Abstract
NifS and NifU (encoded by nifS and nifU) are generally dedicated to biogenesis of the nitrogenase Fe–S cluster in diazotrophs. However, nifS and nifU are not found in N2-fixing Paenibacillus strains, and the mechanisms involved in Fe–S cluster biosynthesis of nitrogenase is not clear. Here, we found that the genome of Paenibacillus polymyxa WLY78 contains the complete sufCDSUB operon, a partial sufC2D2B2 operon, a nifS-like gene, two nifU-like genes (nfuA-like and yutI), and two iscS genes. Deletion and complementation studies showed that the sufC, sufD, and sufB genes of the sufCDSUB operon, and nifS-like and yutI genes were involved in the Fe–S cluster biosynthesis of nitrogenase. Heterologous complementation studies demonstrated that the nifS-like gene of P. polymyxa WLY78 is interchangeable with Klebsiella oxytoca nifS, but P. polymyxa WLY78 SufCDB cannot be functionally replaced by K. oxytoca NifU. In addition, K. oxytoca nifU and Escherichia coli nfuA are able to complement the P. polymyxa WLY78 yutI mutant. Our findings thus indicate that the NifS-like and SufCDB proteins are the specific sulfur donor and the molecular scaffold, respectively, for the Fe–S cluster formation of nitrogenase in P. polymyxa WLY78. YutI can be an Fe–S cluster carrier involved in nitrogenase maturation in P. polymyxa WLY78.
1. Introduction
Iron–sulfur (Fe–S) clusters are contained in a diverse group of proteins called Fe–S proteins, which participate in a wide variety of cellular processes, such as nitrogen fixation, respiration, DNA repair, and gene regulation [1,2,3,4]. So far, three pathways for Fe–S cluster assembly identified in bacteria are the nitrogen fixation (Nif) system, the iron sulfur cluster (Isc) system, and the sulfur formation (Suf) system [5]. The Nif system was initially discovered in the maturation of nitrogenase in Azotobacter vinelandii [6]. Nitrogenase that catalyzes biological nitrogen fixation comprises two components, the Fe protein and the MoFe protein, and both of these are Fe–S proteins. Genetic and biochemical analysis has revealed that the products of nifU and nifS are specifically required for activation of both Fe and MoFe proteins [7,8]. Subsequent studies have suggested that NifS is a cysteine desulfurase that catalyzes the production of sulfur from L-cysteine, while NifU provides a molecular scaffold for the assembly and transfer of the Fe–S cluster to the components of nitrogenase [9,10,11].
In addition to the Nif system that is specifically responsible for the synthesis of the Fe–S clusters of nitrogenase, Isc and Suf are required for a broad range of cell functions [5]. Escherichia coli and closely related enterobacteria possess both Isc and Suf systems, encoded by the iscRSUA-hscBA-fdx-iscX operon and the sufABCDSE operon, respectively. The Isc system functions under normal growth conditions, whereas the Suf system operates under stress conditions such as iron starvation and oxygen limitation [12,13,14]. In the Isc machinery, IscS is a cysteine desulfurase that serves as a sulfur donor and IscU is a scaffold protein. HscA/HscB (molecular chaperones) and ferredoxin (Fdx) interact with IscU containing the Fe–S cluster and facilitate the cluster transfer from IscU to target proteins. IscA plays a role in the transfer of the Fe–S cluster from IscU to target proteins, while IscR is a negative regulator [15,16,17]. In the Suf system, SufS is a cysteine desulfurase and SufE serves as a sulfur shuttle protein that interacts with SufS. The three proteins SufB, SufC, and SufD associate as a SufBCD complex that functions as a scaffold. Thus, SufSE produces sulfur and SufCDB as a scaffold assembles an Fe–S cluster. SufA is an IscA homologue and acts as an Fe–S cluster-specific carrier to target proteins [18,19,20,21].
In the Gram-positive model of the bacterium Bacillus subtilis, Fe–S cluster assembly is mediated by the Suf system consisting of a sufCDSUB operon and a distant sufA [22]. B. subtilis SufS functions as a cysteine desulfurase, and SufBCD is a scaffold, both of which are functionally similar to the corresponding E. coli Suf components [23]. B. subtilis SufU interacts with SufS and is involved in sulfur transfer from SufS to SufBCD, just as SufE is involved in the E. coli Suf system. B. subtilis SufSU is interchangeable with E. coli SufSE but not with IscSU [20,24].
In addition to the three distinct Nif, Suf, and Isc systems, some Fe–S cluster biosynthetic genes have been identified. For example, E. coli NfuA binds a 4Fe–4S cluster and transfers this cluster to the target protein and acts as a scaffold/chaperone for damaged Fe–S proteins under oxidative stress and iron starvation conditions [25,26,27]. E. coli CsdA is the third cysteine desulfurase except for IscS and SufS, and it is also engaged in two separate sulfur transfer pathways by recruiting Suf components and by interacting with CsdE and CsdL participates [28].
Paenibacillus polymyxa WLY78 is a Gram-positive, facultative anaerobic, endospore-forming, N2-fixing bacterium. The genome of this bacterium contains a minimal and compact nif gene cluster composed of nine genes (nifB nifH nifD nifK nifE nifN nifX hesA nifV) [29]. This minimal nif cluster has great potential use in engineering nitrogen fixation into non-N2-fixing organisms [30]. Our recent study demonstrated that the minimal nif gene cluster enables E. coli to synthesize the catalytically active nitrogenase, but the specific activity of the enzyme expressed in E. coli was only approximately 10% of that observed in Paenibacillus [29]. Compared to the nif gene clusters of A. vinelandii and Klebsiella oxytoca, the P. polymyxa WLY78 nif cluster lacks nifS and nifU. Thus, we attempted to identify whether nifS-like and nifU-like or other Fe–S cluster biosynthetic genes are responsible for the Fe–S cluster assembly of nitrogenase.
Here, we searched the genome of P. polymyxa WLY78 and found that there are an entire sufCDSUB operon, a partial sufC2D2B2 operon, a partial isc system (iscSR), and a single iscS (iscS2), one nifS-like, and two nifU-like genes (nfuA-like and yutI). Mutation and complementation analysis of these putative Fe–S assembly genes revealed that SufCDB and NifS-like proteins are essential for biosynthesis of nitrogenase. Furthermore, heterologous complementation studies demonstrated that K. oxytoca nifS is able to restore the nitrogenase activity of the nifS-like mutant of P. polymyxa WLY78, but K. oxytoca nifU cannot complement any of the sufC, sufD, and sufB mutants. We also demonstrated that yutI is involved in nitrogen fixation, but other genes are not. Our study not only reveals the distribution and functions of the Fe–S cluster biosynthetic genes involved in nitrogen fixation in P. polymyxa WLY78, but also provides insight into the mechanisms of Fe–S cluster assembly.
2. Results
2.1. The Putative Fe–S Cluster Biosynthetic Genes in the Genome of P. polymyxa WLY78
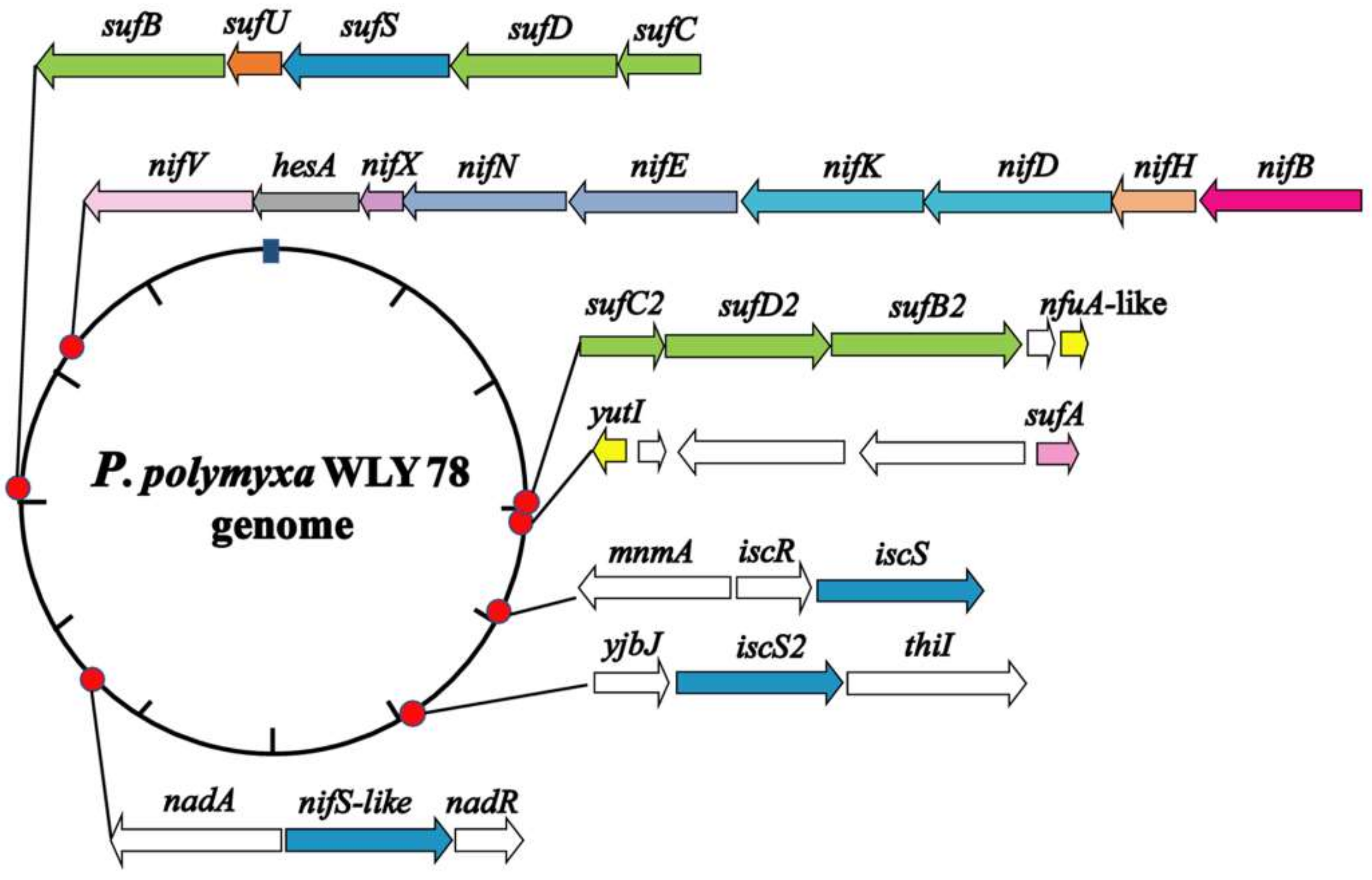
We searched the genome of P. polymyxa WLY78 by using MetalPredator (http://metalweb.cerm.unifi.it/tools/metalpredator/, accessed on 23 February 2020) and identified 118 genes encoding putative Fe–S cluster-containing proteins and Fe–S cluster biosynthetic genes (Table S1). As shown in Figure 1, the Fe–S cluster biosynthetic genes in P. polymyxa WLY78 include an entire Suf system (sufCDSUB), a sufA gene, a partial Suf system (sufC2D2B2), a partial Isc system (iscSR), a single iscS2 gene, and a nifS-like and two nifU-like (nfuA-like and yutI) genes.
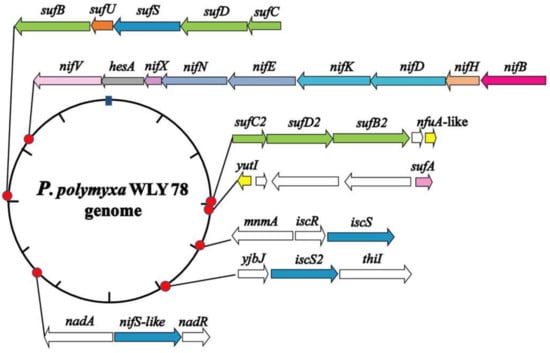
Figure 1.
Genomic location of the Paenibacillus polymyxa WLY78 nif gene cluster and the putative genes involved in Fe–S cluster biogenesis. The schematic diagram representation shows the relative location, size, and orientation of the genes, and relevant flanking genes.
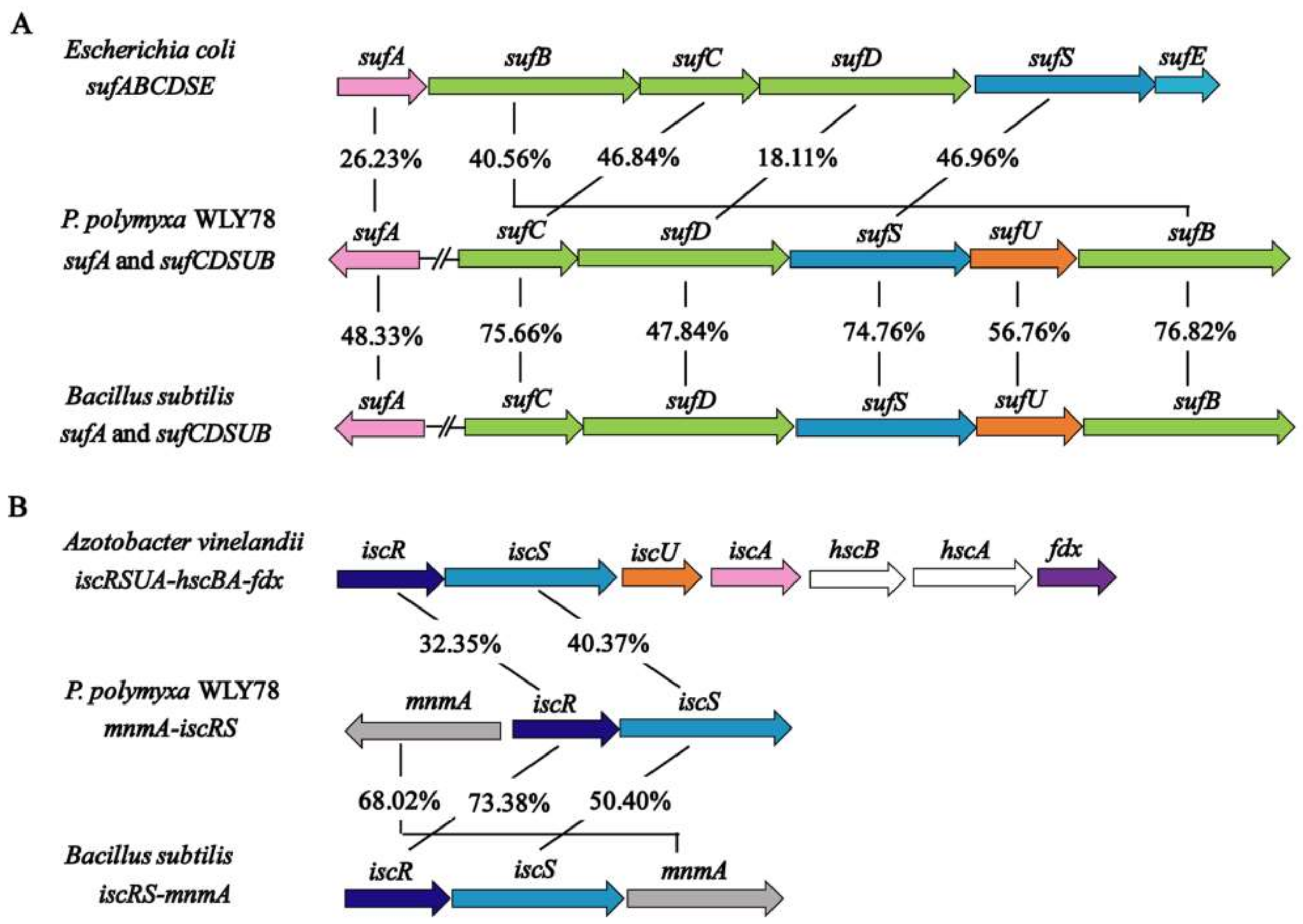
In the genome of P. polymyxa WLY78, sufC, sufD, sufS, sufU, and sufB (sufCDSUB) are tightly arranged with the same transcriptional direction. However, sufA is not in the sufCDSUB cluster of P. polymyxa WLY78, which differs from the suf operon arrangement in E. coli. The current analysis using BLAST alignment showed that the SufA protein of P. polymyxa WLY78 shares 26.23% and 48.33% identities with E. coli and B. subtilis SufA proteins, respectively. The complete sufCDSUB operon of P. polymyxa WLY78 shows a similar arrangement as that of B. subtilis. The SufCDSUB proteins showed 18.11–46.96% identity with their corresponding components from E. coli Suf and 47.84–76.82% identity with their corresponding components from B. subtilis Suf at the amino acid level (Figure 2A). In addition, P. polymyxa WLY78 carries a partial suf operon (here designated as sufC2D2B2) whose predicted products SufC2, SufD2, and SufB2 showed 84.88%, 62.27%, and 84.73% identities with their corresponding components of the entire Suf system, respectively.
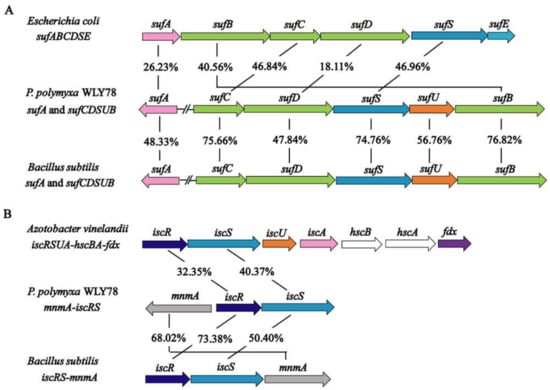
Figure 2.
Comparison of the P. polymyxa WLY78 suf and isc cluster with other bacteria. (A) Comparison of the P. polymyxa sufCDSUB operon and the sufA gene with Escherichia coli and Bacillus subtilis suf operons and sufA genes. (B) Comparison of the P. polymyxa iscRS operon with the Azotobacter vinelandii and B. subtilis isc operons. For gene products with similar sequences, their amino acid identity (%) is indicated.
The genome of P. polymyxa WLY78 contains an iscR gene and two iscS genes. Of the two iscS genes, one is linked to iscR as a dicistronic iscSR operon and the other (here designated as iscS2) is located elsewhere. The iscRS operon of P. polymyxa WLY78 has a similar organization as that of B. subtilis. However, compared to the isc operon (iscRSUA-hscBA-fdx) of A. vinelandii and E. coli, the P. polymyxa WLY78 isc operon (iscRS) lacks iscUA and other genes (Figure 2B).
Unlike A. vinelandii and K. oxytoca, P. polymyxa WLY78 does not have nifS and nifU but contains a nifS-like gene and two nifU-like genes (nfuA-like and yutI) that are not associated with the nif gene cluster (nifB nifH nifD nifK nifE nifN nifX hesA nifV). P. polymyxa WLY78 has a nifS-like gene encoding a 397-amino-acid protein, which showed 28.77% and 28.36% identity with NifS of A. vinelandii and K. oxytoca, respectively (Figure 3). Similar to A. vinelandii and K. oxytoca NifS, the NifS-like protein of P. polymyxa WLY78 has conserved residues that may be involved in pyridoxal-phosphate (PLP) binding and invariable residues Cys that may be involved in substrate binding. A protein alignment revealed that the amino acid sequences derived from two NifU-like proteins with a single domain (NfuA-like and YutI) have high similarity with those of the C-terminal domain of NifU from A. vinelandii and K. oxytoca and the C-terminal domain of NfuA from E. coli and A. vinelandii (Figure 4). Moreover, they have a strict conservation of the CXXC motif, which is the predicted site for Fe–S cluster assembly.
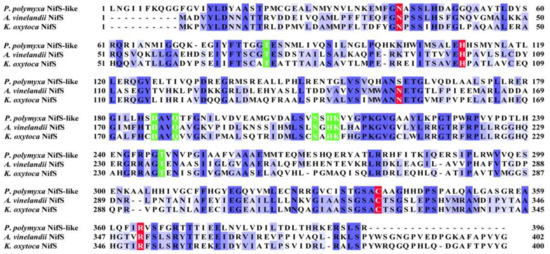
Figure 3.
Comparison of amino acid sequences of the NifS-like protein from P. polymyxa WLY78 and NifS from A. vinelandii and K. oxytoca. Amino acid sequences were aligned using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 25 March 2021). Conserved residues involved in pyridoxal-phosphate (PLP) binding and activity are indicated by a green background. Invariable cysteine and other residues involved in substrate binding are indicated by a red background.

Figure 4.
Comparison of NifU-like proteins (YutI and NfuA-like) from P. polymyxa, NifU from A. vinelandii, NifU from K. oxytoca, and NfuA from E. coli. Protein sequences were aligned using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 25 March 2021). Conserved CXXC motifs are highlighted with a red background.
2.2. NifS-Like Protein Is Essential for Nitrogenase Synthesis
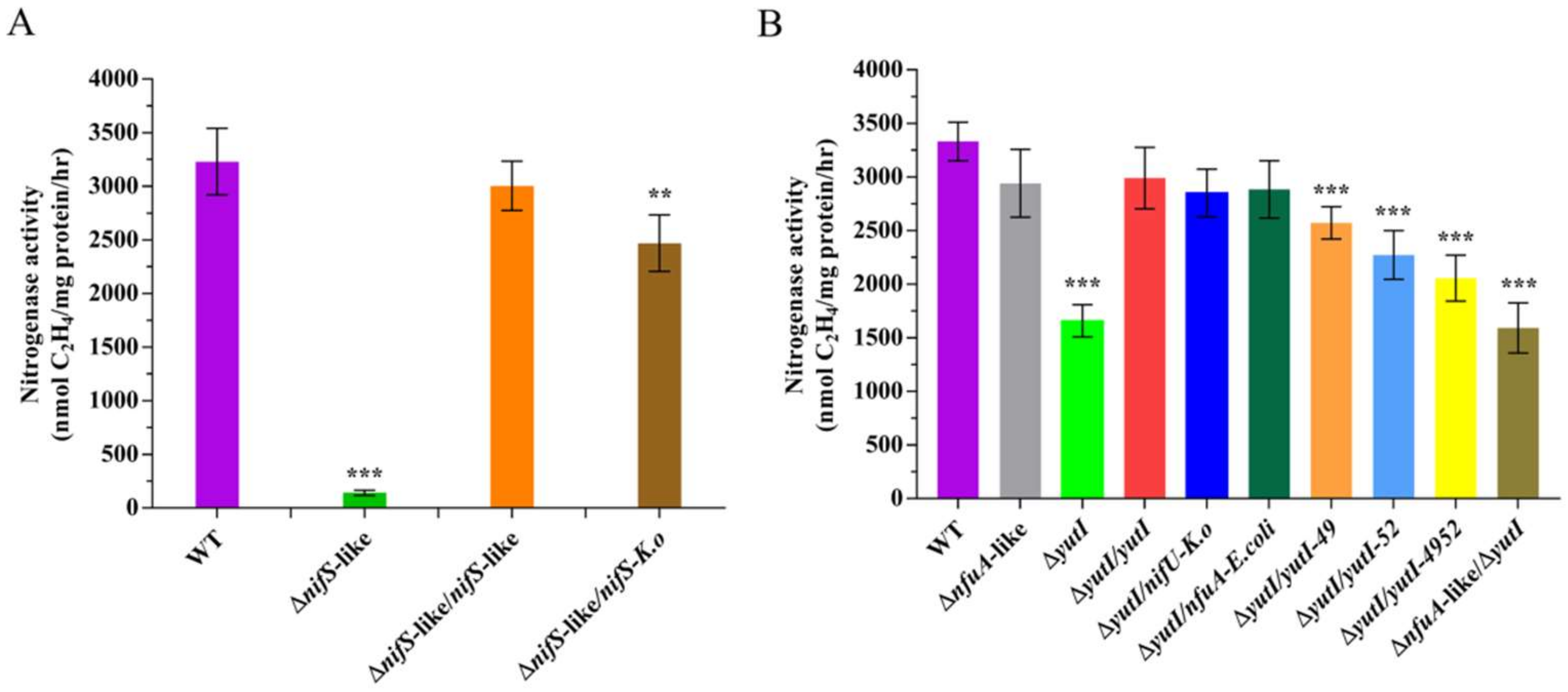
To determine whether the NifS-like protein is involved in the Fe–S cluster assembly of nitrogenase in P. polymyxa WLY78, we constructed an in-frame deletion mutant (ΔnifS-like) and a complementation strain (ΔnifS-like/nifS-like). In comparison with wild-type P. polymyxa WLY78, the ΔnifS-like mutant exhibited nearly no activity under the nitrogen-limited condition, indicating that nifS-like is required for nitrogen fixation. Complementation of ΔnifS-like with the nifS-like gene carried in the plasmid was able to restore nitrogenase activity, suggesting that a change in nitrogenase activity was due solely to the deletion of nifS-like (Figure 5A). Moreover, heterologous complementation of the ΔnifS-like mutant with K. oxytoca nifS partially restored the effect of nifS-like mutation. These data suggest that the nifS-like gene of P. polymyxa WLY78 and the nifS gene of K. oxytoca are similar in function.
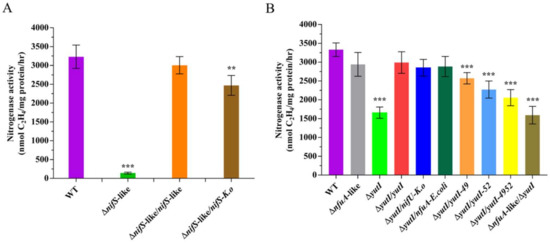
Figure 5.
Effect of disruption of nifS-like and nifU-like genes on nitrogenase activity. (A) The nitrogenase activities of wild-type (WT), ΔnifS-like (deletion mutant), ΔnifS-like/nifS-like (complementation strain), and ΔnifS/nifS-K.o (complementation by K. oxytoca nifS). (B) Nitrogenase activities of wild-type (WT), ΔnfuA and ΔyutI (nfuA-like and yutI deletion mutant), ΔyutI/yutI (yutI complementation strain), ΔyutI/nifU (complementation by K. oxytoca nifU), ΔyutI/nfuA-E. coli (complementation by E. coli nfuA), ΔyutI/yutI-49, ΔyutI/yutI-52, ΔyutI/yutI-4952 (cysteine variants of the YutI complementation strain), and ΔnfuA-like/ΔyutI (nfuA-like and yutI double mutant). The nitrogenase activities were assayed by the C2H2 reduction method and expressed in nmol C2H4/mg protein/hr. The nitrogenase activity of the WT strain was used as a control. Results are representative of at least three independent experiments. Error bars indicate the SD. ** p < 0.01; *** p < 0.001.
2.3. NifU-Like Protein (YutI) Is Involved in Nitrogen Fixation, but NfuA-Like Protein Is Not
Sequence analysis indicates the presence of two genes (nfuA-like and yutI genes), encoding for 85 and 81 amino acids, respectively, that share similarity with the C-terminus of the Fe–S scaffold protein NifU and the Fe–S carrier NfuA in A. vinelandii (Figure 4). The current analysis using the BLAST alignment showed that NfuA-like and YutI proteins encoded by nfuA-like and yutI genes have 61.73% identity. In this study, the in-frame deletion mutants ΔnfuA-like and ΔyutI were constructed. The ΔnfuA-like mutant showed almost similar nitrogenase activity as wild-type P. polymyxa WLY78, suggesting that the nfuA-like gene is not involved in nitrogen fixation (Figure 5B).
The activity of the ΔyutI mutant was approximately 50% that in wild-type P. polymyxa WLY78 (Figure 5B). Complementation experiments showed that the yutI gene carried on the plasmid was able to restore the nitrogenase activity of the ΔyutI mutant. The data suggest that P. polymyxa WLY78 yutI is involved in nitrogen fixation. Deletion of both nfuA and yutI (ΔnfuA-like/ΔyutI double mutant) did not result in a further decline in nitrogenase activity, suggesting that NfuA-like has a limited ability to serve the function of YutI in nitrogen fixation. Furthermore, heterologous complementation studies showed that K. oxytoca nifU and the E. coli nfuA gene were able to restore the nitrogenase activity of the ΔyutI mutant, suggesting that P. polymyxa yutI performs similar functions as K. oxytoca nifU and E. coli nfuA.
To investigate whether the two conserved cysteine residues (Cys-49 and Cys-52) in the YutI protein play roles in Fe–S assembly, the ΔyutI mutant was complemented with the mutated yutI gene whose coding product Cys-49 or Cys-52 or both cysteine residues were replaced with alanine residue(s). As shown in Figure 5B, the YutI protein with mutated Cys-49 or Cys-52, especially with the two mutated cysteine residues, could not complement the yutI mutant as well as the wild-type yutI gene did. These results indicate that the conserved Cys-49 and Cys-52 in the YutI protein are responsible for the Fe–S cluster binding of nitrogenase.
2.4. IscS Is Not Required for Nitrogen Fixation
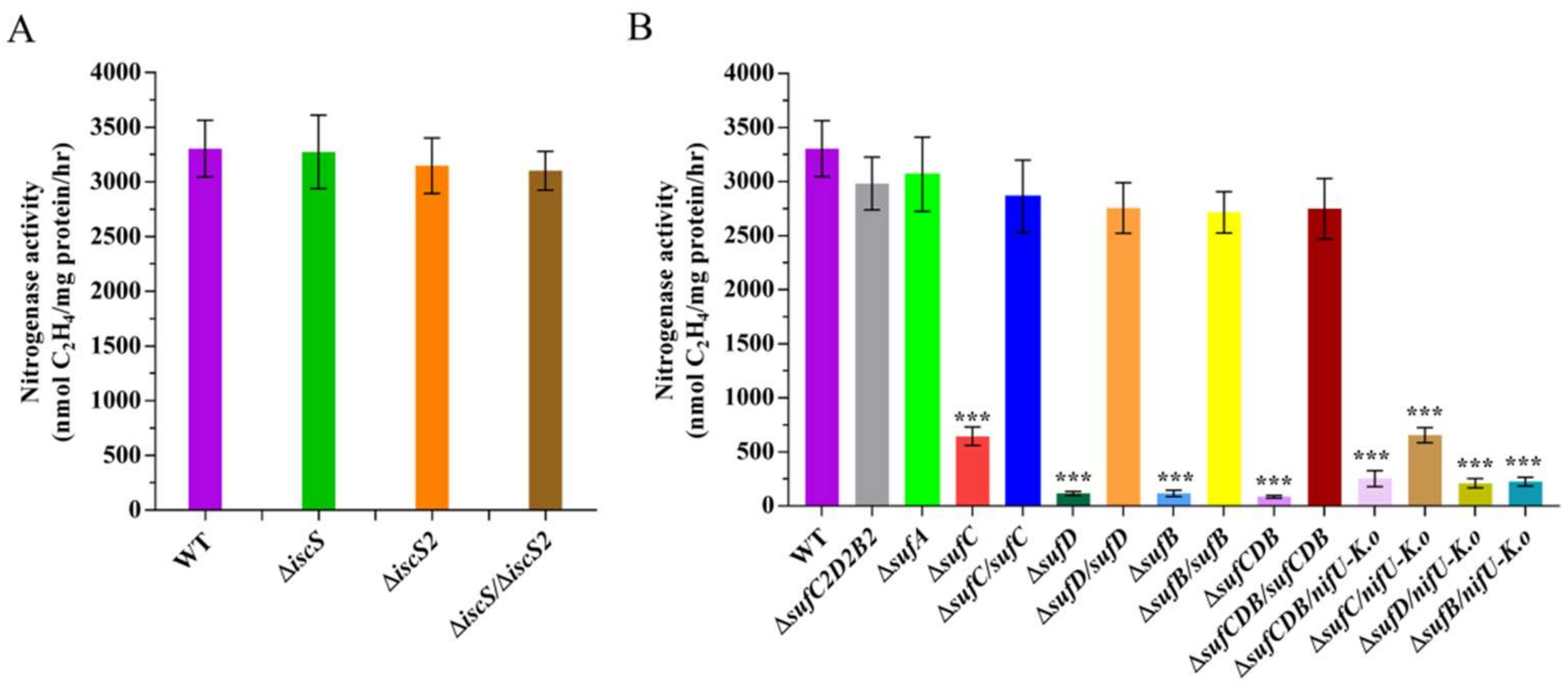
Although P. polymyxa WLY78 contains two iscS genes (iscS and iscS2), no IscU homologues have been identified. Amino acid sequence analysis reveals that IscS and IscS2 of P. polymyxa have 35% identity. However, only IscS has conserved the SSGSACTS sequence, which represents the feature of the IscS protein (Figure S1). We constructed in-frame deletion mutants ΔiscS and ΔiscS2 and the double mutant ΔiscS/ΔiscS2 and found that nitrogenase activities were similar in all iscS mutants and the wild-type strain under the N2-fixing condition (Figure 6A), suggesting that both IscS and IscS2 proteins are not required for nitrogen fixation in P. polymyxa WLY78.
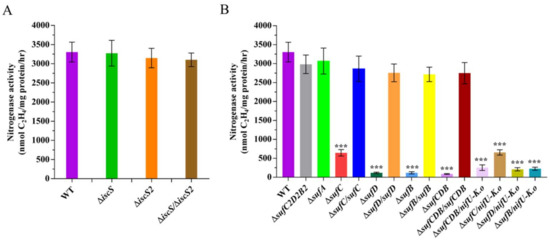
Figure 6.
Effect of disruption of iscS and suf genes on nitrogenase activities. (A) Nitrogenase activities of wild-type (WT), ΔiscS (deletion mutant), ΔiscS2 (deletion mutant), and ΔiscS/ΔiscS2 (double deletion mutant). (B) Nitrogenase activities of wild-type (WT), ΔsufC2D2B2 (deletion mutant), ΔsufA (deletion mutant), ΔsufC (deletion mutant), ΔsufC/sufC (complementation strain), ΔsufD (deletion mutant), ΔsufD/sufD (complementation strain), ΔsufB (deletion mutant), ΔsufB/sufB (complementation strain), ΔsufCDB (deletion mutant), ΔsufCDB/sufCDB (complementation strain), and ΔsufCDB/nifU-K.o, ΔsufC/nifU-K.o, ΔsufD/nifU-K.o, ΔsufB/nifU-K.o (complementation by K. oxytoca nifU). The nitrogenase activities of these strains were assayed by the C2H2 reduction method and expressed in nmol C2H4/mg protein/hr. The nitrogenase activity of the WT strain was used as a control. Results are representative of at least three independent experiments. Error bars indicate the SD. *** p < 0.001.
2.5. The sufCDSUB Operon Is Required for Nitrogenase, but the sufB2C2D2 Operon and sufA Gene Is Not
In B. subtilis, there is only the Suf pathway for Fe–S generation and the knockout of suf genes (any one of sufCDSUB) is lethal [23]. However, P. polymyxa WLY78 contains another partial suf operon (sufC2D2B2) due to which the inactivation of sufCDB does not lead to lethality. Attempts to knock out sufSU in P. polymyxa WLY78 were unsuccessful, suggesting that sufSU might exist to maintain basic levels of Fe–S assembly for survival. In this study, the deletion mutants, including ΔsufC, ΔsufD, ΔsufB, ΔsufCDB, ΔsufC2D2B2, and ΔsufA, were successfully disrupted. qRT-PCR further revealed that the messenger RNA (mRNA) levels of other suf genes in the sufCDSUB operon were not affected by one suf gene mutation (Figure S2), suggesting that the deletion did not generate a polarity effect.
The nitrogenase activities of these suf mutants and wild-type P. polymyxa WLY78 were comparatively analyzed (Figure 6B). The suf gene deletion mutants (ΔsufC, ΔsufD, ΔsufB, and ΔsufCDB) showed a significant decrease in nitrogenase activity compared to the wild-type strain, while ΔsufB2C2D2 and ΔsufA mutants exhibited almost similar activity as the wild-type strain. Furthermore, complementation of ΔsufC, ΔsufD, and ΔsufB mutants with the corresponding P. polymyxa WLY78 suf gene restored nitrogenase activity to the wild-type level, suggesting that sufCDB genes are essential for the Fe–S cluster assembly of nitrogenase in P. polymyxa WLY78. We also tried to complement with K. oxytoca nifU and found that nifU could not restore the nitrogenase activity of any suf mutants (Figure 6B). These results indicate that K. oxytoca NifU cannot replace SufCDB in P. polymyxa WLY78.
3. Discussion
Nitrogenase is a complex metalloenzyme, and the Fe–S clusters of nitrogenase play a critical function in electron transfer and in the reduction of substrates driven by the free energy liberated from Mg-ATP hydrolysis [6,11,31]. In the N2-fixing model bacteria A. vinelandii and K. oxytoca, NifS and NifU are specifically responsible for the Fe–S cluster assembly of nitrogenase, and nifU and nifS usually are clustered with other nif genes [32,33]. However, the nif clusters of P. polymyxa WLY78 and other N2-fixing Paenibacillus species do not have nifS and nifU [34]. In this study, we report that nifS-like, nifU-like (yutI), and sufCDB genes are involved in the Fe–S cluster assembly of nitrogenase in P. polymyxa WLY78.
The NifS protein was involved in providing sulfur to the iron–sulfur clusters of nitrogenase [35]. In this study, we revealed that the nifS-like gene of P. polymyxa WLY78 is involved in nitrogen fixation. The NifS-like protein from P. polymyxa WLY78 and NifS from K. oxytoca and A. vinelandii have common characteristics, which contain conserved residues known to be essential for activity, substrate recognition, and PLP binding. However, P. polymyxa WLY78 nifS-like exhibits two features different from A. vinelandii nifS and K. oxytoca nifS. One is that each of A. vinelandii nifS and K. oxytoca nifS is linked together with nifU as a nifSU operon that is located in a large nif gene cluster [32,33], while P. polymyxa WLY78 nifS-like is not linked together with any nif genes. The other feature is that there is a C-terminal extension consisting of 20-21 amino acids, including the consensus sequence SPL(W/Y)(E/D)(M/L)X(K/Q)XG(I/V)D(L/I)XX(/V)XWXXX in A. vinelandii NifS and K. oxytoca NifS [36], while this extension is absent in the NifS-like protein of P. polymyxa WLY78. Deletion of the A. vinelandii nifS gene did not lead to complete loss of nitrogenase activity, suggesting that the housekeeping Fe–S cluster biosynthetic system Isc could weakly replace NifS function [6,37]. We deduce that NifS activity could be replaced at low levels by the housekeeping Fe–S protein SufSU in P. polymyxa WLY78, since loss of NifS-like function does not completely eliminate the capacity for nitrogen fixation. K. oxytoca nifS could complement the P. polymyxa WLY78 nifS-like mutant, which further confirmed that the NifS-like protein is the specific S donor for the assembly of Fe–S clusters for nitrogenase in P. polymyxa WLY78.
NifU was shown as an Fe–S scaffold protein constructed of several domains for maturation of the nitrogenase [38]. The NfuA protein (known as NifU-like proteins) with sequence similarity to the C-terminal domain of NifU was found to be highly conserved among different bacteria [27]. P. polymyxa WLY78 possess two NifU proteins (NfuA-like and YutI) that share significant sequence homology to the C-terminus domain of the NifU family and include the conserved CXXC motif. Such sequence conservation indicates a likely role for NfuA-like and YutI in Fe–S protein maturation in P. polymyxa WLY78. In A. vinelandii, the NfuA protein contains the N-terminal A-type domain and the C-terminal Nfu-type domain, and functional loss of the Nfu-type domain has no obvious effect on nitrogenase maturation [39]. In this study, YutI has only one Nfu domain, which was found to be required for nitrogen fixation in P. polymyxa WLY78, and deletion of yutI results in lower nitrogenase activity. The conserved cysteine residues in the CXXC motif were shown to be functionally important for NfuA in E. coli, A. vinelandii, and Pseudomonas aeruginosa [26,39,40,41]. Accordingly, our results demonstrate that the Cys-49 and Cys-52 residues of the CXXC motif are essential for YutI to carry out its fixating nitrogen function in vivo. Previous studies have shown that the elevated expression of NifU could replace the function of NfuA in A. vinelandii [39]. The heterologous complementation assays performed in this study provide compelling evidence that the NifU of K. oxytoca and the NfuA of E. coli are functionally interchangeable with the YutI of P. polymyxa WLY78. The NfuA as an Fe–S cluster carrier was able to accept the Fe–S cluster from scaffold proteins IscU or SufBCD in vitro [27]. Therefore, the YutI of P. polymyxa WLY78 could potentially be a stand-alone carrier protein, playing a role similar to NfuA to accept the Fe–S cluster from the scaffold protein, as described in other microorganisms.
The cysteine desulfurase IscS is a highly conserved and essential component of the Isc system that serves as a sulfur donor for Fe–S cluster biogenesis [16]. In the present work, we showed that the iscS and iscS2 of P. polymyxa WLY78 are not required for nitrogen fixation. The result is in line with our previous work that showed that the iscSR system cannot increase any activity of E. coli 78-7 carrying a Paenibacillus nif gene operon [42]. In addition, the Isc system is used for the maturation of other Fe–S proteins but not nitrogenase in A. vinelandii [6,43]. Inactivation of iscS led to no defects in Fe–S metabolism, suggesting that IscS is not used for housekeeping Fe–S protein maturation in P. polymyxa WLY78. However, in B. subtilis, the iscS gene could not be deleted, indicating that IscS participates in essential cellular processes [44].
Besides the NifSU-like and Isc system, the Suf system (the sufCDSUB operon) is highly conserved in N2-fixing Paenibacillus strains [34]. The Suf system has been characterized in several organisms such as E. coli [14,45], B. subtilis [23], Staphylococcus aureus [46], and Synechocystis [47]. Mutations in the suf operon can have severe consequences: for instance, disruption of sufCDSUB or its individual gene is lethal for B. subtilis [23]. The P. polymyxa suf genes had not been characterized before; therefore, it was not clear what impact the disruption of suf would have on the metabolism and viability. Directly obtaining the sufCDSUB operon mutant of P. polymyxa WLY78 was unsuccessful, which places the Suf component as key proteins in Fe–S cluster metabolism. This is in accordance with the fact the Suf Fe–S cluster biosynthetic system is essential for Gram-positive bacteria viability. However, in this study, we disrupted single sufC, sufD, and sufB genes of the suf operon successfully, showing that they are required for nitrogen fixation. This is in line with our previous work that the transcript abundances of sufCDSUB was up-regulated in N2-fixing condition [48] and the suf (sufCDSUB) operon can increase nitrogenase activity of E. coli 78-7 from 10% to 20% [42]. sufS and sufU could not be deleted, suggesting a critical role of these proteins for the survival of P. polymyxa WLY78. Our finding that the sufCDB mutant phenotype could not be rescued by the nifU of K. oxytoca demonstrated that SufCDB is not interchangeable with the NifU of P. polymyxa WLY78. RT-PCR analysis indicated that nifU was transcribed in these complementation strains (Figure S3). We considered one possible explanation for the inability of NifU to replace functionally SufCDB. Namely, that NifU does not productively interact with SufSU and that such a specific interaction might be required for maturation of Fe–S proteins. Taken together, we conclude that P. polymyxa WLY78 sufCDB plays an important role in the Fe–S cluster assembly of nitrogenase.
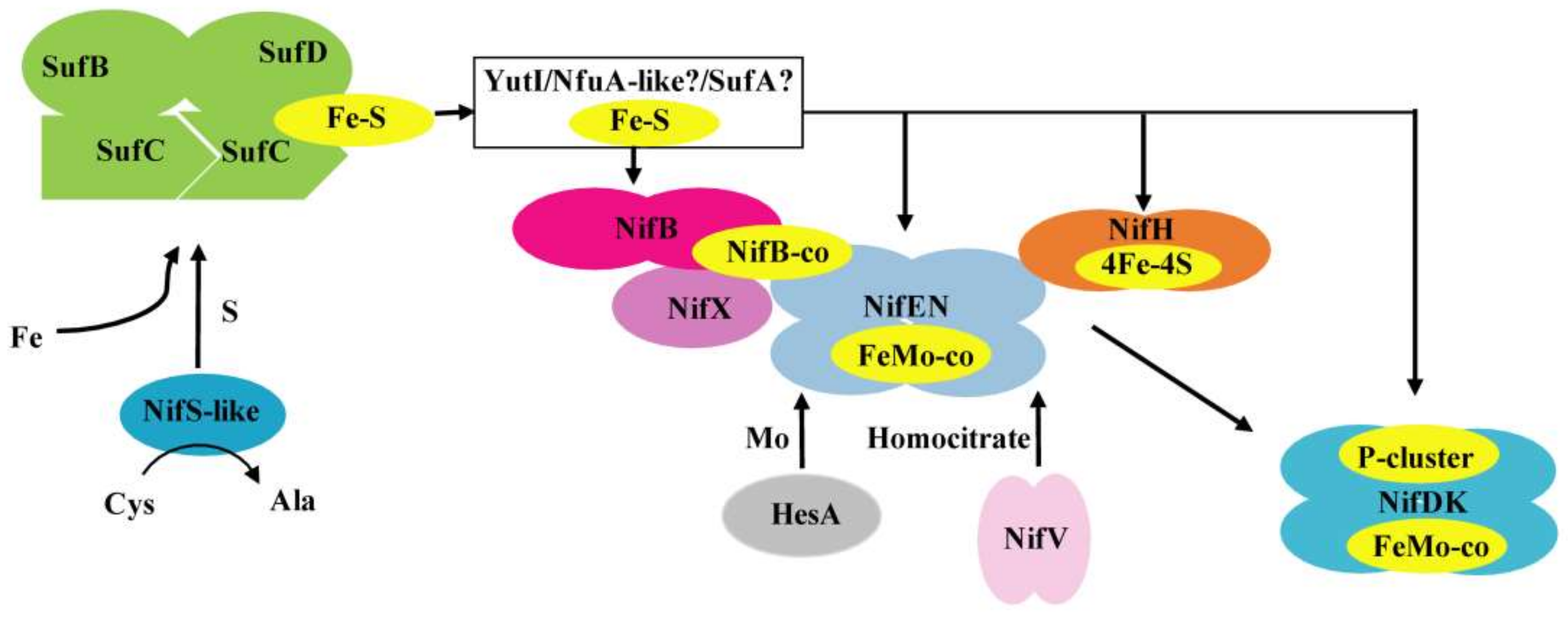
Deletion of nifS-like, yutI, or sufBCD genes leads to a decrease in nitrogenase activity. One reason is insufficiency to supply the Fe–S clusters necessary for nitrogenase maturation. It is also possible that the effect of deletion on the synthesis of nitrogenase might be direct or indirect by affecting nif transcription and Nif expression, as described in K. oxytoca [49]. qRT-PCR revealed that the transcription levels of nifH and nifD in these mutants exhibited a 2–5-fold decrease compared to the wild-type strain (Figure S4A). Western blot analysis with the protein extracts also demonstrated that mutations affected the amounts of both the Fe protein (NifH) and the MoFe protein (NifD) (Figure S4B). According to our results, we proposed the mechanisms involved in the Fe–S cluster biosynthesis of nitrogenase in P. polymyxa WLY78 (Figure 7). The Fe–S cluster assembles on a scaffold protein SufCDB, which receives sulfur from a cysteine desulfurase NifS-like and iron from an as yet unidentified source. Then, the pre-formed Fe–S cluster is transferred to a carrier protein YutI, which delivers it to the final apo-nitrogenase. Further studies are required to determine how these Fe–S proteins are involved in the formation and maturation of nitrogenase. Homologues of NifS and NifU have been identified in non-nitrogen-fixing organisms such as E. coli, yeast, and plants, leading to the proposal that attempts to engineer eukaryotic species for heterologous nitrogen fixation activity can incorporate an endogenous native Fe–S cluster.
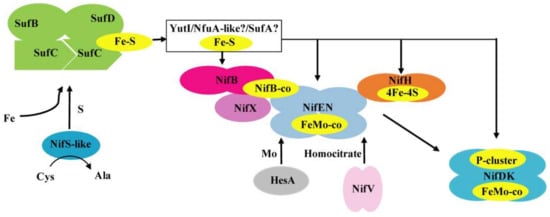
Figure 7.
A model of Fe–S cluster biosynthesis of nitrogenase in P. polymyxa WLY78. The SufBCD complex acts as a scaffold complex receiving sulfur from NifS-like and iron from a still unknown donor. YutI (possibly work with SufA or/and NfuA-like) is as an Fe–S carrier that may receive an Fe–S cluster from the SufBCD scaffold and transfer it to nitrogenase component proteins. The model shows in yellow the Fe–S cluster. The 4Fe–4S cluster is inserted into apo-NifH to activate it. Simple Fe–S clusters were reconstituted on NifB and are converted into NifB-co. NifEN binds NifB-co and converts it into FeMo-co. HesA and NifV provide Mo and homocitrate. NifX may assist NifB, NifEN, and NifDK in FeMo-co precursor trafficking. The FeMo-co and in situ assembly of the P-cluster are inserted into apo-NifDK to form holo-NifDK.
4. Materials and Methods
4.1. Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are summarized in Table S2. P. polymyxa strains and E. coli strains were routinely grown in LB (per liter contains 10 g of NaCl, 5 g of yeast, and 10 g of tryptone) or LD medium (per liter contains 5 g of NaCl, 5 g of yeast, and 10 g of tryptone) at 30 °C with shaking. For assays of nitrogenase activity, P. polymyxa strains were grown in nitrogen-limited media under anaerobic conditions. The nitrogen-limited media contained (per liter) 0.4 g of Na2HPO4, 3.4 g of KH2PO4, 26 mg of CaCl2·2H2O, 30 mg of MgSO4, 0.3 mg of MnSO4, 36 mg of ferric citrate, 7.6 mg of Na2MoO4·2H2O, 10 mg of p-aminobenzoic acid, 5 µg of biotin, 2 mM glutamate, and 4 g of glucose as the carbon source. E. coli strains JM109 were used for routine cloning. Thermo-sensitive vector pRN5101 [50] was used for gene disruption in P. polymyxa WLY78. The shuttle vector pHY300PLK was used for the complementation experiment. When appropriate, antibiotics were added in the following concentrations for maintenance of plasmids: 100 μg/mL of ampicillin, 12.5 μg/mL of tetracycline, and 5 μg/mL of erythromycin.
4.2. Construction of ΔnifS-Like, ΔnfuA-Like, ΔyutI, ΔnfuA-Like/ΔyutI, ΔiscS, ΔiscS2, ΔiscS/ΔiscS2, ΔsufC, ΔsufD, ΔsufB, ΔsufCDB, ΔsufC2D2B2, and ΔsufA Mutants
The in-frame-deletion mutants ΔnifS-like, ΔnfuA-like, ΔyutI, ΔnfuA-like/ΔyutI, ΔiscS, ΔiscS2, ΔsufC, ΔsufD, ΔsufB, ΔsufA, and ΔsufC2D2B2 were constructed through homologous recombination with pRN5101. The upstream (ca. 1 kb) and downstream fragments (ca.1.0 kb) flanking the coding region of iscS, iscS2, nifS-like, nfuA-like, yutI, sufC, sufD, sufB, sufCD, sufA, and sufC2D2B2 were separately PCR-amplified from the genomic DNA of P. polymyxa WLY78. The primers used for these PCR amplifications are listed in Table S3. The two fragments flanking each coding region of iscS, iscS2, nifS-like, nfuA-like, yutI, sufC, sufD, sufB, sufCD, sufA, and sufC2D2B2 were then fused with the Hind III/BamH I-digested pRN5101 vector using the Gibson assembly master mix (New England Biolabs, Ipswich, USA), generating the recombinant plasmids pRDiscS, pRDiscS2, pRDnifS, pRDnfuA, pRDyutI, pRDsufC, pRDsufD, pRDsufB, pRDsufCD, pRDsufA, and pRDsufC2D2B2. Then, each of these recombinant plasmids was transformed into P. polymyxa WLY78, and the double-crossover transformants ΔiscS, ΔiscS2, ΔnifS-like, ΔnfuA-like, ΔyutI, ΔsufC, ΔsufD, ΔsufB, ΔsufCD, ΔsufA, and ΔsufC2D2B2 were selected from the initial Emr transformants after several rounds of nonselective growth at 39 °C and confirmed by PCR amplification and sequencing analysis. Further, pRDnfuA, pRDiscS2, and pRDsufCD were transformed into ΔnfuA-like, ΔiscS, and ΔsufB, respectively, to generate double mutants ΔnfuA-like/ΔyutI, ΔiscS/ΔiscS2, and ΔsufCDB.
4.3. Construction of Plasmids for Complementation of P. polymyxa WLY78 Mutants
Complementation of ΔnifS-like, ΔyutI, ΔsufC, ΔsufD, ΔsufB, and ΔsufCDB was performed. For complementation of ΔnifS-like, a 1578 bp DNA fragment containing the coding region of nifS-like and its own promoter was PCR-amplified from the genomic DNA of P. polymyxa WLY78. For complementation of the ΔyutI mutant, a 626 bp DNA fragment containing the coding region of yutI and its own promoter was PCR-amplified. For complementation of the ΔsufC mutant, a 1173 bp DNA fragment carrying the sufC coding region and its own promoter was PCR-amplified. For complementation of ΔsufD and ΔsufB mutants, a 390 bp promoter region of the sufCDSUB operon and their respective coding region were PCR-amplified. For complementation of the ΔsufCDB mutant, a 2501 bp DNA fragment containing the coding region of sufCD and its promoter, and a 1398 bp nifB coding sequence was PCR-amplified. These fragments were digested with BamHI/HindIII and ligated into the vector pHY300PLK, generating vectors pHYnifS, pHYyutI, pHYsufC, pHYsufD, pHYsufB, and pHYsufCDB. Each of these recombinant plasmids was correspondingly transformed into its mutants, and tetracycline-resistant (Tcr) transformants were selected and confirmed by PCR and sequencing.
The substituted forms of YutI were constructed by the PCR-based mutagenesis method. The pairs of mutagenic primer were designed to be complementary to each other and to span the substituted site. To produce the YutIC49A variant, two separate PCR reactions were conducted with P. polymyxa WLY78 as the template and by using the primer sets P-yutI-F/YutIC49A-R and YutIC49A-F/C-yutI-R. The two PCR products were then fused with the Hind III/BamHI-digested pHY300PLK vector, yielding the complementary plasmid pHYyutI-C49A, and then the plasmid was transformed into ΔyutI. The same procedure was to obtain pHYyutI-C52A and pHYyutI-C49/52A. The accuracy of mutagenesis was verified by DNA sequencing.
For heterologous complementation assays, the nifS gene was PCR-amplified from the genomic DNA of K. oxytoca M5a1, being under the control of the nifS-like promoter of P. polymyxa WLY78, and cloned to the plasmid pHY300PLK, generating the vector pHYnifS (K.o). This vector was transformed into a ΔnifS-like mutant, yielding the heterologous complementation strain ΔnifS-like/nifS-K.o. An 829 bp DNA fragment containing the coding region of nifU from K. oxytoca M5a1 and a 365 bp promoter region of yutI were PCR-amplified and assembled, generating the vector pHYnifU. Then, the constructed vector pHYnifU was transformed into the ΔyutI, ΔsufC, ΔsufD, ΔsufB, and ΔsufCDB mutants, yielding the heterologous complementation strain ΔyutI/nifU-K.o, ΔsufC/nifU-K.o, ΔsufD/nifU-K.o, ΔsufB/nifU-K.o, and ΔsufCDB/nifU-K.o, respectively. Similarly, a 591 bp DNA fragment containing the coding region of nfuA from E. coli and a 365 bp promoter region of yutI were PCR-amplified and assembled, generating the vector pHYnfuA (E. coli). Then it was transformed into the ΔyutI mutant, yielding the heterologous complementation strain ΔyutI/nfuA-E. coli.
4.4. Acetylene Reduction Assays of Nitrogenase Activity
Acetylene reduction assays were performed, as described previously, to measure nitrogenase activity [29]. P. polymyxa WLY78 and its mutant strains were grown overnight in LD medium. The cultures were collected by centrifugation, washed three times with sterilized water, and then resuspended in a nitrogen-limited medium containing 2 mM glutamate as a nitrogen source to a final OD600 of 0.2–0.4. Then, 4 mL of the culture was transferred to a 25 mL test tube, and the test tube was sealed with a rubber stopper. The headspace (21 mL) in the tube was then evacuated and replaced with argon gas. Then, 2.1 mL of C2H2 (10% of the headspace volume) was injected into the test tube. Cultures were incubated at 30 °C. C2H4 production was analyzed by gas chromatography. The nitrogenase activity was expressed in nmol C2H4/mg protein/h. The nitrogenase activity assays were measured at least three times, and the error bars were evaluated as the standard deviation. Statistical analyses were performed using SPSS software version 20 (SPSS Inc., Chicago, IL, USA). One-way ANOVA was employed to check the significant differences between strains. Means of different strains were compared using the least significant difference (LSD) at the 0.05, 0.01, or 0.001 level of probability.
4.5. qRT-PCR and RT-PCR Analysis of Gene Expression
For quantitative real-time-PCR (qRT-PCR), cultures of P. polymyxa WLY78, ∆nifS-like, ∆yutI, ∆sufCDB, ∆sufC, and ∆sufD were grown under N2-fixing conditions (2 mM glutamate and without O2) and harvested after 8 h of incubation. To detect the expression of the nifU gene, total RNA was extracted from ∆sufC, ∆sufD, ∆sufB, ∆sufCDB, ∆sufC/nifU-K.o, ∆sufD/nifU-K.o, ∆sufB/nifU-K.o, and ∆sufCDB/nifU-K.o. Total RNA was isolated using TRIzol (Takara Bio, Tokyo, Japan). The possibility of contamination of genomic DNA was eliminated by digestion with RNase-free DNase I (Takara Bio, Tokyo, Japan). The integrity and size distribution of the RNA were verified by agarose gel electrophoresis, and the concentrations were determined spectrophotometrically. Synthesis of cDNA was carried out using RT Prime Mix according to the manufacturer’s specifications (Takara Bio, Tokyo, Japan). cDNA (0.4 µg) was used for qRT-PCR. The relative transcript levels of sufD, sufS, sufU, and sufB were determined with 16S rDNA as a control by the SYBR Premix Ex Taq (Tli RNaseH Plus) kit (Takara Bio, Tokyo, Japan). Primers for sufC, sufD, sufS, sufU, sufB, nifH, nifD, nifU, and 16S rDNA used for RT-PCR or qRT-PCR are listed in Table S3.
4.6. Western Blot Assays for NifH and NifD Expression
Cultures of P. polymyxa WLY78, ∆nifS-like, ∆sufCDB, and ∆yutI were grown under N2-fixing conditions and harvested after 20 h of incubation. Cells were collected and disrupted in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole) by sonication on ice. Cell debris were removed by centrifugation, and 40 µg of total proteins was analyzed by SDS-PAGE (10% acrylamide) and immunoblotting. Proteins were blotted onto a nitrocellulose (NC) membrane, and the Fe and MoFe proteins were detected using polyclonal anti-NifH and anti-NifD, respectively. Binding of antibodies was visualizedby enhanced chemiluminescence (ECL) (ComWin Biotech, Beijing, China).
5. Conclusions
In P. polymyxa WLY78, the sufC, sufD, and sufB genes of the suf (sufCDSUB) operon, the nifS-like gene, and the yutI gene are required for the Fe–S cluster biosynthesis of nitrogenase. The cysteine desulfurase NifS-like catalyzes the production of sulfur, and SufCDB proteins provide a molecular scaffold for the assembly of the Fe–S cluster. YutI is an Fe–S carrier that is involved in nitrogen fixation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22073771/s1: Figure S1: Comparison of IscS and IscS2 from P. polymyxa, IscS from A. vinelandii, IscS and IscS2 from B. subtilis; Figure S2: The relative transcript levels of suf genes were determined by qRT-PCR; Figure S3: RT-PCR analysis of nifU gene expression in ΔsufCDB/nifU-K.o, ΔsufC/nifU-K.o, ΔsufD/nifU-K.o, ΔsufB/nifU-K.o (complementation by K. oxytoca nifU) and WT (P. polymyxa WLY78); Figure S4: Transcripts of nifHD genes and expression of NifHD proteins. Table S1: P. polymyxa WLY78 proteins predicted to contain Fe-S clusters; Table S2: Bacterial strains and plasmids used in this study; Table S3: Primers used in this study.
Author Contributions
Conceptualization, S.C.; methodology, Q.L., Y.L., and X.L.; formal analysis: Q.L. and S.C.; investigation: Q.L.; writing—original draft preparation, Q.L. and S.C.; writing—review and editing, S.C. and Q.L.; project administration: S.C.; funding acquisition: S.C. and Q.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (no. 2019YFA0904700) and the National Natural Science Foundation of China (grant no. 32000048).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicale.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Roche, B.; Aussel, L.; Ezraty, B.; Mandin, P.; Py, B.; Barras, F. Reprint of: Iron/sulfur proteins biogenesis in prokaryotes: Formation, regulation and diversity. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1827, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Lill, R. Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017, 292, 12754–12763. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta (BBA) Bioenerg. 2021, 1868, 118863. [Google Scholar] [CrossRef]
- Xu, X.M.; Møller, S.G. Iron–sulfur clusters: Biogenesis, molecular mechanisms, and their functional significance. Antioxid. Redox Signal. 2011, 15, 271–307. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef]
- Johnson, D.; Dos Santos, P.; Dean, D. NifU and NifS are required for the maturation of nitrogenase and cannot replace the function of isc-gene products in Azotobacter vinelandii. Biochem. Soc. Trans. 2005, 33, 90–93. [Google Scholar] [CrossRef]
- Dos Santos, P.C.; Smith, A.D.; Frazzon, J.; Cash, V.L.; Johnson, M.K.; Dean, D.R. Iron-sulfur cluster assembly - NifU-directed activation of the nitrogenase Fe protein. J. Biol. Chem. 2004, 279, 19705–19711. [Google Scholar] [CrossRef]
- Zhao, D.; Curatti, L.; Rubio, L.M. Evidence for nifU and nifS participation in the biosynthesis of the iron-molybdenum cofactor of nitrogenase. J. Biol. Chem. 2007, 282, 37016–37025. [Google Scholar] [CrossRef]
- Sickerman, N.S.; Ribbe, M.W.; Hu, Y. Nitrogenase cofactor assembly: An elemental inventory. Acc. Chem. Res. 2017, 50, 2834–2841. [Google Scholar] [CrossRef]
- Burén, S.; Jiménez-Vicente, E.; Echavarri-Erasun, C.; Rubio, L.M. Biosynthesis of nitrogenase cofactors. Chem. Rev. 2020, 120, 4921–4968. [Google Scholar] [CrossRef]
- Rubio, L.M.; Ludden, P.W. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 2008, 62, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Castro, C.; Saini, A.; Outten, F.W. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 2008, 72, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Tokumoto, U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in Archaea and Plastids. J. Biol. Chem. 2002, 277, 28380–28383. [Google Scholar] [CrossRef] [PubMed]
- Outten, F.W.; Djaman, O.; Storz, G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 2004, 52, 861–872. [Google Scholar] [CrossRef]
- Chandramouli, K.; Johnson, M.K. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemestry 2006, 45, 11087–11095. [Google Scholar] [CrossRef] [PubMed]
- Blanc, B.; Gerez, C.; de Choudens, S.A. Assembly of Fe/S proteins in bacterial systems biochemistry of the bacterial ISC system. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 1436–1447. [Google Scholar]
- Tokumoto, U.; Kitamura, S.; Fukuyama, K.; Takahashi, Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 2004, 136, 199–209. [Google Scholar] [CrossRef]
- Vinella, D.; Brochier-Armanet, C.; Loiseau, L.; Talla, E.; Barras, F. Iron-sulfur (Fe/S) protein biogenesis: Phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009, 5, e1000497. [Google Scholar] [CrossRef]
- Tanaka, N.; Kanazawa, M.; Tonosaki, K.; Yokoyama, N.; Kuzuyama, T.; Takahashi, Y. Novel features of the ISC machinery revealed by characterization of Escherichia coli mutants that survive without iron-sulfur clusters. Mol. Microbiol. 2015, 99, 835–848. [Google Scholar] [CrossRef]
- Outten, F.W. Recent advances in the Suf Fe–S cluster biogenesis pathway: Beyond the Proteobacteria. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1853, 1464–1469. [Google Scholar] [CrossRef]
- Garcia, P.S.; Gribaldo, S.; Py, B.; Barras, F. The SUF system: An ABC ATPase-dependent protein complex with a role in Fe–S cluster biogenesis. Res. Microbiol. 2019, 170, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.C.B. B. subtilis as a model for studying the assembly of Fe-S clusters in Gram-positive bacteria. In Fe-S Cluster Enzymes; Pt, A., David, S.S., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2017; Volume 595, pp. 185–212. [Google Scholar]
- Yokoyama, N.; Nonaka, C.; Ohashi, Y.; Shioda, M.; Terahata, T.; Chen, W.; Sakamoto, K.; Maruyama, C.; Saito, T.; Yuda, E.; et al. Distinct roles for U-type proteins in iron-sulfur cluster bio-synthesis revealed by genetic analysis of the Bacillus subtilis sufCDSUB operon. Mol. Microbiol. 2018, 107, 688–703. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, A.G.; Netz, D.J.A.; Miethke, M.; Pierik, A.J.; Burghaus, O.; Peuckert, F.; Lill, R.; Marahiel, M.A. SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J. Bacteriol. 2010, 192, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Py, B.; Gerez, C.; Huguenot, A.; Vidaud, C.; Fontecave, M.; de Choudens, S.O.; Barras, F. The ErpA/NfuA complex builds an oxidation-resistant Fe-S cluster delivery pathway. J. Biol. Chem. 2018, 293, 7689–7702. [Google Scholar] [CrossRef] [PubMed]
- Angelini, S.; Gerez, C.; Choudens, S.O.-D.; Sanakis, Y.; Fontecave, M.; Barras, F.; Py, B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 2008, 283, 14084–14091. [Google Scholar] [CrossRef]
- Py, B.; Gerez, C.; Angelini, S.; Planel, R.; Vinella, D.; Loiseau, L.; Talla, E.; Brochier-Armanet, C.; Serres, R.G.; Latour, J.-M.; et al. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol. Microbiol. 2012, 86, 155–171. [Google Scholar] [CrossRef]
- Trotter, V.; Vinella, D.; Loiseau, L.; de Choudens, S.O.; Fontecave, M.; Barras, F. The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Mol. Microbiol. 2009, 74, 1527–1542. [Google Scholar] [CrossRef]
- Wang, L.Y.; Zhang, L.H.; Liu, Z.Z.; Zhao, D.H.; Liu, X.M.; Zhang, B.; Xie, J.B.; Hong, Y.Y.; Li, P.F.; Chen, S.F.; et al. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet. 2013, 9, e1003865. [Google Scholar] [CrossRef]
- Li, Q.; Chen, S. Transfer of Nitrogen fixation (nif) Genes to non-diazotrophic hosts. ChemBioChem 2020, 21, 1717–1722. [Google Scholar] [CrossRef]
- Hu, Y.; Ribbe, M.W. Biosynthesis of the Metalloclusters of Molybdenum Nitrogenase. Microbiol. Mol. Biol. Rev. 2011, 75, 664–677. [Google Scholar] [CrossRef]
- Arnold, W.; Rump, A.; Klipp, W.; Priefer, U.B.; Pühler, A. Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae. J. Mol. Biol. 1988, 203, 715–738. [Google Scholar] [CrossRef]
- Jacobson, M.R.; E Brigle, K.; Bennett, L.T.; Setterquist, R.A.; Wilson, M.S.; Cash, V.L.; Beynon, J.; Newton, W.E.; Dean, D.R. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J. Bacteriol. 1989, 171, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.-B.; Du, Z.; Bai, L.; Tian, C.; Zhang, Y.; Xie, J.-Y.; Wang, T.; Liu, X.; Chen, X.; Cheng, Q.; et al. Comparative genomic analysis of N2-fixing and non-N2-fixing Paenibacillus spp.: Organization, evolution and expression of the nitrogen fixation genes. PLoS Genet. 2014, 10, e1004231. [Google Scholar] [CrossRef] [PubMed]
- Yuvaniyama, P.; Agar, J.N.; Cash, V.L.; Johnson, M.K.; Dean, D.R. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. USA 2000, 97, 599–604. [Google Scholar] [CrossRef]
- Ali, V.; Shigeta, Y.; Tokumoto, U.; Takahashi, Y.; Nozaki, T. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 2004, 279, 16863–16874. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.R.; Cash, V.L.; Weiss, M.C.; Laird, N.F.; Newton, W.E.; Dean, D.R. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Genet. Genom. 1989, 219, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.M.; Dempsey, A.; Tan, K.T.; Liew, C.C. A modular domain of NifU, a nitrogen fixation cluster protein, is highly conserved in evolution. J. Mol. Evol. 1996, 43, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Naik, S.G.; O’Carroll, I.P.; Huynh, B.-H.; Dean, D.R.; Johnson, M.K.; Dos Santos, P.C. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 2008, 283, 14092–14099. [Google Scholar] [CrossRef]
- Romsang, A.; Duang-Nkern, J.; Saninjuk, K.; Vattanaviboon, P.; Mongkolsuk, S. Pseudomonas aeruginosa nfuA: Gene regulation and its physiological roles in sustaining growth under stress and anaerobic conditions and maintaining bacterial virulence. PLoS ONE 2018, 13, e0202151. [Google Scholar] [CrossRef]
- McCarthy, E.L.; Rankin, A.N.; Dill, Z.R.; Booker, S.J. The A-type domain in Escherichia coli NfuA is required for regenerating the auxiliary [4Fe–4S] cluster in Escherichia coli lipoyl synthase. J. Biol. Chem. 2019, 294, 1609–1617. [Google Scholar] [CrossRef]
- Li, X.-X.; Liu, Q.; Liu, X.-M.; Shi, H.-W.; Chen, S.-F. Using synthetic biology to increase nitrogenase activity. Microb. Cell Factories 2016, 15, 1–11. [Google Scholar] [CrossRef]
- Dos Santos, P.C.; Johnson, D.C.; Ragle, B.E.; Unciuleac, M.-C.; Dean, D.R. Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J. Bacteriol. 2007, 189, 2854–2862. [Google Scholar] [CrossRef]
- Black, K.A.; Dos Santos, P.C. Abbreviated pathway for biosynthesis of 2-thiouridine in Bacillus subtilis. J. Bacteriol. 2015, 197, 1952–1962. [Google Scholar] [CrossRef]
- Bühning, M.; Valleriani, A.; Leimkühler, S. The role of SufS is restricted to Fe–S cluster biosynthesis in Escherichia coli. Biochemistry 2017, 56, 1987–2000. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.A.; Al-Tameemi, H.M.; Mashruwala, A.A.; Rosario-Cruz, Z.; Chauhan, U.; Sause, W.E.; Torres, V.J.; Belden, W.J.; Boyd, J.M. The Suf iron-sulfur cluster biosynthetic system is essential in Staphylococcus aureus, and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect. Immun. 2017, 85, e00100-17. [Google Scholar] [CrossRef] [PubMed]
- Zang, S.-S.; Jiang, H.-B.; Song, W.-Y.; Chen, M.; Qiu, B.-S. Characterization of the sulfur-formation (suf) genes in Synechocystis sp. PCC 6803 under photoautotrophic and heterotrophic growth conditions. Planta 2017, 246, 927–938. [Google Scholar] [CrossRef]
- Shi, H.-W.; Wang, L.-Y.; Li, X.-X.; Liu, X.-M.; Hao, T.-Y.; He, X.-J.; Chen, S.-F. Genome-wide transcriptome profiling of nitrogen fixation in Paenibacillus sp. WLY78. BMC Microbiol. 2016, 16, 1–10. [Google Scholar] [CrossRef]
- Roberts, G.P.; MacNeil, T.; MacNeil, D.; Brill, W.J. Regulation and characterization of protein products coded by the nif (nitrogen fixation) genes of Klebsiella pneumoniae. J. Bacteriol. 1978, 136, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Villafane, R.; Bechhofer, D.H.; Narayanan, C.S.; Dubnau, D. Replication control genes of plasmid pE194. J. Bacteriol. 1987, 169, 4822–4829. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).