MicroRNAs and Long Noncoding RNAs as Novel Therapeutic Targets in Estrogen Receptor-Positive Breast and Ovarian Cancers
Abstract
:1. Introduction
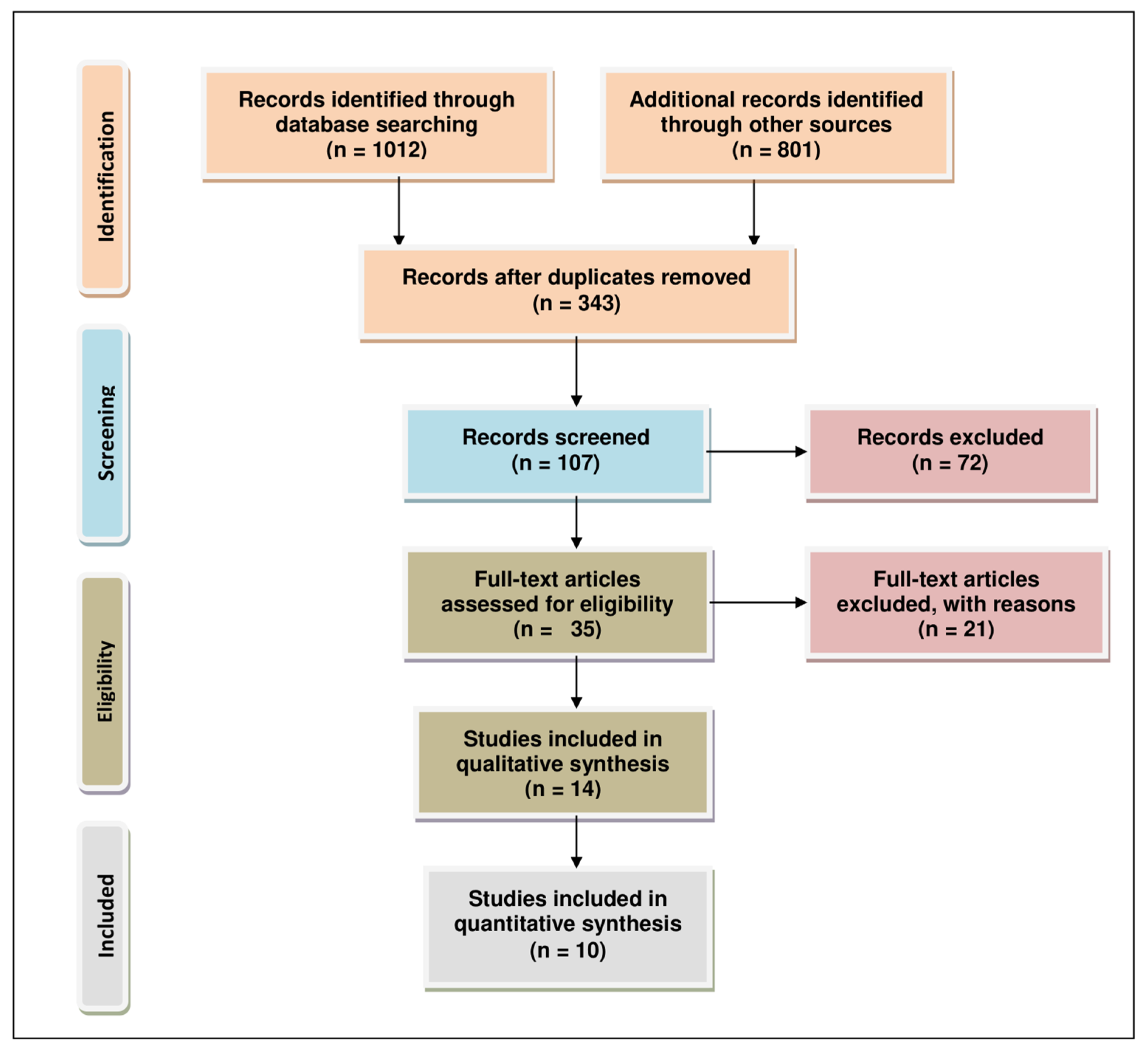
2. Methodology for Literature Search and Selection
3. Noncoding RNAs (miRNAs and lncRNA) and Their Aberrant Expression in ER-Positive Breast Cancer
3.1. Aromatase Associated with miRNAs Aberrantly Expressed in Breast Cancer
3.1.1. Lethal-7f
3.1.2. MIR125B1
3.1.3. MIR27B
3.1.4. MIR155
3.1.5. MIR221 and MIR222
3.1.6. MIR128-1
3.2. Aromatase Associated with miRNAs Aberrantly Expressed in Breast Cancer
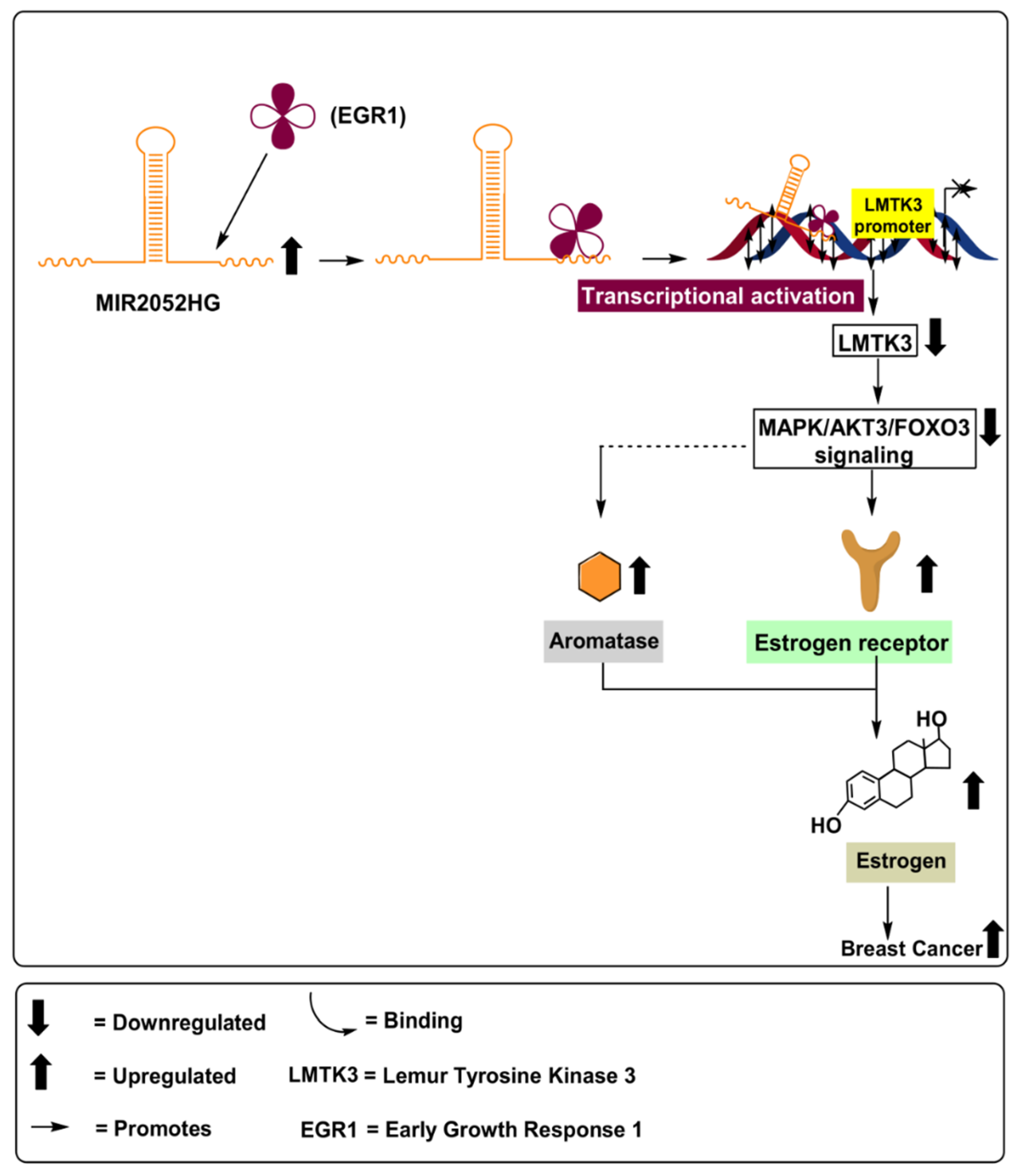
MIR2052HG
4. MiRNAs and Their Aberrant Expression in ER-Positive Ovarian Cancer
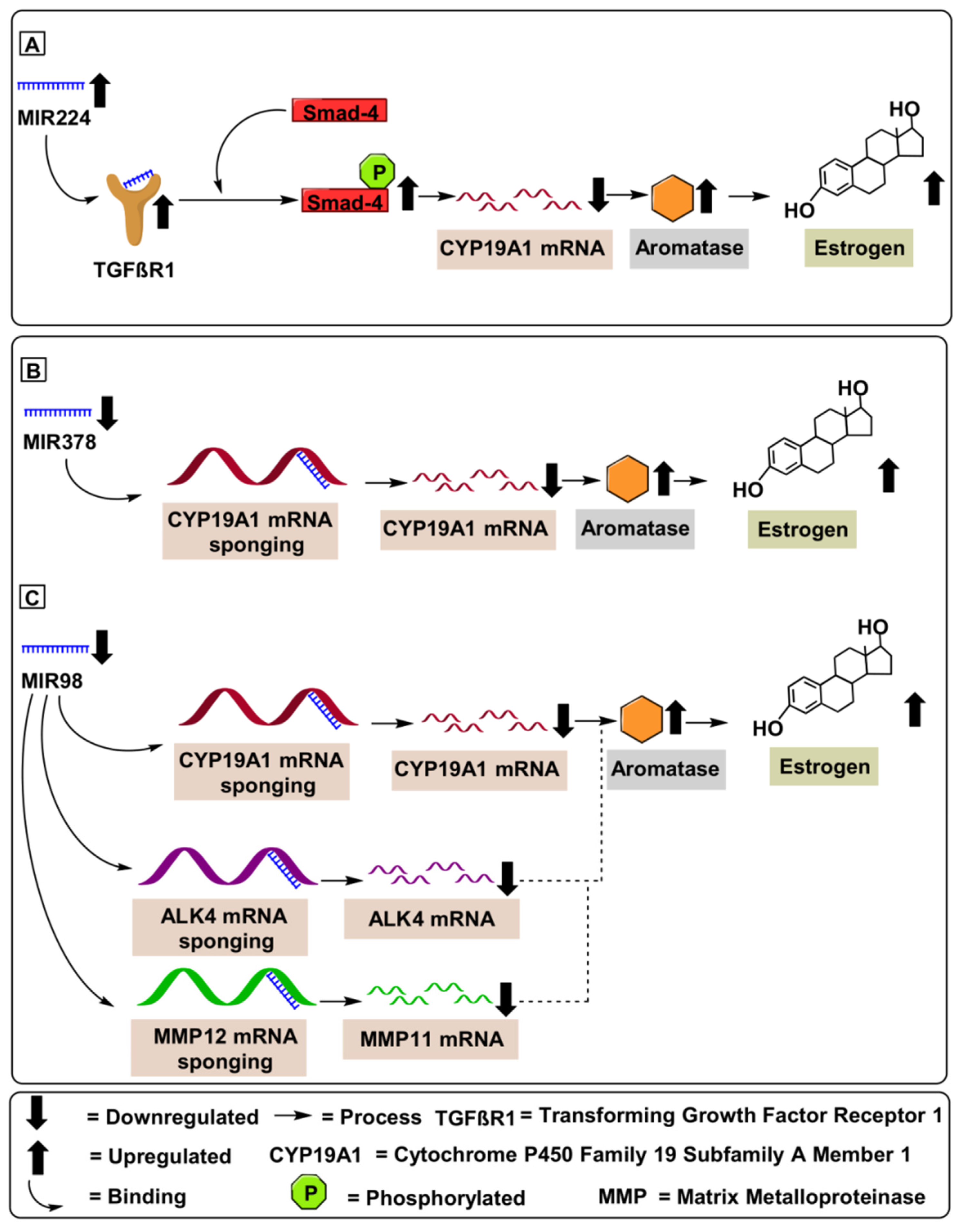
4.1. MIR224
4.2. MIR378
4.3. MIR98
5. Mechanisms of Various Delivery Methods for Therapeutic miRNAs and Future Direction for Efficacious Use in the Clinical Setting
5.1. Approaches for Systemic Delivery of Therapeutic ncRNAs
5.2. Suppression of Oncogenic ncRNAs
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Norman, G.; Rice, S.; Spackman, E.; Stirk, L.; Danso-Appiah, A.; Suh, D.; Palmer, S.; Eastwood, A. Trastuzumab for the treatment of HER2-positive metastatic adenocarcinoma of the stomach or gastro-oesophageal junction. Health Technol. Assess 2011, 15 (Suppl. 1), 33–42. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Menon, U.; Karpinskyj, C.; Gentry-Maharaj, A. Ovarian Cancer Prevention and Screening. Obstet. Gynecol. 2018, 131, 909–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najim, O.; Seghers, S.; Sergoynne, L.; Van Gaver, H.; Papadimitriou, K.; Wouters, K.; Trinh, X.B.; Huizing, M.T.; Tjalma, W. The association between type of endocrine therapy and development of estrogen receptor-1 mutation(s) in patients with hormone-sensitive advanced breast cancer: A systematic review and meta-analysis of randomized and non-randomized trials. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188315. [Google Scholar] [CrossRef]
- Ribas, R.; Pancholi, S.; Rani, A.; Schuster, E.; Guest, S.K.; Nikitorowicz-Buniak, J.; Simigdala, N.; Thornhill, A.; Avogadri-Connors, F.; Cutler, R.E., Jr.; et al. Targeting tumour re-wiring by triple blockade of mTORC1, epidermal growth factor, and oestrogen receptor signalling pathways in endocrine-resistant breast cancer. Breast Cancer Res. 2018, 20, 44. [Google Scholar] [CrossRef]
- Osawa, Y.; Higashiyama, T.; Fronckowiak, M.; Yoshida, N.; Yarborough, C. Aromatase. J. Steroid Biochem. 1987, 27, 781–789. [Google Scholar]
- Simpson, E.R.; Clyne, C.; Rubin, G.; Boon, W.C.; Robertson, K.; Britt, K.; Speed, C.; Jones, M. Aromatase—A brief overview. Annu. Rev. Physiol. 2002, 64, 93–127. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, L.; Shangguan, A.J.; Bulun, S.E. Aromatase expression and regulation in breast and endometrial cancer. J. Mol. Endocrinol. 2016, 57, R19–R33. [Google Scholar] [CrossRef] [Green Version]
- Bulun, S.E.; Chen, D.; Lu, M.; Zhao, H.; Cheng, Y.; Demura, M.; Yilmaz, B.; Martin, R.; Utsunomiya, H.; Thung, S.; et al. Aromatase excess in cancers of breast, endometrium and ovary. J. Steroid Biochem. Mol. Biol. 2007, 106, 81–96. [Google Scholar] [CrossRef] [Green Version]
- Berry, J. Are all aromatase inhibitors the same? A review of controlled clinical trials in breast cancer. Clin. Ther. 2005, 27, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, A.; Bacolla, A.; Vasquez, K.M.; Jain, A. Long non-coding RNA: A new paradigm for lung cancer. Mol. Carcinog. 2015, 54, 1235–1251. [Google Scholar] [CrossRef]
- Tamang, S.; Acharya, V.; Roy, D.; Sharma, R.; Aryaa, A.; Sharma, U.; Khandelwal, A.; Prakash, H.; Vasquez, K.M.; Jain, A. SNHG12: An LncRNA as a Potential Therapeutic Target and Biomarker for Human Cancer. Front. Oncol. 2019, 9, 901. [Google Scholar] [CrossRef]
- Barwal, T.S.; Sharma, U.; Vasquez, K.M.; Prakash, H.; Jain, A. A panel of circulating long non-coding RNAs as liquid biopsy biomarkers for breast and cervical cancers. Biochimie 2020, 176, 62–70. [Google Scholar] [CrossRef]
- Pandey, A.D.; Goswami, S.; Shukla, S.; Das, S.; Ghosal, S.; Pal, M.; Bandyopadhyay, B.; Ramachandran, V.; Basu, N.; Sood, V.; et al. Correlation of altered expression of a long non-coding RNA, NEAT1, in peripheral blood mononuclear cells with dengue disease progression. J. Infect. 2017, 75, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.H.; Rhodes, A. Estrogen and progesterone receptors in breast cancer. Future Oncol. 2014, 10, 2293–2301. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Mungenast, F.; Thalhammer, T. Estrogen biosynthesis and action in ovarian cancer. Front. Endocrinol. 2014, 5, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cairns, J.; Ingle, J.N.; Kalari, K.R.; Shepherd, L.E.; Kubo, M.; Goetz, M.P.; Weinshilboum, R.M.; Wang, L. The lncRNA MIR2052HG regulates ERalpha levels and aromatase inhibitor resistance through LMTK3 by recruiting EGR1. Breast Cancer Res. 2019, 21, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingle, J.N.; Xie, F.; Ellis, M.J.; Goss, P.E.; Shepherd, L.E.; Chapman, J.W.; Chen, B.E.; Kubo, M.; Furukawa, Y.; Momozawa, Y.; et al. Genetic Polymorphisms in the Long Noncoding RNA MIR2052HG Offer a Pharmacogenomic Basis for the Response of Breast Cancer Patients to Aromatase Inhibitor Therapy. Cancer Res. 2016, 76, 7012–7023. [Google Scholar] [CrossRef] [Green Version]
- Ghimenti, C.; Mello-Grand, M.; Grosso, E.; Scatolini, M.; Regolo, L.; Zambelli, A.; Chiorino, G. Regulation of aromatase expression in breast cancer treated with anastrozole neoadjuvant therapy. Exp. Ther. Med. 2013, 5, 902–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katchy, A.; Williams, C. Profiling of estrogen-regulated microRNAs in breast cancer cells. J. Vis. Exp. 2014, 84, e51285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spear, R.; Boytard, L.; Blervaque, R.; Chwastyniak, M.; Hot, D.; Vanhoutte, J.; Lamblin, N.; Amouyel, P.; Pinet, F. Let-7f: A New Potential Circulating Biomarker Identified by miRNA Profiling of Cells Isolated from Human Abdominal Aortic Aneurysm. Int. J. Mol. Sci. 2019, 20, 5499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Selvamani, A.; Sathyan, P.; Miranda, R.C.; Sohrabji, F. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS ONE 2012, 7, e32662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussing, I.; Slack, F.J.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Deng, Q.; Sun, L.; Wang, T.; Yang, Z.; Chen, H.; Guo, L.; Liu, Y.; Ma, Y.; Guo, N.; et al. A Her2-let-7-beta2-AR circuit affects prognosis in patients with Her2-positive breast cancer. BMC Cancer 2015, 15, 832. [Google Scholar] [CrossRef] [Green Version]
- Shibahara, Y.; Miki, Y.; Onodera, Y.; Hata, S.; Chan, M.S.; Yiu, C.C.; Loo, T.Y.; Nakamura, Y.; Akahira, J.; Ishida, T.; et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J. Pathol. 2012, 227, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.Y.; Liang, X.S.; Liu, Y.; Wang, C.Y.; Pang, D. Decrease of let-7f in low-dose metronomic Paclitaxel chemotherapy contributed to upregulation of thrombospondin-1 in breast cancer. Int. J. Biol. Sci. 2015, 11, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Shao, X.; Huang, P.; Shi, L.; Lei, L.; Cao, W.; Chen, Z.; Wang, X.; Zheng, Y. MicroRNA and LncRNA Expression Profiles in Human Estrogen Receptor Positive Breast Cancer. Clin. Lab. 2019, 65, 180340. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Y.; Wang, Z.; Zhang, Z.; Lu, M.; Wang, Y. Long Non-coding RNA LINC01787 Drives Breast Cancer Progression via Disrupting miR-125b Generation. Front. Oncol. 2019, 9, 1140. [Google Scholar] [CrossRef] [Green Version]
- Banno, K.; Yanokura, M.; Iida, M.; Adachi, M.; Nakamura, K.; Nogami, Y.; Umene, K.; Masuda, K.; Kisu, I.; Nomura, H.; et al. Application of microRNA in diagnosis and treatment of ovarian cancer. BioMed Res. Int. 2014, 2014, 232817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilquin, P.; Donini, C.F.; Villedieu, M.; Grisard, E.; Corbo, L.; Bachelot, T.; Vendrell, J.A.; Cohen, P.A. MicroRNA-125b upregulation confers aromatase inhibitor resistance and is a novel marker of poor prognosis in breast cancer. Breast Cancer Res. 2015, 17, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilquin, P.; Villedieu, M.; Grisard, E.; Ben Larbi, S.; Ghayad, S.E.; Heudel, P.E.; Bachelot, T.; Corbo, L.; Treilleux, I.; Vendrell, J.A.; et al. Molecular characterization of anastrozole resistance in breast cancer: Pivotal role of the Akt/mTOR pathway in the emergence of de novo or acquired resistance and importance of combining the allosteric Akt inhibitor MK-2206 with an aromatase inhibitor. Int. J. Cancer 2013, 133, 1589–1602. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Cai, A.; Calloway, C.L.; Xu, Y.F.; Zhang, R.; Fung, K.M.; Ding, W.Q. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: Evidence from a CRISPR/Cas9 deletion study. BMC Cancer 2019, 19, 642. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Ni, J.; Yang, F.; Huang, L.; Deng, H.; Wu, Y.; Ding, X.; Tang, J. Promising therapeutic role of miR-27b in tumor. Tumour Biol. 2017, 39, 1010428317691657. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Zou, Z.; Nie, P.; Kou, X.; Wu, B.; Wang, S.; Song, Z.; He, J. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. 2016, 7, e2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Yu, Y.; Zhu, M.; Jing, W.; Yu, M.; Chai, H.; Liang, C.; Tu, J. Inhibition of miR-155, a therapeutic target for breast cancer, prevented in cancer stem cell formation. Cancer Biomark. 2018, 21, 383–392. [Google Scholar] [CrossRef]
- Bacci, M.; Giannoni, E.; Fearns, A.; Ribas, R.; Gao, Q.; Taddei, M.L.; Pintus, G.; Dowsett, M.; Isacke, C.M.; Martin, L.A.; et al. miR-155 Drives Metabolic Reprogramming of ER+ Breast Cancer Cells Following Long-Term Estrogen Deprivation and Predicts Clinical Response to Aromatase Inhibitors. Cancer Res. 2016, 76, 1615–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Shang, S.; Hua, F.; Hu, Z.W. The regulation of beta-catenin activity and function in cancer: Therapeutic opportunities. Oncotarget 2017, 8, 33972–33989. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Fu, W.; Wo, L.; Shu, X.; Liu, F.; Li, C. miR-128 and its target genes in tumorigenesis and metastasis. Exp. Cell Res. 2013, 319, 3059–3064. [Google Scholar] [CrossRef] [Green Version]
- Masri, S.; Liu, Z.; Phung, S.; Wang, E.; Yuan, Y.C.; Chen, S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res. Treat. 2010, 124, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, D.; Perry, J.R.; Hernandez, D.; Corsi, A.M.; Stevens, K.; Rafferty, I.; Lauretani, F.; Murray, A.; Gibbs, J.R.; Paolisso, G.; et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 2008, 4, e1000072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, T.; Suzuki, Y.; Nishikawa, T.; Otsuki, T.; Sugiyama, T.; Irie, R.; Wakamatsu, A.; Hayashi, K.; Sato, H.; Nagai, K.; et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004, 36, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Sahibzada, M.U.K.; Sadiq, A.; Khan, S.; Faidah, H.S.; Naseemullah; Khurram, M.; Amin, M.U.; Haseeb, A. Fabrication, characterization and in vitro evaluation of silibinin nanoparticles: An attempt to enhance its oral bioavailability. Drug Des. Dev. Ther. 2017, 11, 1453–1464. [Google Scholar] [CrossRef] [Green Version]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta 2009, 1792, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.X.; Huang, X.F.; Shao, Q.; Huang, M.Y.; Deng, L.; Wu, Q.L.; Zeng, Y.X.; Shao, J.Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008, 14, 2348–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Tang, S.; Le, S.Y.; Lu, R.; Rader, J.S.; Meyers, C.; Zheng, Z.M. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 2008, 3, e2557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 2007, 120, 1046–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Dai, G.; Yu, L.; Hu, Q.; Chen, J.; Guo, W. miR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci. Rep. 2018, 8, 606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galardi, S.; Mercatelli, N.; Giorda, E.; Massalini, S.; Frajese, G.V.; Ciafre, S.A.; Farace, M.G. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007, 282, 23716–23724. [Google Scholar] [CrossRef] [Green Version]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Croce, C.M.; Condorelli, G. miR221/222 in cancer: Their role in tumor progression and response to therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef]
- Rao, X.; Di Leva, G.; Li, M.; Fang, F.; Devlin, C.; Hartman-Frey, C.; Burow, M.E.; Ivan, M.; Croce, C.M.; Nephew, K.P. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011, 30, 1082–1097. [Google Scholar] [CrossRef] [Green Version]
- Morin, P.J. β-catenin signaling and cancer. Bioessays 1999, 21, 1021–1030. [Google Scholar] [CrossRef]
- Qian, P.; Banerjee, A.; Wu, Z.S.; Zhang, X.; Wang, H.; Pandey, V.; Zhang, W.J.; Lv, X.F.; Tan, S.; Lobie, P.E.; et al. Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 2012, 72, 6036–6050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Chen, S.; Zeng, J. TGFbeta signaling: A complex role in tumorigenesis (Review). Mol. Med. Rep. 2018, 17, 699–704. [Google Scholar] [PubMed] [Green Version]
- Saluja, R.; Kumar, A.; Jain, M.; Goel, S.K.; Jain, A. Role of Sphingosine-1-Phosphate in Mast Cell Functions and Asthma and Its Regulation by Non-Coding RNA. Front. Immunol. 2017, 8, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, U.; Barwal, T.S.; Acharya, V.; Tamang, S.; Vasquez, K.M.; Jain, A. Cancer Susceptibility Candidate 9 (CASC9): A Novel Targetable Long Noncoding RNA in Cancer Treatment. Transl. Oncol. 2020, 13, 100774. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Barwal, T.S.; Malhotra, A.; Pant, N.; Vivek; Dey, D.; Gautam, A.; Tuli, H.S.; Vasquez, K.M.; Jain, A. Long non-coding RNA TINCR as potential biomarker and therapeutic target for cancer. Life Sci. 2020, 257, 118035. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, M.; Lin, S.; Guo, Y.; Dai, Z.; Liu, K.; Yang, P.; Dai, C.; Zhu, Y.; Zheng, Y.; et al. The Impact of lncRNA Dysregulation on Clinicopathology and Survival of Breast Cancer: A Systematic Review and Meta-analysis. Mol. Ther. Nucleic Acids 2018, 12, 359–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wentzensen, N.; Poole, E.M.; Trabert, B.; White, E.; Arslan, A.A.; Patel, A.V.; Setiawan, V.W.; Visvanathan, K.; Weiderpass, E.; Adami, H.O.; et al. Ovarian Cancer Risk Factors by Histologic Subtype: An Analysis from the Ovarian Cancer Cohort Consortium. J. Clin. Oncol. 2016, 34, 2888–2898. [Google Scholar] [CrossRef] [PubMed]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Tew, W.P. Ovarian cancer in the older woman. J. Geriatr. Oncol. 2016, 7, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [PubMed] [Green Version]
- Modugno, F.; Laskey, R.; Smith, A.L.; Andersen, C.L.; Haluska, P.; Oesterreich, S. Hormone response in ovarian cancer: Time to reconsider as a clinical target? Endocr. Relat. Cancer 2012, 19, R255–R279. [Google Scholar] [CrossRef] [Green Version]
- Langdon, S.P.; Herrington, C.S.; Hollis, R.L.; Gourley, C. Estrogen Signaling and Its Potential as a Target for Therapy in Ovarian Cancer. Cancers 2020, 12, 1647. [Google Scholar] [CrossRef]
- Chan, H.J.; Petrossian, K.; Chen, S. Structural and functional characterization of aromatase, estrogen receptor, and their genes in endocrine-responsive and -resistant breast cancer cells. J. Steroid Biochem. Mol. Biol. 2016, 161, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Hurteau, J.A.; Brady, M.F.; Darcy, K.M.; McGuire, W.P.; Edmonds, P.; Pearl, M.L.; Ivanov, I.; Tewari, K.S.; Mannel, R.S.; Zanotti, K.; et al. Randomized phase III trial of tamoxifen versus thalidomide in women with biochemical-recurrent-only epithelial ovarian, fallopian tube or primary peritoneal carcinoma after a complete response to first-line platinum/taxane chemotherapy with an evaluation of serum vascular endothelial growth factor (VEGF): A Gynecologic Oncology Group Study. Gynecol. Oncol. 2010, 119, 444–450. [Google Scholar] [PubMed] [Green Version]
- Musahl, A.S.; Huang, X.; Rusakiewicz, S.; Ntini, E.; Marsico, A.; Kroemer, G.; Kepp, O.; Orom, U.A. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene 2015, 34, 5046–5054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Bi, T.; Qu, Z.; Jiang, J.; Cui, S.; Wang, Y. Expression of miR-224-5p is associated with the original cisplatin resistance of ovarian papillary serous carcinoma. Oncol. Rep. 2014, 32, 1003–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lite, C.; Ahmed, S.; Santosh, W.; Seetharaman, B. Prenatal exposure to bisphenol-A altered miRNA-224 and protein expression of aromatase in ovarian granulosa cells concomitant with elevated serum estradiol levels in F1 adult offspring. J. Biochem. Mol. Toxicol. 2019, 33, e22317. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Linher-Melville, K.; Yang, B.B.; Wu, D.; Li, J. Micro-RNA378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology 2011, 152, 3941–3951. [Google Scholar] [CrossRef] [Green Version]
- Wolfram, C. The benefits of professional credentialing. J. Intraven. Nurs. 1991, 14, 362–363. [Google Scholar]
- Zhang, X.; Huang, S.; Guo, J.; Zhou, L.; You, L.; Zhang, T.; Zhao, Y. Insights into the distinct roles of MMP-11 in tumor biology and future therapeutics (Review). Int. J. Oncol. 2016, 48, 1783–1793. [Google Scholar] [CrossRef] [Green Version]
- Panda, H.; Chuang, T.D.; Luo, X.; Chegini, N. Endometrial miR-181a and miR-98 expression is altered during transition from normal into cancerous state and target PGR, PGRMC1, CYP19A1, DDX3X, and TIMP3. J. Clin. Endocrinol. Metab. 2012, 97, E1316–E1326. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Zhu, L.; Li, L.; Kang, C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol. Biol. Lett. 2017, 22, 12. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.; Toms, D.; Shen, W.; Li, J. MicroRNA-378 regulates oocyte maturation via the suppression of aromatase in porcine cumulus cells. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E525–E534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebrun, J.J.; Takabe, K.; Chen, Y.; Vale, W. Roles of pathway-specific and inhibitory Smads in activin receptor signaling. Mol. Endocrinol. 1999, 13, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Pucci, C.; Martinelli, C.; Ciofani, G. Innovative approaches for cancer treatment: Current perspectives and new challenges. Ecancermedicalscience 2019, 13, 961. [Google Scholar] [CrossRef] [PubMed]
- Masoud, V.; Pages, G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef] [Green Version]
- Nagini, S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anticancer Agents Med. Chem. 2017, 17, 152–163. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, Y.; Peng, H.; Chen, Y.; Zhu, P.; Huang, Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv. Drug Deliv. Rev. 2015, 81, 142–160. [Google Scholar] [CrossRef]
- Szebeni, J.; Muggia, F.; Gabizon, A.; Barenholz, Y. Activation of complement by therapeutic liposomes and other lipid excipient-based therapeutic products: Prediction and prevention. Adv. Drug Deliv. Rev. 2011, 63, 1020–1030. [Google Scholar] [CrossRef]
- Szebeni, J.; Moghimi, S.M. Liposome triggering of innate immune responses: A perspective on benefits and adverse reactions. J. Liposome Res. 2009, 19, 85–90. [Google Scholar] [CrossRef]
- Sharma, S.; Rajendran, V.; Kulshreshtha, R.; Ghosh, P.C. Enhanced efficacy of anti-miR-191 delivery through stearylamine liposome formulation for the treatment of breast cancer cells. Int. J. Pharm. 2017, 530, 387–400. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gai, C.; Liu, C.; Wu, X.; Yu, M.; Zheng, J.; Zhang, W.; Lv, S.; Li, W. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020, 11, 751. [Google Scholar] [CrossRef]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef]
- Ma, D.; Tian, S.; Baryza, J.; Luft, J.C.; DeSimone, J.M. Reductively Responsive Hydrogel Nanoparticles with Uniform Size, Shape, and Tunable Composition for Systemic siRNA Delivery in Vivo. Mol. Pharm. 2015, 12, 3518–3526. [Google Scholar] [CrossRef] [Green Version]
- Gong, N.; Teng, X.; Li, J.; Liang, X.J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef]
- Peng, Z.H.; Xie, Y.; Wang, Y.; Li, J.; Oupicky, D. Dual-Function Polymeric HPMA Prodrugs for the Delivery of miRNA. Mol. Pharm. 2017, 14, 1395–1404. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Sharifi, M.; Bagheri, M. Knockdown of Long Noncoding RNA Plasmacytoma Variant Translocation 1 with Antisense Locked Nucleic Acid GapmeRs Exerts Tumor-Suppressive Functions in Human Acute Erythroleukemia Cells Through Downregulation of C-MYC Expression. Cancer Biother. Radiopharm. 2019, 34, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, G.; Reigadas, S.; Ittig, D.; Arzumanov, A.; Andreola, M.L.; Leumann, C.; Toulme, J.J.; Gait, M.J. Tricyclo-DNA containing oligonucleotides as steric block inhibitors of human immunodeficiency virus type 1 tat-dependent trans-activation and HIV-1 infectivity. Oligonucleotides 2007, 17, 54–65. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Sahu, A.; Asangani, I.A.; Cao, Q.; Patel, L.; Vergara, I.A.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| S. No. | Dysregulated miRNA/lncRNA | Chromosomal Location | Patient Sample | Cell Lines Used | Animal Model Used | References |
|---|---|---|---|---|---|---|
| Tumor-suppressive | ||||||
| 1 | Lethal-7f | 9q22.32 | 3 Frozen human breast cancer tissues | MCF-7 & SK-BR-3 | N.A. | [24,25,29] |
| Oncogenic | ||||||
| 2 | MIR125B1 | 21q221.31 | 65 Human breast cancer tissues and equivalent number of control samples | MCF-7 (Dosing: 1, 3 and 5 µm letrozole) | N.A. | [31,34] |
| 3 | MIR27B | 9q22.32 | 53 Human breast cancer tissues and 19 healthy tissues | MCF-7 & TAM-1 | N.A. | [36,37,38] |
| 4 | MIR155 | 21q21.3 | N.A. | MCF-7 & ZR75-1 | Ncr foxhed nude mice 6 to 8 weeks old given a 1 mg/kg of letrozole for 21 days | [39,40] |
| 5 | MIR221/222 | Xp11.3 | N.A. | MCF-7 (Dosing: 100 nM fulvestrant) | N.A. | [41,42] |
| 6 | MIR128-1 | 2q21.3 | N.A. | MCF-7 | N.A. | [43,44] |
| 7 | MIR2052HG | 8q21.11 | 5221 Breast cancer blood sample | N.A. | N.A. | [45,46,47] |
| S. No. | Dysregulated miRNA/lncRNA | Chromosomal Location | Patient Samples | Cell Lines Used | Animal Model Used | References |
|---|---|---|---|---|---|---|
| Oncogenic | ||||||
| 1 | MIR224 | Xq28 | N.A. | KGN cell lines | Three-months-old nulliparous rats | [74,75] |
| Tumor Suppressive | ||||||
| 2 | MIR378 | 5q32 | N.A. | KGN cell lines | Porcine ovaries | [76,77] |
| 3 | MIR98 | Xp11.22 | Tissues from 52 ovarian cancer patients and 15 healthy control samples | Ishikawa cells | N.A. | [78,79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barwal, T.S.; Sharma, U.; Bazala, S.; Singh, I.; Jain, M.; Prakash, H.; Shekhar, S.; Sandberg, E.N.; Bishayee, A.; Jain, A. MicroRNAs and Long Noncoding RNAs as Novel Therapeutic Targets in Estrogen Receptor-Positive Breast and Ovarian Cancers. Int. J. Mol. Sci. 2021, 22, 4072. https://doi.org/10.3390/ijms22084072
Barwal TS, Sharma U, Bazala S, Singh I, Jain M, Prakash H, Shekhar S, Sandberg EN, Bishayee A, Jain A. MicroRNAs and Long Noncoding RNAs as Novel Therapeutic Targets in Estrogen Receptor-Positive Breast and Ovarian Cancers. International Journal of Molecular Sciences. 2021; 22(8):4072. https://doi.org/10.3390/ijms22084072
Chicago/Turabian StyleBarwal, Tushar Singh, Uttam Sharma, Sonali Bazala, Ipsa Singh, Manju Jain, Hridayesh Prakash, Shashank Shekhar, Elise N. Sandberg, Anupam Bishayee, and Aklank Jain. 2021. "MicroRNAs and Long Noncoding RNAs as Novel Therapeutic Targets in Estrogen Receptor-Positive Breast and Ovarian Cancers" International Journal of Molecular Sciences 22, no. 8: 4072. https://doi.org/10.3390/ijms22084072
APA StyleBarwal, T. S., Sharma, U., Bazala, S., Singh, I., Jain, M., Prakash, H., Shekhar, S., Sandberg, E. N., Bishayee, A., & Jain, A. (2021). MicroRNAs and Long Noncoding RNAs as Novel Therapeutic Targets in Estrogen Receptor-Positive Breast and Ovarian Cancers. International Journal of Molecular Sciences, 22(8), 4072. https://doi.org/10.3390/ijms22084072








