The Epstein-Barr Virus Lytic Protein BMLF1 Induces Upregulation of GRP78 Expression through ATF6 Activation
Abstract
1. Introduction
2. Results
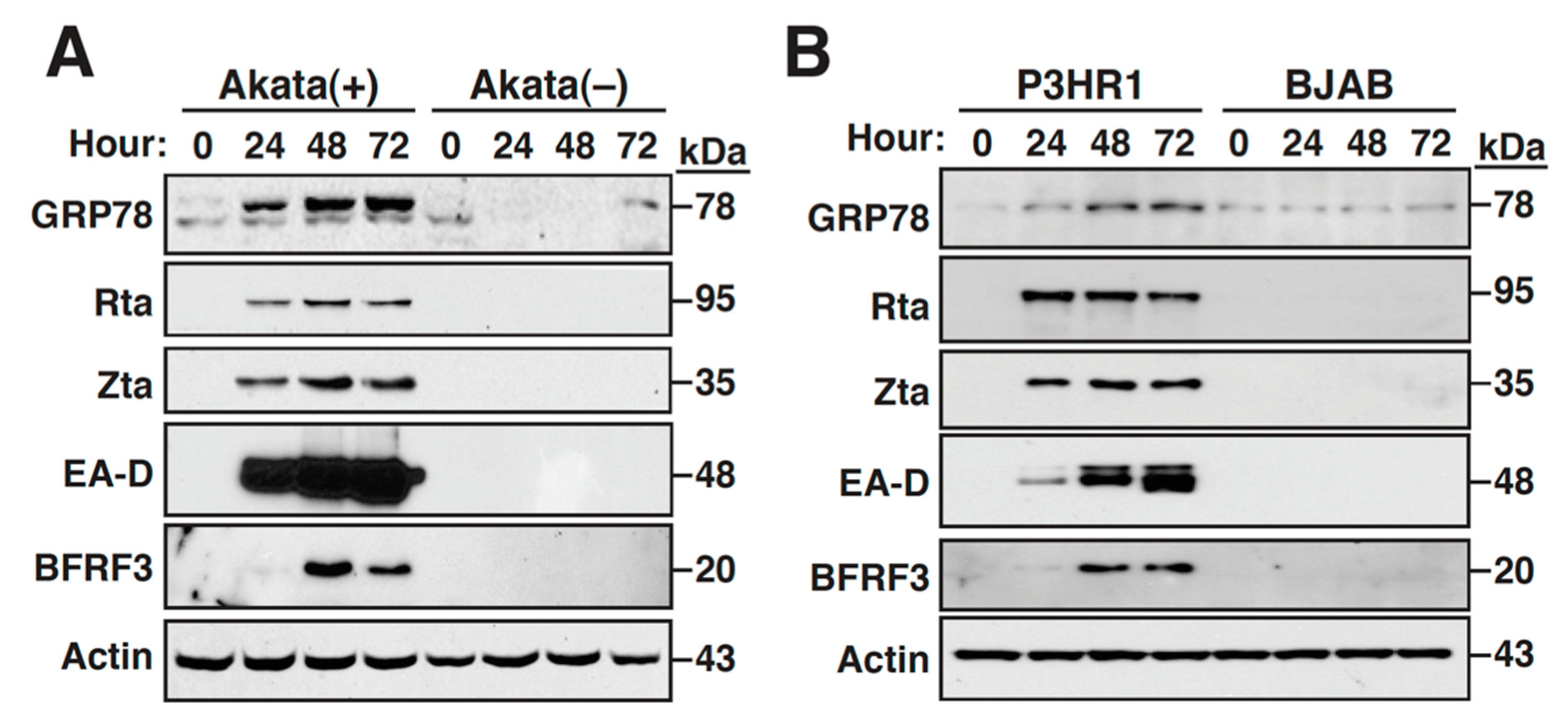
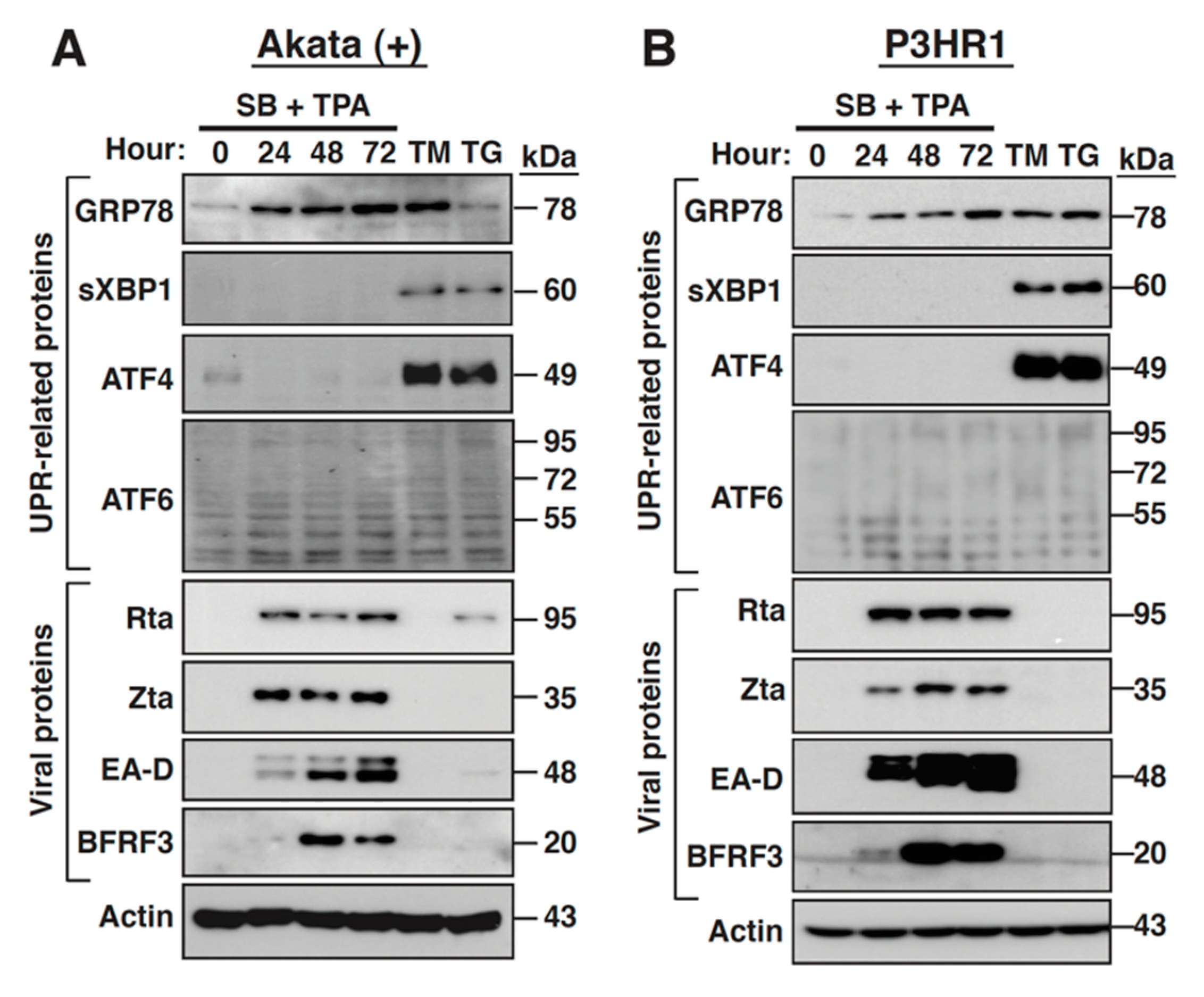
2.1. The Expression of GRP78 Is Upregulated during the EBV Lytic Cycle
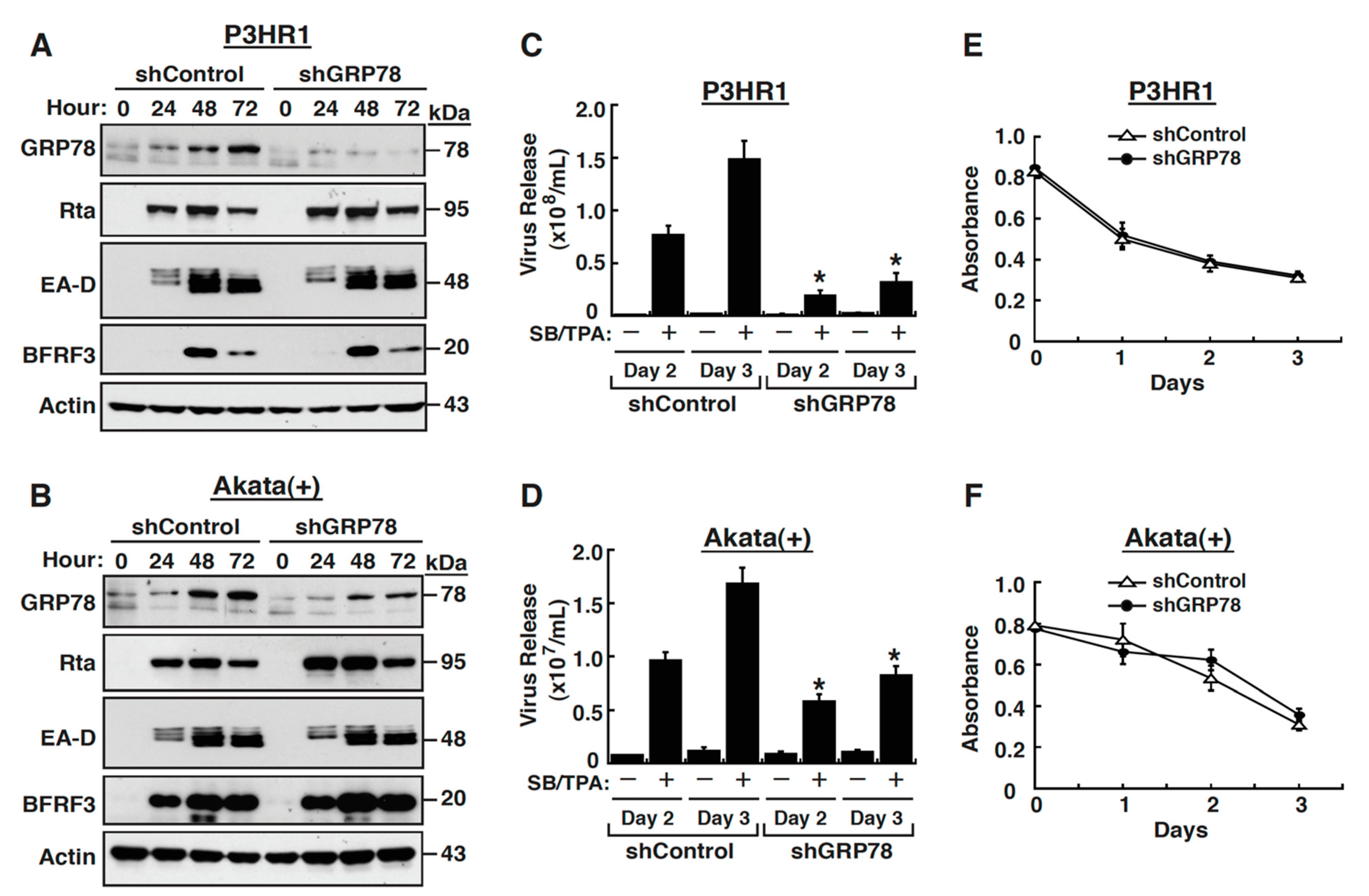
2.2. GRP78 Upregulation Is Important for Completion of the EBV Lytic Cycle
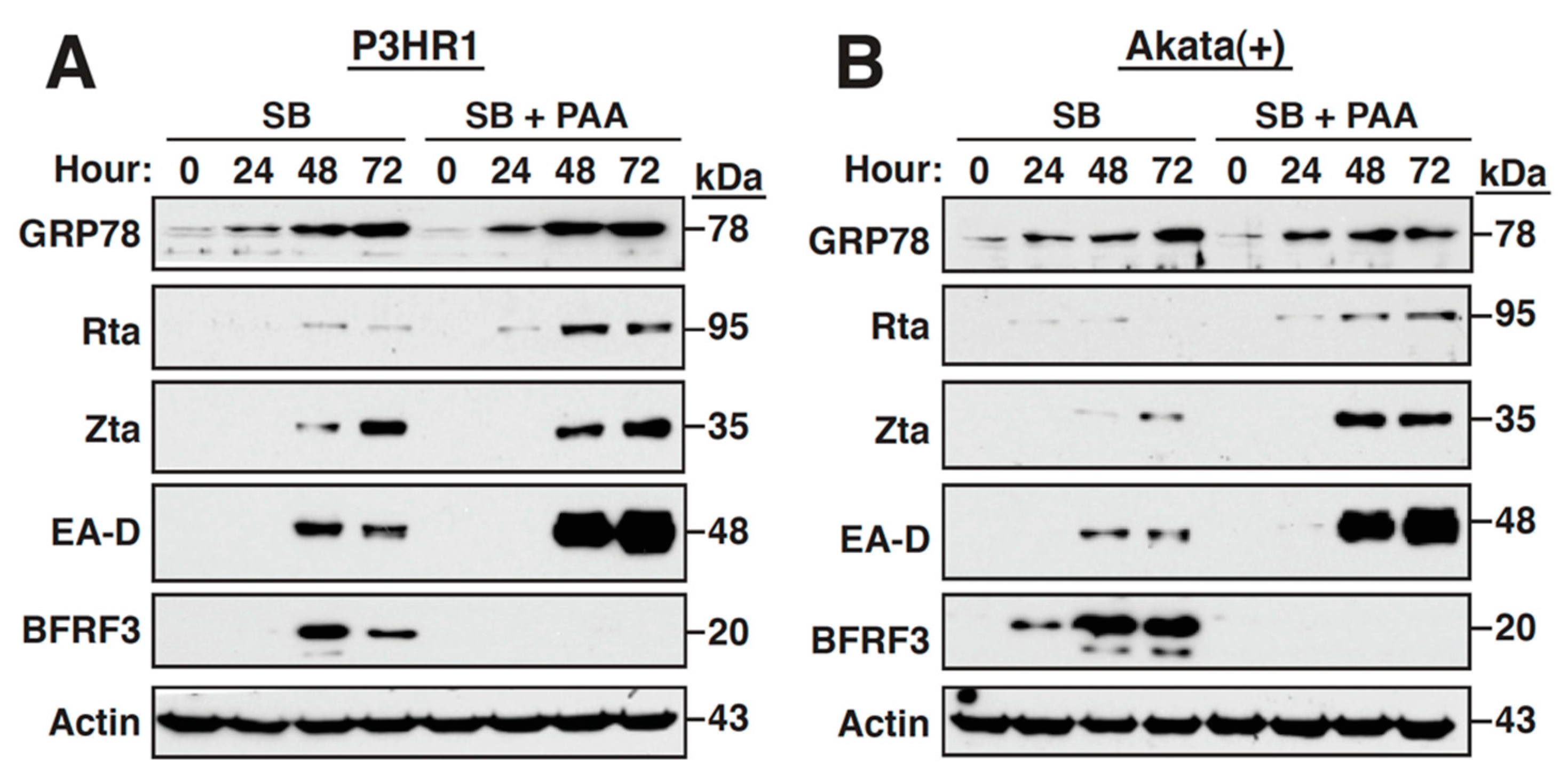
2.3. Inhibition of the Viral DNA Synthesis and Late Gene Expression Has No Effect on the Sustained GRP78 Upregulation
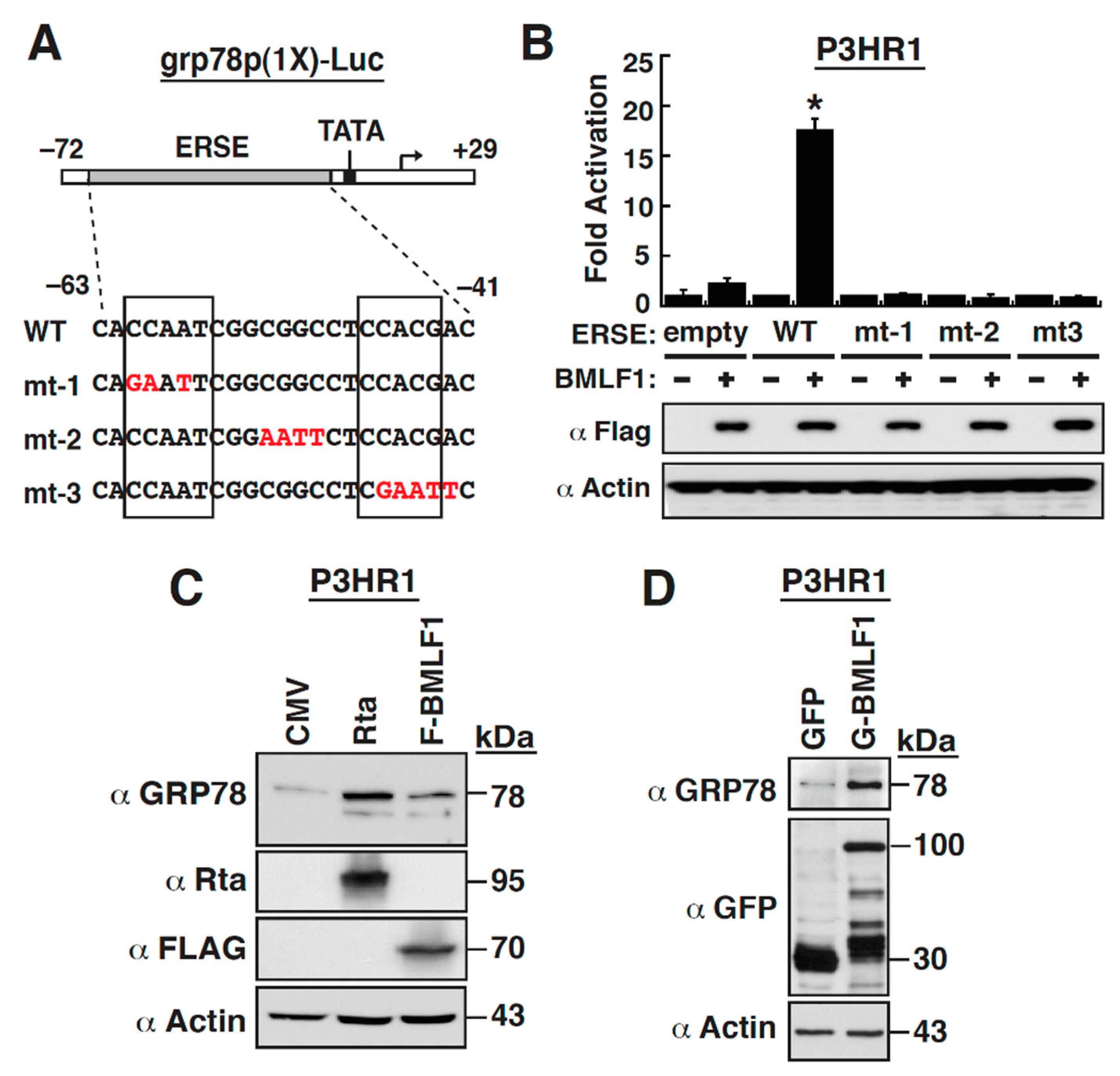
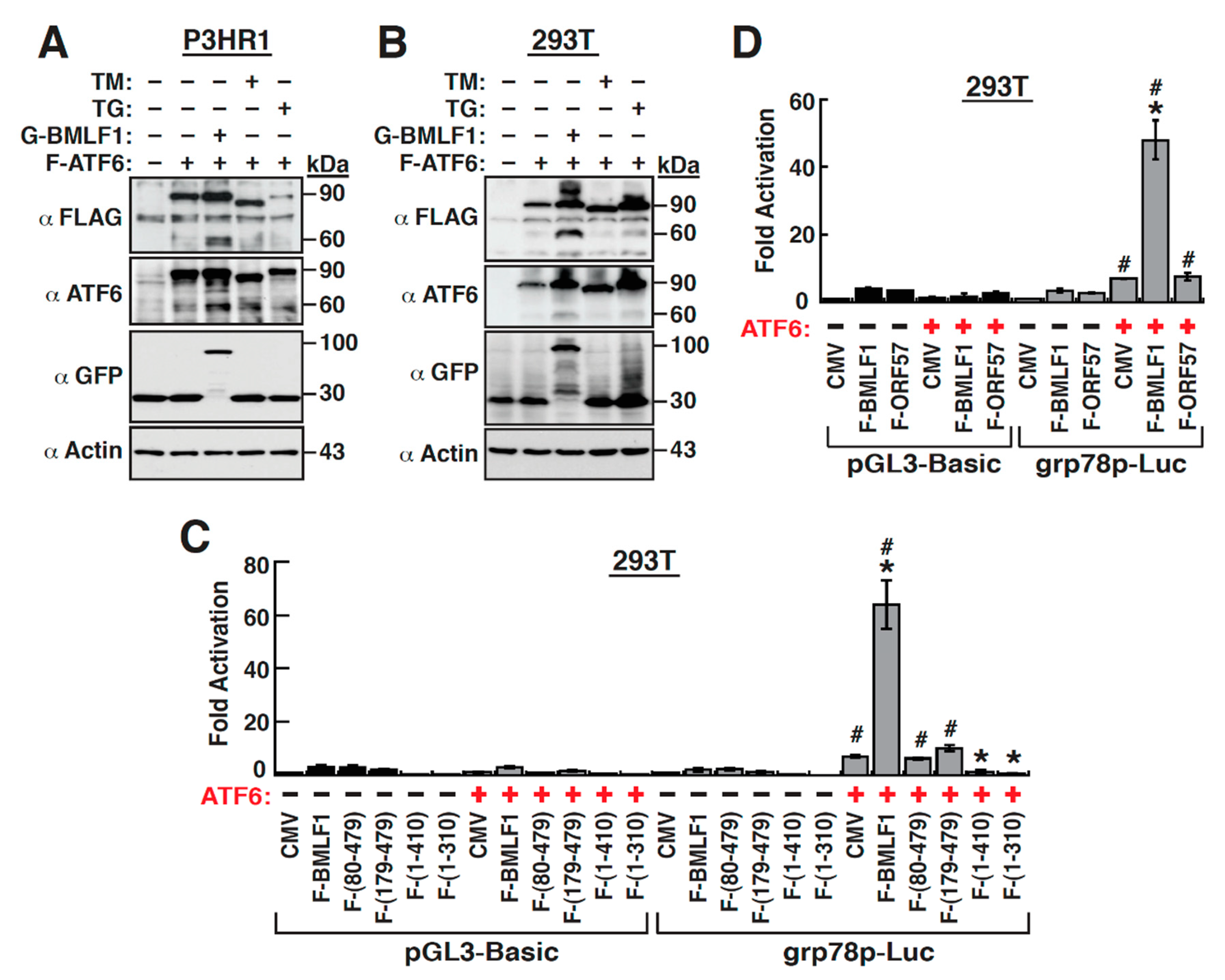
2.4. The Early Lytic Protein BMLF1 Sufficiently Activates the GRP78 Promoter in Lymphoma Cell Lines
2.5. The ER Stress Response Element within the GRP78 Gene Promoter Is Essential for BMLF1-Mediated Activation
2.6. Mapping of the BMLF1 Regions Responsible for the GRP78 Promoter Activation
2.7. The PERK- and IRE-1-Mediated UPR Signaling Pathways Are Not Activated during the EBV Lytic Cycle
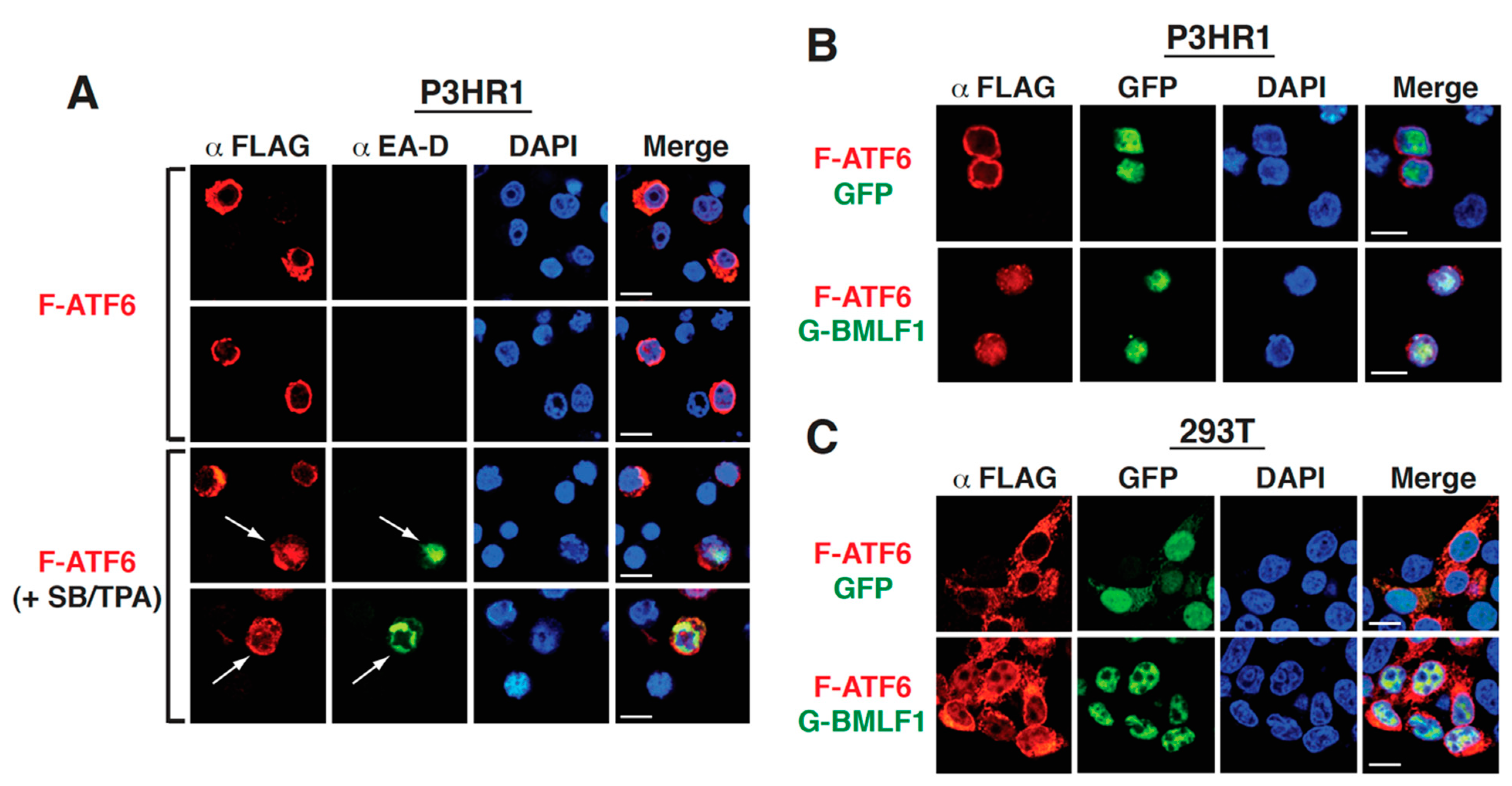
2.8. The Upregulation of GRP78 Expression by BMLF1 Is Mediated through ATF6 Activation
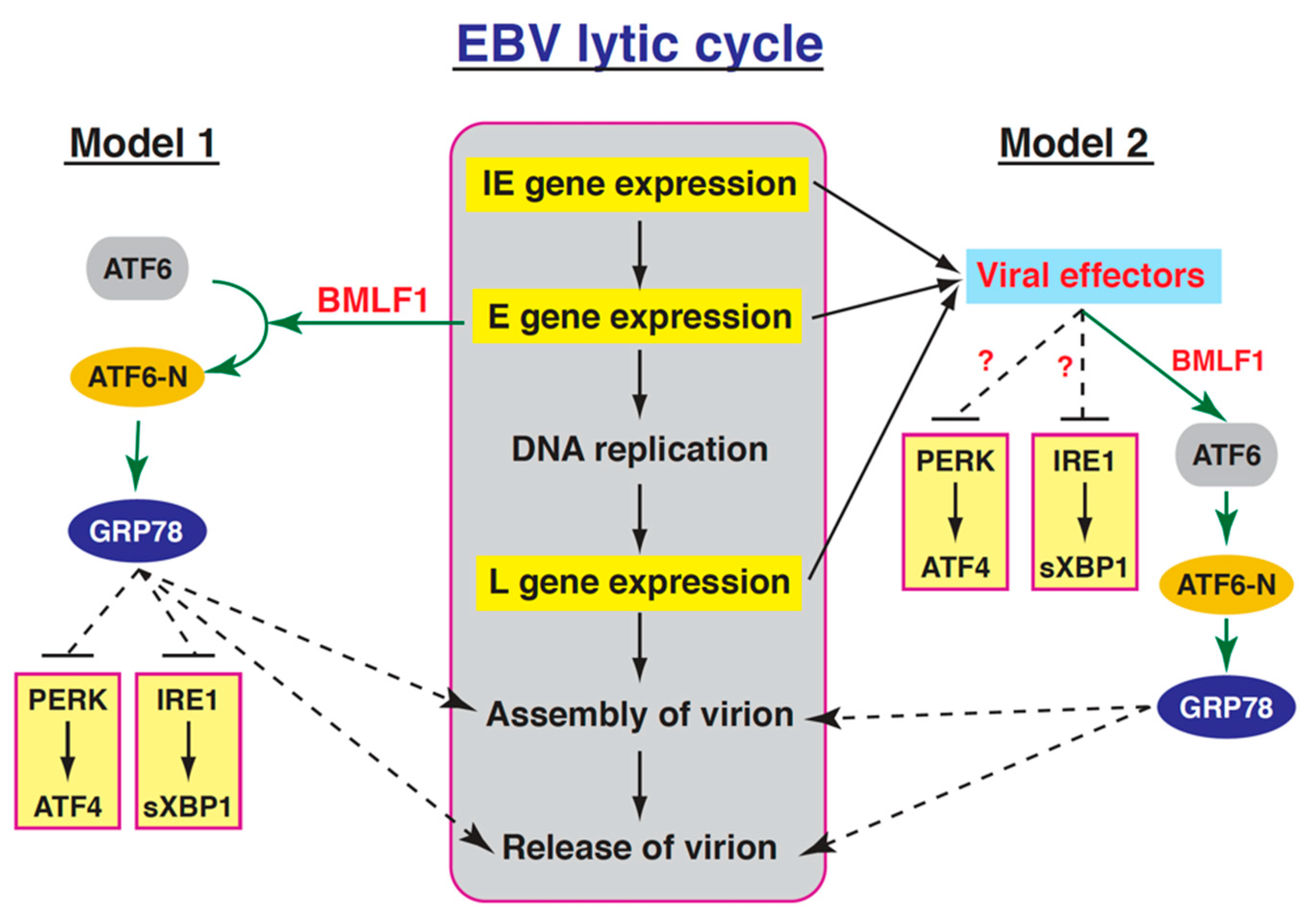
3. Discussion
3.1. Expression and Function of GRP78 in the EBV Lytic Cycle
3.2. Relationships between the Three UPR Branches and the EBV Lytic Cycle
3.3. Activation of the GRP78 Promoter by BMLF1 through ATF6
4. Materials and Methods
4.1. Cell Cultures and Chemical Reagents
4.2. DNA Transfection
4.3. Plasmid Construction
4.4. Western Blot Analysis
4.5. Lentivirus-Based Knockdown
4.6. Quantitative Determination of Viral Particles
4.7. Cell Viability Assay
4.8. Luciferase Assays
4.9. Confocal Immunofluorescence Analysis
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| EBV | Epstein-Barr virus |
| BMLF1 | BamHI-M leftward reading frame 1 |
| ATF6 | activating transcription factor 6 |
| GRP78 | 78-kDa glucose-regulated protein |
| UPR | unfolded protein response |
| ER | Endoplasmic reticulum |
| PERK | PKR-like ER kinase |
| IRES | inositol-requiring enzyme 1 |
| BiP | binding immunoglobulin protein |
| HSPA5 | heat shock protein family A member 5 |
| HSP70 | heat shock protein 70 kDa |
| eIF2a | eukaryotic translation initiation factor 2α |
| ATF4 | activating transcription factor 4 |
| XBP1 | X-box binding protein 1 |
| GRP94 | 94-kDa glucose-regulated protein |
| PDI | protein disulfide isomerase |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| SB | sodium butyrate |
| TPA | 12-O-tetradecanoylphorbol-13-acetate |
| Zta | Z transcriptional transactivator |
| Rta | R transcriptional transactivator |
| PAA | phosphonoacetic acid |
| ERSE | ER stress response element |
| GFP | green fluorescence protein |
| TM | tunicamycin |
| TG | thapsigargin |
| HCMV | human cytomegalovirus |
| VZV | varicella-zoster virus |
References
- Schröder, M.; Kaufman, R.J. ER stress and the unfolded protein response. Mutat. Res. Mol. Mech. Mutagen. 2005, 569, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nat. Cell Biol. 2016, 529, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S. The glucose-regulated proteins: Stress induction and clinical applications. Trends Biochem. Sci. 2001, 26, 504–510. [Google Scholar] [CrossRef]
- Wang, M.; Wey, S.; Zhang, Y.; Ye, R.; Lee, A.S. Role of the Unfolded Protein Response Regulator GRP78/BiP in Development, Cancer, and Neurological Disorders. Antioxid. Redox Signal. 2009, 11, 2307–2316. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Bertolotti, A.; Zeng, H.; Ron, D. Perk Is Essential for Translational Regulation and Cell Survival during the Unfolded Protein Response. Mol. Cell 2000, 5, 897–904. [Google Scholar] [CrossRef]
- Lee, A.-H.; Iwakoshi, N.N.; Glimcher, L.H. XBP-1 Regulates a Subset of Endoplasmic Reticulum Resident Chaperone Genes in the Unfolded Protein Response. Mol. Cell. Biol. 2003, 23, 7448–7459. [Google Scholar] [CrossRef]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Davé, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER Stress Induces Cleavage of Membrane-Bound ATF6 by the Same Proteases that Process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Yoshida, H.; Haze, K.; Yanagi, H.; Yura, T.; Mori, K. Identification of the cis-Acting Endoplasmic Reticulum Stress Response Element Responsible for Transcriptional Induction of Mammalian Glucose-regulated Proteins. J. Biol. Chem. 1998, 273, 33741–33749. [Google Scholar] [CrossRef]
- Chan, S.W. The unfolded protein response in virus infections. Front. Microbiol. 2014, 5, 518. [Google Scholar] [CrossRef]
- Jheng, J.R.; Ho, J.Y.; Horng, J.T. ER stress, autophagy, and RNA viruses. Front. Microbiol. 2014, 5, 388. [Google Scholar] [CrossRef] [PubMed]
- Johnston, B.P.; McCormick, C. Herpesviruses and the Unfolded Protein Response. Viruses 2019, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kong, L.; Yu, X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit. Rev. Microbiol. 2013, 41, 150–164. [Google Scholar] [CrossRef]
- Komano, J.; Sugiura, M.; Takada, K. Epstein-Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt’s lymphoma cell line Akata. J. Virol. 1998, 72, 9150–9156. [Google Scholar] [CrossRef]
- Young, L.S.; Dawson, C.W. Epstein-Barr virus and nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J. Epstein-Barr virus and Hodgkin’s lymphoma. Herpes 2006, 13, 12–16. [Google Scholar] [PubMed]
- Takada, K. Epstein-Barr virus and gastric carcinoma. Mol. Pathol. 2000, 53, 255–261. [Google Scholar] [CrossRef]
- Thompson, M.P.; Kurzrock, R. Epstein-Barr Virus and Cancer. Clin. Cancer Res. 2004, 10, 803–821. [Google Scholar] [CrossRef] [PubMed]
- Miller, G. The switch between EBV latency and replication. Yale J. Boil. Med. 1989, 62, 205–213. [Google Scholar]
- Münz, C. Latency and lytic replication in Epstein–Barr virus-associated oncogenesis. Nat. Rev. Genet. 2019, 17, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.N.; Dong, D.L.; Hayward, G.S.; Hayward, S.D. The Epstein-Barr virus Zta transactivator: A member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 1990, 64, 3358–3369. [Google Scholar] [CrossRef] [PubMed]
- Farrell, P.J.; Rowe, D.T.; Rooney, C.M.; Kouzarides, T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989, 8, 127–132. [Google Scholar] [CrossRef]
- Gruffat, H.; Manet, E.; Rigolet, A.; Sergeant, A. The enhancer factor R of Epstein-Barr virus (EBV) Is a sequence-specific DNA binding protein. Nucleic Acids Res. 1990, 18, 6835–6843. [Google Scholar] [CrossRef]
- Gruffat, H.; Sergeant, A. Characterization of the DNA-binding site repertoire for the Epstein—Barr virus transcription factor R. Nucleic Acids Res. 1994, 22, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Ragoczy, T.; Heston, L.; Miller, G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 1998, 72, 7978–7984. [Google Scholar] [CrossRef]
- Zalani, S.; Holley-Guthrie, E.; Kenney, S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 1996, 93, 9194–9199. [Google Scholar] [CrossRef]
- Juillard, F.; Bazot, Q.; Mure, F.; Tafforeau, L.; Macri, C.; Rabourdin-Combe, C.; Lotteau, V.; Manet, E.; Gruffat, H. Epstein–Barr virus protein EB2 stimulates cytoplasmic mRNA accumulation by counteracting the deleterious effects of SRp20 on viral mRNAs. Nucleic Acids Res. 2012, 40, 6834–6849. [Google Scholar] [CrossRef]
- Thompson, J.; Verma, D.; Li, D.; Mosbruger, T.; Swaminathan, S. Identification and Characterization of the Physiological Gene Targets of the Essential Lytic Replicative Epstein-Barr Virus SM Protein. J. Virol. 2016, 90, 1206–1221. [Google Scholar] [CrossRef]
- Verma, D.; Thompson, J.; Swaminathan, S. Spironolactone blocks Epstein–Barr virus production by inhibiting EBV SM protein function. Proc. Natl. Acad. Sci. USA 2016, 113, 3609–3614. [Google Scholar] [CrossRef]
- Hiriart, E.; Bardouillet, L.; Manet, E.; Gruffat, H.; Penin, F.; Montserret, R.; Farjot, G.; Sergeant, A. A Region of the Epstein-Barr Virus (EBV) mRNA Export Factor EB2 Containing an Arginine-rich Motif Mediates Direct Binding to RNA. J. Biol. Chem. 2003, 278, 37790–37798. [Google Scholar] [CrossRef]
- Hiriart, E.; Farjot, G.; Gruffat, H.; Nguyen, M.V.C.; Sergeant, A.; Manet, E. A Novel Nuclear Export Signal and a REF Interaction Domain Both Promote mRNA Export by the Epstein-Barr Virus EB2 Protein. J. Biol. Chem. 2003, 278, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Mure, F.; Panthu, B.; Zanella-Cléon, I.; Delolme, F.; Manet, E.; Ohlmann, T.; Gruffat, H. Epstein-Barr Virus Protein EB2 Stimulates Translation Initiation of mRNAs through Direct Interactions with both Poly(A)-Binding Protein and Eukaryotic Initiation Factor 4G. J. Virol. 2017, 92, 3. [Google Scholar] [CrossRef] [PubMed]
- Ricci, E.P.; Mure, F.; Gruffat, H.; Decimo, D.; Medina-Palazon, C.; Ohlmann, T.; Manet, E. Translation of intronless RNAs is strongly stimulated by the Epstein–Barr virus mRNA export factor EB2. Nucleic Acids Res. 2009, 37, 4932–4943. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.J.; Hung, C.H.; Wang, S.S.; Tsai, P.H.; Shih, Y.J.; Chen, L.Y.; Huang, H.Y.; Wei, L.H.; Yen, J.B.; Lin, C.L.; et al. Identification and characterization of two novel spliced genes located in the orf47-orf46-orf45 gene locus of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2014, 88, 10092–10109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Al Masud, H.M.A.; Watanabe, T.; Yoshida, M.; Sato, Y.; Goshima, F.; Kimura, H.; Murata, T. Epstein-Barr Virus BKRF4 Gene Product Is Required for Efficient Progeny Production. J. Virol. 2017, 91, e00975-17. [Google Scholar] [CrossRef]
- Li, W.W.; Hsiung, Y.; Zhou, Y.; Roy, B.; Lee, A.S. Induction of the mammalian GRP78/BiP gene by Ca2+ depletion and formation of aberrant proteins: Activation of the conserved stress-inducible grp core promoter element by the human nuclear factor YY1. Mol. Cell. Biol. 1997, 17, 54–60. [Google Scholar] [CrossRef]
- Roy, B.; Li, W.W.; Lee, A.S. Calcium-sensitive transcriptional activation of the proximal CCAAT regulatory element of the grp78/BiP promoter by the human nuclear factor CBF/NF-Y. J. Biol. Chem. 1996, 271, 28995–29002. [Google Scholar] [CrossRef]
- Juillard, F.; Hiriart, E.; Sergeant, N.; Vingtdeux-Didier, V.; Drobecq, H.; Sergeant, A.; Manet, E.; Gruffat, H. Epstein-Barr Virus Protein EB2 Contains an N-Terminal Transferable Nuclear Export Signal That Promotes Nucleocytoplasmic Export by Directly Binding TAP/NXF1. J. Virol. 2009, 83, 12759–12768. [Google Scholar] [CrossRef]
- Han, Z.; Swaminathan, S. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 2006, 80, 5251–5260. [Google Scholar] [CrossRef]
- Buchkovich, N.J.; Maguire, T.G.; Paton, A.W.; Paton, J.C.; Alwine, J.C. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J. Virol. 2009, 83, 11421–11428. [Google Scholar] [CrossRef]
- Buchkovich, N.J.; Maguire, T.G.; Yu, Y.; Paton, A.W.; Paton, J.C.; Alwine, J.C. Human Cytomegalovirus Specifically Controls the Levels of the Endoplasmic Reticulum Chaperone BiP/GRP78, Which Is Required for Virion Assembly. J. Virol. 2007, 82, 31–39. [Google Scholar] [CrossRef]
- Carpenter, J.E.; Grose, C. Varicella-zoster virus glycoprotein expression differentially induces the unfolded protein response in infected cells. Front. Microbiol. 2014, 5, 322. [Google Scholar] [CrossRef] [PubMed]
- Ou, D.S.-C.; Lee, S.-B.; Chu, C.-S.; Chang, L.-H.; Chung, B.-C.; Juan, L.-J. Transcriptional activation of endoplasmic reticulum chaperone GRP78 by HCMV IE1-72 protein. Cell Res. 2011, 21, 642–653. [Google Scholar] [CrossRef]
- Johnston, B.P.; Pringle, E.S.; McCormick, C. KSHV activates unfolded protein response sensors but suppresses downstream transcriptional responses to support lytic replication. PLoS Pathog. 2019, 15, e1008185. [Google Scholar] [CrossRef]
- Isler, J.A.; Skalet, A.H.; Alwine, J.C. Human Cytomegalovirus Infection Activates and Regulates the Unfolded Protein Response. J. Virol. 2005, 79, 6890–6899. [Google Scholar] [CrossRef] [PubMed]
- Bhende, P.M.; Dickerson, S.J.; Sun, X.; Feng, W.-H.; Kenney, S.C. X-Box-Binding Protein 1 Activates Lytic Epstein-Barr Virus Gene Expression in Combination with Protein Kinase D. J. Virol. 2007, 81, 7363–7370. [Google Scholar] [CrossRef] [PubMed]
- Heston, L.; Rabson, M.; Brown, N.; Miller, G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 1982, 295, 160–163. [Google Scholar] [CrossRef]
- Takada, K.; Horinouchi, K.; Ono, Y.; Aya, T.; Osato, T.; Takahashi, M.; Hayasaka, S. An Epstein-Barr virus-producer line Akata: Establishment of the cell line and analysis of viral DNA. Virus Genes 1991, 5, 147–156. [Google Scholar] [CrossRef]
- Shimizu, N.; Tanabe-Tochikura, A.; Kuroiwa, Y.; Takada, K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: Malignant phenotypes of BL cells are dependent on EBV. J. Virol. 1994, 68, 6069–6073. [Google Scholar] [CrossRef]
- Menezes, J.; Leibold, W.; Klein, G.; Clements, G. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine 1975, 22, 276–284. [Google Scholar]
- Graham, F.L.; Russell, W.C.; Smiley, J.; Nairn, R. Characteristics of a Human Cell Line Transformed by DNA from Human Adenovirus Type 5. J. Gen. Virol. 1977, 36, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Ragoczy, T.; Miller, G. Role of the epstein-barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 1999, 73, 9858–9866. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-W.; Hung, C.-H.; Wang, S.-S.; Yen, J.-B.; Liu, A.-C.; Hung, Y.-H.; Chang, P.-J. Expression and regulation of the BKRF2, BKRF3 and BKRF4 genes of Epstein-Barr virus. Virus Res. 2018, 256, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-H.; Kuo, C.-W.; Chang, L.-K.; Hung, C.-C.; Chang, T.-H.; Liu, S.-T. Assembly of Epstein-Barr Virus Capsid in Promyelocytic Leukemia Nuclear Bodies. J. Virol. 2015, 89, 8922–8931. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.-W.; Wang, S.-S.; Hung, C.-H.; Hung, Y.-H.; Lin, C.-L.; Chang, P.-J. The Epstein-Barr Virus Lytic Protein BMLF1 Induces Upregulation of GRP78 Expression through ATF6 Activation. Int. J. Mol. Sci. 2021, 22, 4024. https://doi.org/10.3390/ijms22084024
Chen L-W, Wang S-S, Hung C-H, Hung Y-H, Lin C-L, Chang P-J. The Epstein-Barr Virus Lytic Protein BMLF1 Induces Upregulation of GRP78 Expression through ATF6 Activation. International Journal of Molecular Sciences. 2021; 22(8):4024. https://doi.org/10.3390/ijms22084024
Chicago/Turabian StyleChen, Lee-Wen, Shie-Shan Wang, Chien-Hui Hung, Ya-Hui Hung, Chun-Liang Lin, and Pey-Jium Chang. 2021. "The Epstein-Barr Virus Lytic Protein BMLF1 Induces Upregulation of GRP78 Expression through ATF6 Activation" International Journal of Molecular Sciences 22, no. 8: 4024. https://doi.org/10.3390/ijms22084024
APA StyleChen, L.-W., Wang, S.-S., Hung, C.-H., Hung, Y.-H., Lin, C.-L., & Chang, P.-J. (2021). The Epstein-Barr Virus Lytic Protein BMLF1 Induces Upregulation of GRP78 Expression through ATF6 Activation. International Journal of Molecular Sciences, 22(8), 4024. https://doi.org/10.3390/ijms22084024




