sun, H.; Yang, Y.; Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 2012, 3, 1152. [Google Scholar] [CrossRef] [PubMed]
- Broothaerts, W.; Mitchell, H.J.; Weir, B.; Kaines, S.; Smith, L.M.A.; Yang, W.; Mayer, J.E.; Roa-Rodríguez, C.; Jefferson, R.A. Gene transfer to plants by diverse species of bacteria. Nature 2005, 433, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Dunning Hotopp, J.C.; Clark, M.E.; Oliveira, D.C.; Foster, J.M.; Fischer, P.; Munoz Torres, M.C.; Giebel, J.D.; Kumar, N.; Ishmael, N.; Wang, S.; et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 2007, 317, 1753–1756. [Google Scholar] [CrossRef] [PubMed]
- Dunning Hotopp, J.C. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011, 27, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Wybouw, N.; Dermauw, W.; Tirry, L.; Stevens, C.; Grbic, M.; Feyereisen, R.; Van Leeuwen, T. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife 2014, 3, e02365. [Google Scholar] [CrossRef]
- Ricard, G.; McEwan, N.R.; Dutilh, B.E.; Jouany, J.P.; Macheboeuf, D.; Mitsumori, M.; McIntosh, F.M.; Michalowski, T.; Nagamine, T.; Nelson, N.; et al. Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genom. 2006, 7, 22. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Richards, T.A.; Leonard, G.; Soanes, D.M.; Talbot, N.J. Gene transfer into the fungi. Fungal Biol. Rev. 2011, 25, 98–110. [Google Scholar] [CrossRef]
- Finnegan, D.J. Transposable elements in eukaryotes. In International Review of Cytology; Bourne, G.H., Danielli, J.F., Jeon, K.W., Eds.; Academic Press: Cambridge, MA, USA, 1985; Volume 93, pp. 281–326. [Google Scholar]
- Schaack, S.; Gilbert, C.; Feschotte, C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 2010, 25, 537–546. [Google Scholar] [CrossRef]
- El Baidouri, M.; Carpentier, M.C.; Cooke, R.; Gao, D.; Lasserre, E.; Llauro, C.; Mirouze, M.; Picault, N.; Jackson, S.A.; Panaud, O. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014, 24, 831–838. [Google Scholar] [CrossRef]
- Lin, X.; Faridi, N.; Casola, C. An ancient transkingdom horizontal transfer of penelope-like retroelements from arthropods to conifers. Genome Biol. Evol. 2016, 8, 1252–1266. [Google Scholar] [CrossRef]
- Betancur, R.R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Orti, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed]
- Haberle, R.C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 2008, 66, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Weng, M.L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2014, 31, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef]
- Lee, H.L.; Jansen, R.K.; Chumley, T.W.; Kim, K.J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007, 24, 1161–1180. [Google Scholar] [CrossRef]
- Feng, Y.L.; Wicke, S.; Li, J.W.; Han, Y.; Lin, C.S.; Li, D.Z.; Zhou, T.T.; Huang, W.C.; Huang, L.Q.; Jin, X.H. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 2016, 8, 2164–2175. [Google Scholar] [CrossRef]
- Park, I.; Yang, S.; Kim, W.J.; Noh, P.; Lee, H.O.; Moon, B.C. Authentication of herbal medicines Dipsacus asper and Phlomoides umbrosa using DNA barcodes, chloroplast genome, and sequence characterized amplified region (SCAR) marker. Molecules 2018, 23, 1748. [Google Scholar] [CrossRef]
- Liu, H.; Xia, M.; Xiao, Q.; Fang, J.; Wang, A.; Chen, S.; Zhang, D. Characterization of the complete chloroplast genome of Linnaea borealis, a rare, clonal self-incompatible plant. Mitochondrial DNA Part B 2020, 5, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, J.; Shimada, H.; Whittier, R.; Ishibashi, T.; Sakamoto, M.; Mori, M.; Kondo, C.; Honji, Y.; Sun, C.-R.; Meng, B.-Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. General Genet. MGG 1989, 217, 185–194. [Google Scholar] [CrossRef]
- Yang, J.Y.; Pak, J.-H.; Kim, S.-C. The complete plastome sequence of Rubus takesimensis endemic to Ulleung Island, Korea: Insights into molecular evolution of anagenetically derived species in Rubus (Rosaceae). Gene 2018, 668, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kang, G.-H.; Pak, J.-H.; Kim, S.-C. Characterization and comparison of two complete plastomes of Rosaceae species (Potentilla dickinsii var. glabrata and Spiraea insularis) endemic to Ulleung Island, Korea. Int. J. Mol. Sci. 2020, 21, 4933. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.-S.; Yang, J.Y.; Kim, S.-C. Complete chloroplast genome of Ulleung Island endemic flowering cherry, Prunus takesimensis (Rosaceae), in Korea. Mitochondrial DNA Part B 2018, 3, 274–275. [Google Scholar] [CrossRef]
- Park, J.-S.; Jin, D.-P.; Park, J.-W.; Choi, B.-H. Complete chloroplast genome of Fagus multinervis, a beech species endemic to Ulleung Island in South Korea. Mitochondrial DNA Part B 2019, 4, 1698–1699. [Google Scholar] [CrossRef]
- Yang, J.Y.; Pak, J.-H.; Kim, S.-C. Chloroplast genome of critically endangered Cotoneaster wilsonii (Rosaceae) endemic to Ulleung Island, Korea. Mitochondrial DNA Part B 2019, 4, 3892–3893. [Google Scholar] [CrossRef] [PubMed]
- Gil, H.-Y.; Kim, S.-C. The plastome sequence of Ulleung Rowan, Sorbus ulleungensis (Rosaceae), a new endemic species on Ulleung Island, Korea. Mitochondrial DNA Part B 2018, 3, 284–285. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.S.; Lee, Y.M.; Mun, J.-H.; Kim, J.-H. Molecular markers for phylogenetic applications derived from comparative plastome analysis of Prunus species. J. Syst. Evol. 2019, 57, 15–22. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Chase, M.W. Independent degradation in genes of the plastid ndh gene family in species of the orchid genus Cymbidium (Orchidaceae; Epidendroideae). PLoS ONE 2017, 12, e0187318. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef] [PubMed]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1. 0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]




| Gene | Asterales | Dipsacales | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chrysanthemum boreale | Chrysanthemum indicum | Codonopsis lanceolata | Diplostephium hartwegii | Helianthus annuus 1 | Helianthus annuus 2 | Helianthus annuus 3 | Helianthus annuus 4 | Helianthus annuus 5 | Helianthus annuus 6 | Lactuca saligna | Lactuca sativa | Lactuca serriola | Paraprenanthes iversifolia | Platycodon grandiflorus | Valeriana sambucifolia f. dageletiana | |
| atp1 | + a | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| atp4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| atp6 | + | + | + | + | + | + | D b | + | + | + | + | + | + | + | + | + |
| atp8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| atp9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ccmB | + | + | + | + | + | + | + | + | + | + | D | D | D | + | + | + |
| ccmC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ccmFc | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ccmFn | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| cob | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| cox1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| cox2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| cox3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| matR | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| mttB | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad4L | + | D | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad5 | + | + | + | + | + | + | + | + | + | + | D | D | D | D | P c | + |
| nad6 | + | + | P | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad7 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| nad9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| rpl5 | + | + | - d | + | + | + | + | + | + | + | + | + | + | + | - | + |
| rpl10 | + | + | + | + | + | + | + | + | + | + | D | D | D | + | + | + |
| rpl16 | P | P | + | + | + | + | + | + | + | + | + | + | + | + | + | P |
| rps1 | P | + | + | + | - | - | - | - | - | - | - | - | - | P | + | P |
| rps3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| rps4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| rps7 | - | - | + | - | - | - | - | - | - | - | - | - | - | - | + | + |
| rps10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + |
| rps11 | - | P | - | P | + | + | + | + | + | + | P | P | P | P | - | - |
| rps12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| rps13 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | P |
| rps14 | P | P | - | P | P | P | P | P | P | P | P | P | P | P | - | + |
| rps19 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P |
| shd3 | - | - | P | - | - | - | - | - | - | - | - | - | - | - | + | - |
| shd4 | + | + | - | + | P | P | P | + | P | P | P | P | P | P | + | - |
| Taxa | Accession of Mitogenome/Length (bp) | Accession of WGS a | % of Non-Hit Regions | |
|---|---|---|---|---|
| nt b | nt + WGS | |||
| Valeriana sambucifolia f. dageletiana | Present study/1,187,459 | - | 31.7 | - |
| Platycodon grandiflorus | NC_035958/1249,593 | - | 63.5 | - |
| Pinus taeda | NC_039746/1,191,054 | - | 43.3 | - |
| Schisandra sphenanthera | NC_042758/1,101,768 | - | 19.9 | - |
| Cucurbita pepo subsp. pepo | NC_014050/982,833 | NC_036638.1~NC_036657.1 | 46.2 | 21.2 |
| Mangifera indica | CM021857/871,458 | CM021837.1~CM021856.1 | 19.8 | 15.6 |
| Acer yangbiense | CM017774/803,281 | CM017761.1~CM017773.1 | 12.0 | 10.9 |
| Number of Queries | Number of Subjects Against Queries | Gene Transfer | |
|---|---|---|---|
| Percentage of 9025 Plastomes | Percentage of 364 Mitogenomes | ||
| 31 | ≥40 | ≥40 | Plastome → Mitogenome (mitochondrial plastid DNA (MTPT)) |
| 21 | ≥40 | <40 and ≥10 | |
| 16 | ≥40 | <10 and ≥1 | |
| 1 | <40 and ≥10 | <40 and ≥10 | |
| 1 | <40 and ≥10 | <10 and ≥1 | |
| 4 | <10 and ≥1 | ≥40 | Mitogenome → Plastome (plastid mitochondrial DNA (PTMT)) |
| 1 | <10 and ≥1 | <40 and ≥10 | |
| 1 | <10 and ≥1 | <10 and ≥1 | |
| 42 | <1 | ≥40 | Mitogenome → Plastome (PTMT) |
| 7 | <1 | <40 and ≥10 | |
| 17 | <1 | <10 and ≥1 | |
| 7 | <1 | <1 | Nuclear genome → Organelle genomes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.T.; Kim, J.S. Structural Mutations in the Organellar Genomes of Valeriana sambucifolia f. dageletiana (Nakai. ex Maekawa) Hara Show Dynamic Gene Transfer. Int. J. Mol. Sci. 2021, 22, 3770. https://doi.org/10.3390/ijms22073770
Kim HT, Kim JS. Structural Mutations in the Organellar Genomes of Valeriana sambucifolia f. dageletiana (Nakai. ex Maekawa) Hara Show Dynamic Gene Transfer. International Journal of Molecular Sciences. 2021; 22(7):3770. https://doi.org/10.3390/ijms22073770
Chicago/Turabian StyleKim, Hyoung Tae, and Jung Sung Kim. 2021. "Structural Mutations in the Organellar Genomes of Valeriana sambucifolia f. dageletiana (Nakai. ex Maekawa) Hara Show Dynamic Gene Transfer" International Journal of Molecular Sciences 22, no. 7: 3770. https://doi.org/10.3390/ijms22073770
APA StyleKim, H. T., & Kim, J. S. (2021). Structural Mutations in the Organellar Genomes of Valeriana sambucifolia f. dageletiana (Nakai. ex Maekawa) Hara Show Dynamic Gene Transfer. International Journal of Molecular Sciences, 22(7), 3770. https://doi.org/10.3390/ijms22073770







