A Breach in Plant Defences: Pseudomonas syringae pv. actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses
Abstract
:1. Introduction
2. Results
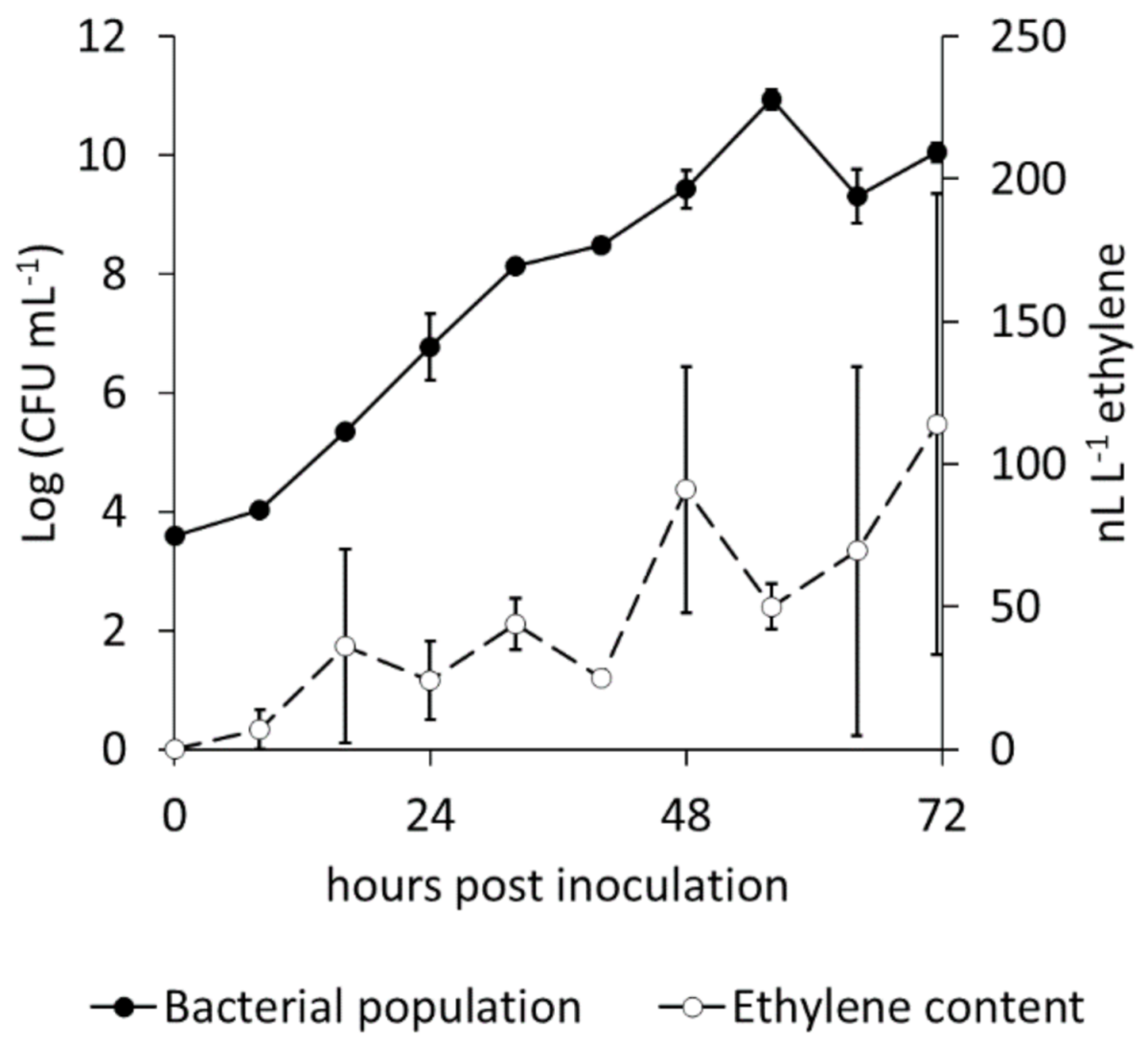
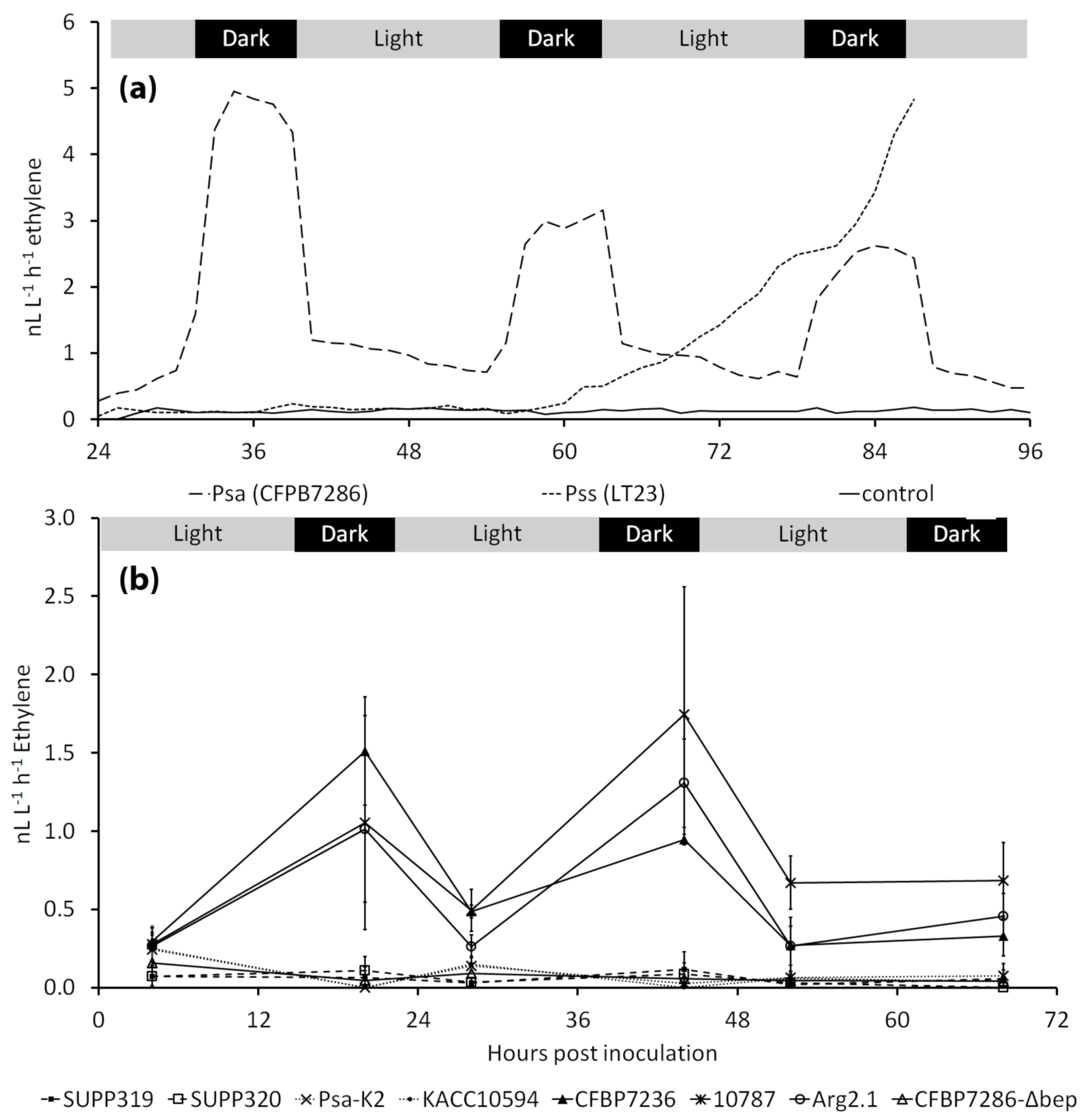
2.1. Ethylene Emission by Pseudomonas syringae pv. actinidiae
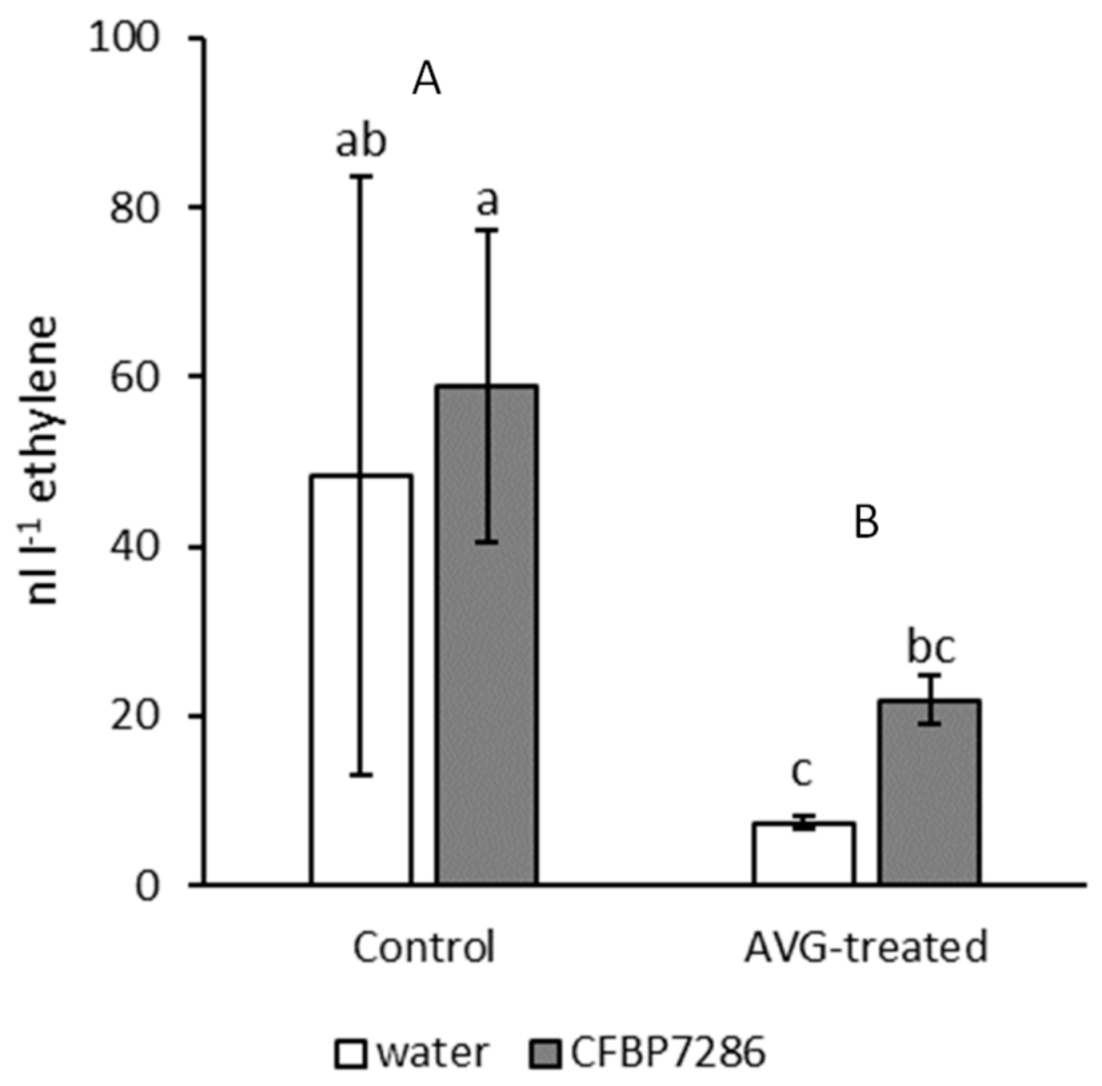
2.2. Modulation of Ethylene Emission in Pseudomonas syringae pv. actinidiae
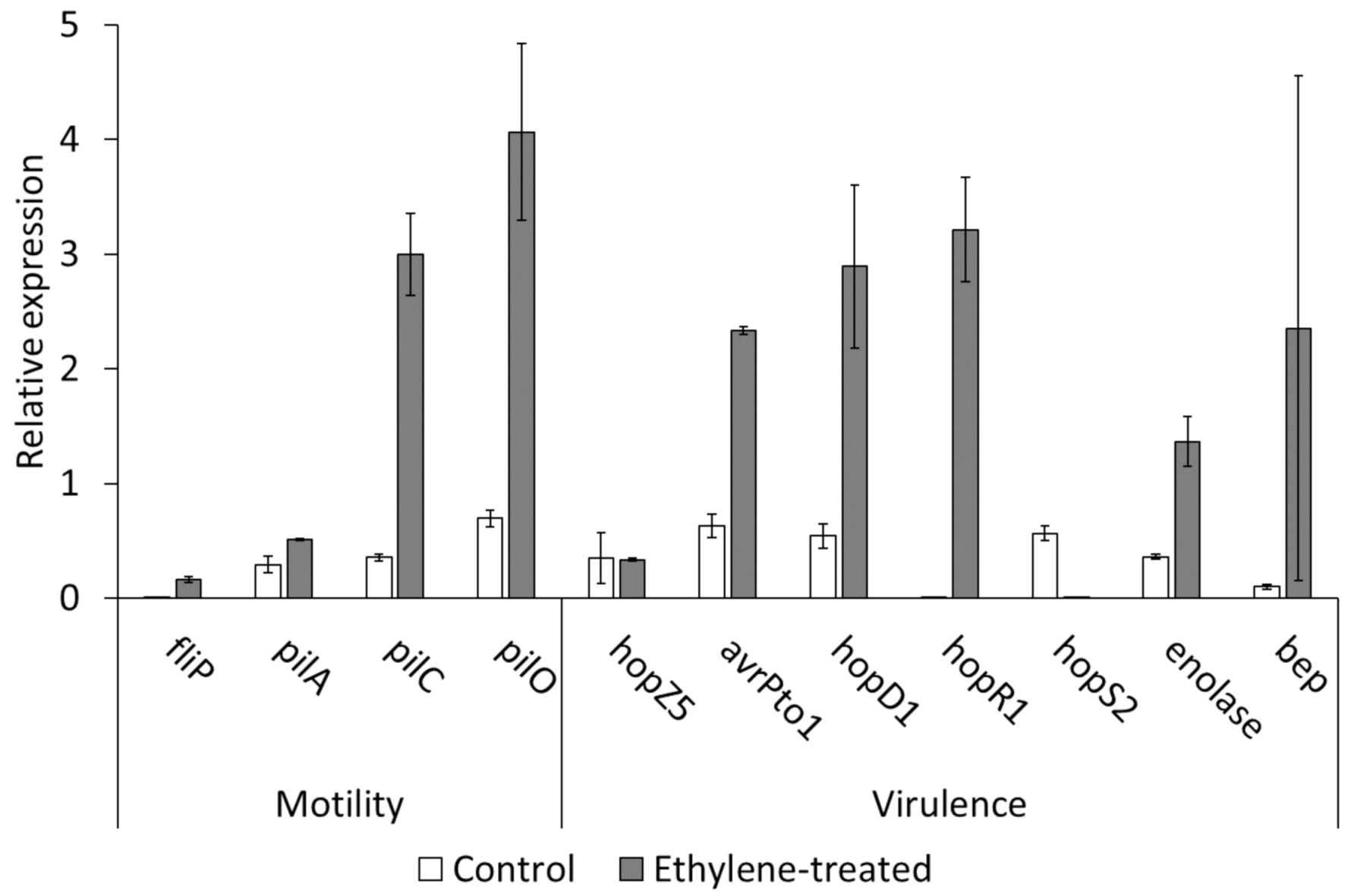
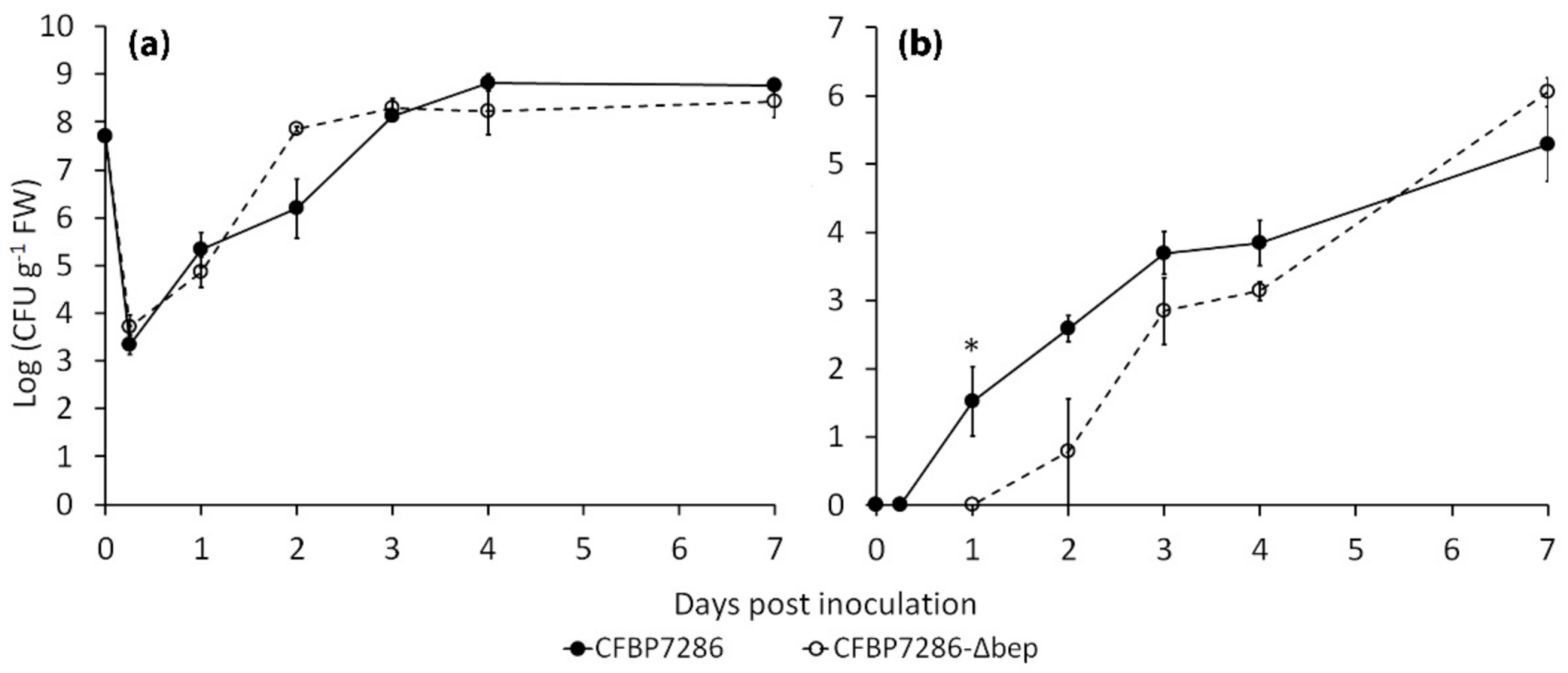
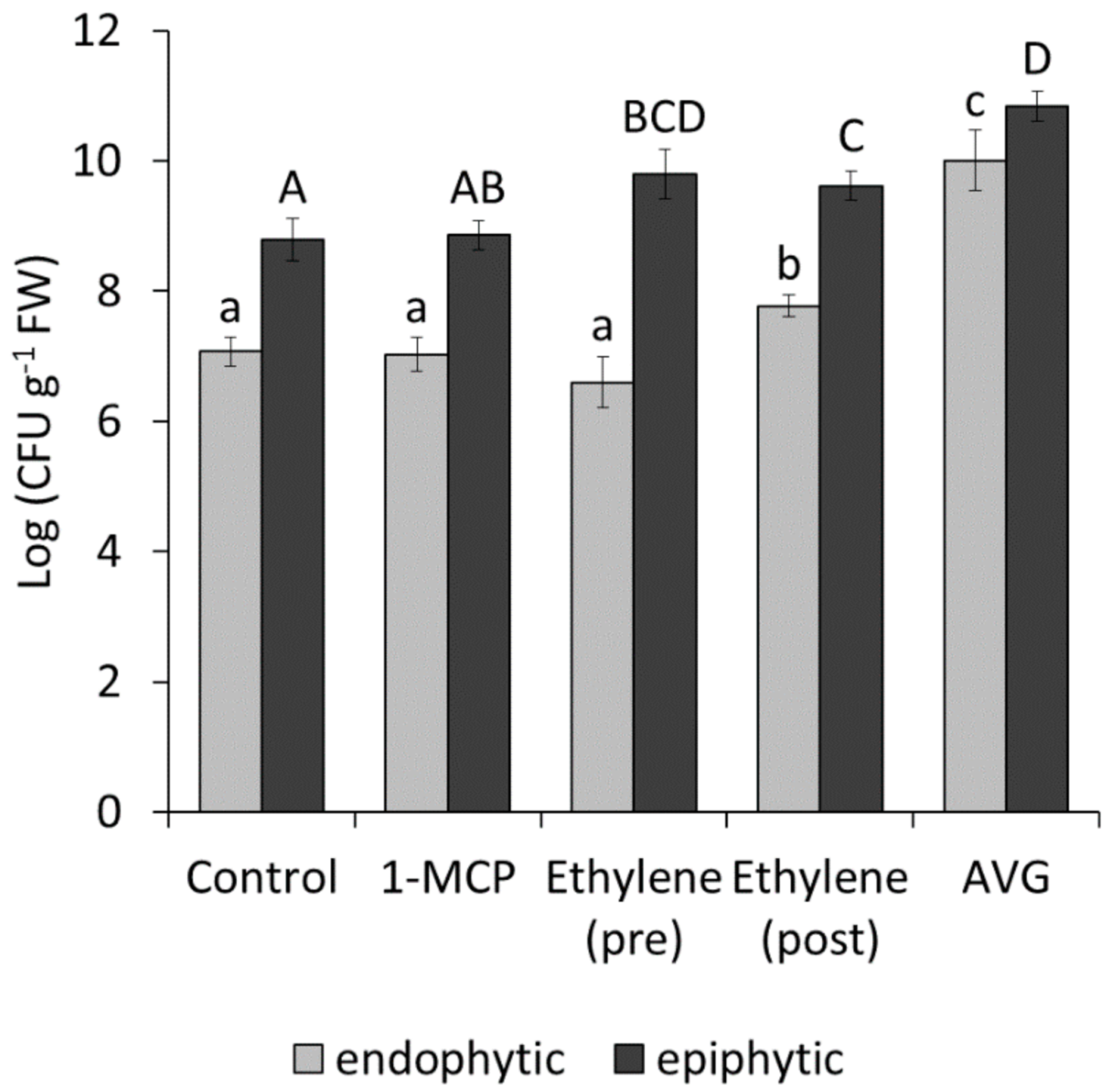
2.3. Effect of Ethylene on Bacterial Growth, Motility, Virulence and Host Plant Colonisation
2.4. Ethylene Emissions from Infected Plants
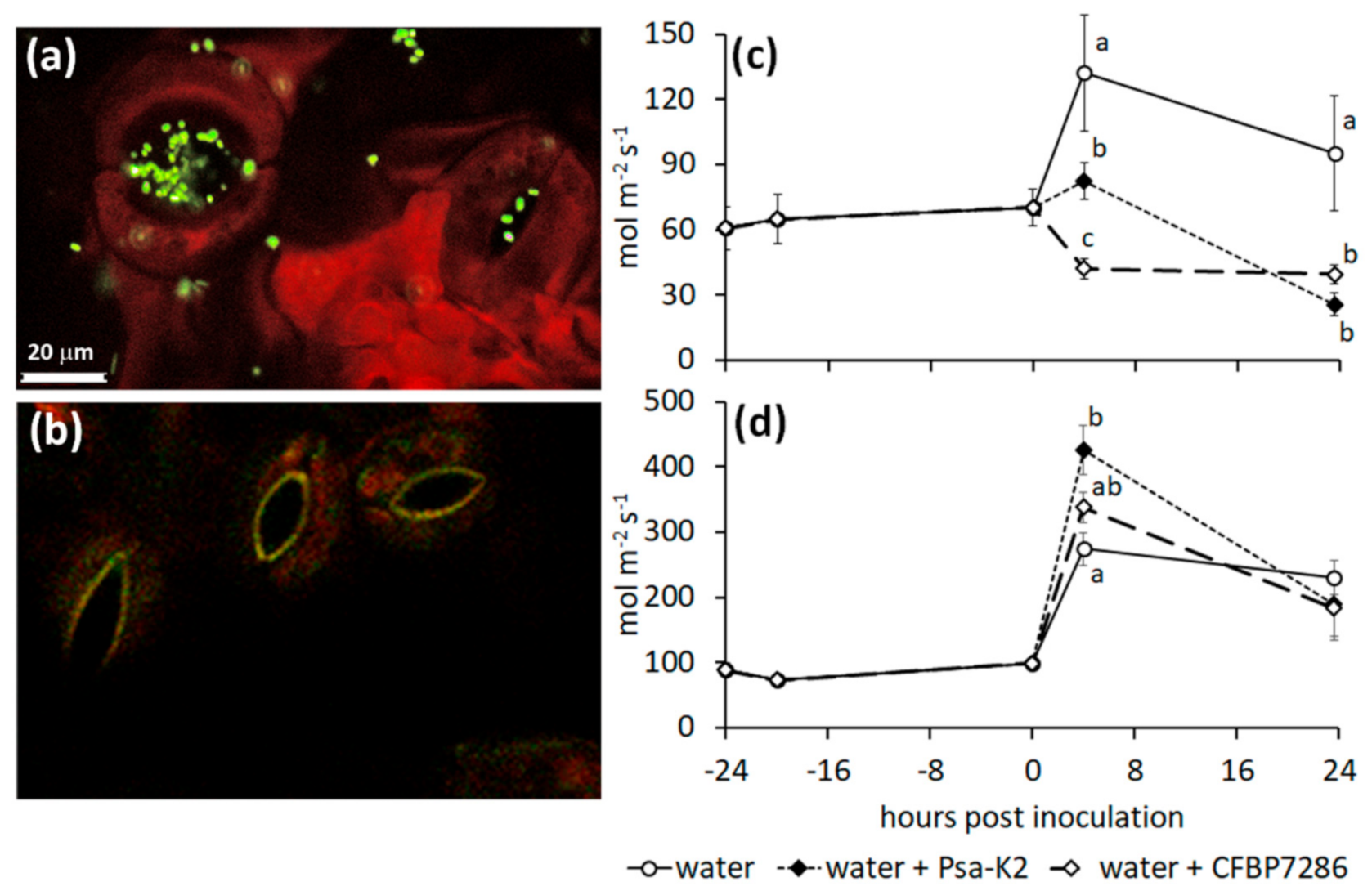
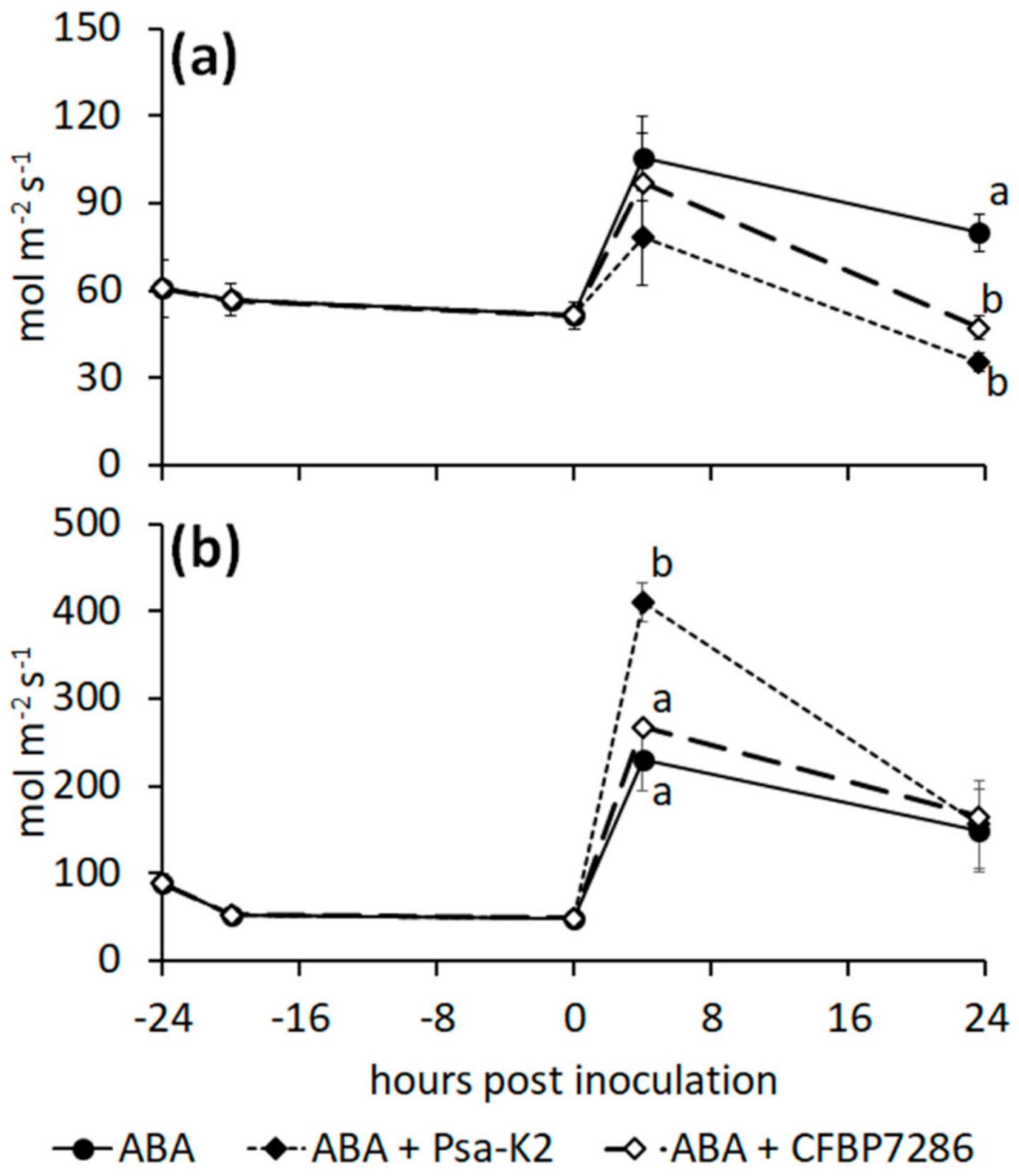
2.5. Effect of Ethylene on the Host–Pathogen Interactions
3. Discussion
3.1. Ethylene Emission by Pseudomonas syringae pv. actinidiae
3.2. Modulation of Ethylene Emission in Pseudomonas syringae pv. actinidiae
3.3. Perception of Exogenous Ethylene by Psa
3.4. Regulation of Stomata Opening by the Plant and the Pathogen
3.5. Role of Ethylene in the Host–Pathogen Interactions
3.6. Conclusions
4. Materials and Methods
4.1. Bacterial Strains, Culture and Inoculation
4.2. Plant Material and Growing Conditions
4.3. Ethylene Measure Systems
4.4. Ethylene Emission by Pseudomonas syringae pv. actinidiae
4.5. Biochemical and Molecular Basis of Ethylene Emission by Pseudomonas syringae pv. actinidiae
4.6. Modulation of Ethylene Emission in Pseudomonas syringae pv. actinidiae
4.7. Effect of Ethylene on Bacterial Growth, Motility, Virulence and Host Plant Colonisation
4.8. Ethylene Emissions from Infected Plants
4.9. Effect of Psa Infection on Ethylene Signalling in Host Plants
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Grant, M.R.; Jones, J.D.G. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 2016, 56, 174–189. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mur, L.A.; Lloyd, A.J.; Cristescu, S.M.; Harren, F.J.; Hall, M.; Smith, A. Biphasic ethylene production during the hypersensitive response in Arabidopsis. Plant Signal. Behav. 2009, 4, 610–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, R.; Su, J.; Meng, X.; Li, S.; Liu, Y.; Xu, J.; Zhang, S. Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol. 2015, 169, 299–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig-Müller, J. Bacteria and fungi controlling plant growth by manipulating auxin: Balance between development and defense. J. Plant Physiol. 2015, 172, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [Green Version]
- Ichinose, Y.; Taguchi, F.; Mukaihara, T. Pathogenicity and virulence factors of Pseudomonas syringae. J. Gen. Plant Pathol. 2013, 79, 285–296. [Google Scholar] [CrossRef]
- Weingart, H.; Ullrich, H.; Geider, K.; Völksch, B. The role of ethylene production in virulence of Pseudomonas syringae pvs. glycinea and phaseolicola. Phytopathology 2001, 91, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Brooks, D.M.; Bender, C.L.; Kunkel, B.N. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 2005, 6, 629–639. [Google Scholar] [CrossRef]
- Zheng, X.-Y.; Spivey, N.W.; Zeng, W.; Liu, P.-P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Geng, X.; Jin, L.; Shimada, M.; Kim, M.G.; Mackey, D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 2014, 240, 1149–1165. [Google Scholar] [CrossRef] [Green Version]
- De Torres-Zabala, M.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Egea, P.R.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef]
- Xiao, F.; He, P.; Abramovitch, R.B.; Dawson, J.E.; Nicholson, L.K.; Sheen, J.; Martin, G.B. The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 2007, 52, 595–614. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Agnew, J.L.; Cohen, J.D.; He, P.; Shan, L.; Sheen, J.; Kunkel, B.N. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 2007, 104, 20131–20136. [Google Scholar] [CrossRef] [Green Version]
- Cunnac, S.; Lindeberg, M.; Collmer, A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr. Opin. Microbiol. 2009, 12, 53–60. [Google Scholar] [CrossRef]
- Ferrante, P.; Scortichini, M. Identification of Pseudomonas syringae pv. actinidiae as causal agent of bacterial canker of yellow kiwifruit (Actinidia chinensis Planchon) in central Italy. J. Phytopathol. 2009, 157, 768–770. [Google Scholar] [CrossRef]
- Donati, I.; Buriani, G.; Cellini, A.; Mauri, S.; Costa, G.; Spinelli, F. New insights on the bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). J. Berry Res. 2014, 4, 53–67. [Google Scholar] [CrossRef] [Green Version]
- Vanneste, J.L. The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 2017, 55, 377–399. [Google Scholar] [CrossRef]
- Ferrante, P.; Scortichini, M. Redefining the global populations of Pseudomonas syringae pv. actinidiae based on pathogenic, molecular and phenotypic characteristics. Plant Pathol. 2014, 64, 51–62. [Google Scholar] [CrossRef]
- Fujikawa, T.; Sawada, H. Genome analysis of the kiwifruit canker pathogen Pseudomonas syringae pv. actinidiae biovar 5. Sci. Rep. 2016, 6, 21399. [Google Scholar] [CrossRef] [Green Version]
- Sawada, H.; Kondo, K.; Nakaune, R. Novel biovar (biovar 6) of Pseudomonas syringae pv. actinidiae causing bacterial canker of kiwifruit (Actinidia deliciosa) in Japan. Jpn. J. Phytopathol. 2016, 82, 101–115. [Google Scholar] [CrossRef] [Green Version]
- Donati, I.; Cellini, A.; Sangiorgio, D.; Vanneste, J.L.; Scortichini, M.; Balestra, G.M.; Spinelli, F. Pseudomonas syringae pv. actinidiae: Ecology, infection dynamics and disease epidemiology. Microb. Ecol. 2020, 80, 81–102. [Google Scholar] [CrossRef]
- Serizawa, S.; Ichikawa, T.; Takikawa, Y.; Tsuyumu, S.; Goto, M. Occurrence of bacterial canker of kiwifruit in Japan: Description of symptoms, isolation of the pathogen and screening of bactericides. Jpn. J. Phytopathol. 1989, 55, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, F.; Donati, I.; Vanneste, J.; Costa, M.; Costa, G. Real time monitoring of the interactions between Pseudomonas syringae pv. actinidiae and Actinidia species. Acta Hortic. 2011, 913, 461–465. [Google Scholar] [CrossRef]
- Han, H.S.; Koh, Y.J.; Hur, J.S.; Jung, J.S. Identification and characterization of coronatine-producing Pseudomonas syringae pv. actinidiae. J. Microbiol. Biotechnol. 2003, 13, 110–118. [Google Scholar]
- Ferrante, P.; Scortichini, M. Molecular and phenotypic features of Pseudomonas syringae pv. actinidiae isolated during recent epidemics of bacterial canker on yellow kiwifruit (Actinidia chinensis) in central Italy. Plant Pathol. 2010, 59, 954–962. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, S.; Davies, W.J. Ozone suppresses soil drying- and abscisic acid (ABA)-induced stomatal closure via an ethylene-dependent mechanism. Plant Cell Environ. 2009, 32, 949–959. [Google Scholar] [CrossRef]
- Cellini, A.; Fiorentini, L.; Buriani, G.; Yu, J.; Donati, I.; Cornish, D.; Novak, B.; Costa, G.; Vanneste, J.; Spinelli, F. Elicitors of the salicylic acid pathway reduce incidence of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidae. Ann. Appl. Biol. 2014, 165, 441–453. [Google Scholar] [CrossRef]
- Ogawa, T.; Takahashi, M.; Fujii, T.; Tazaki, M.; Fukuda, H. The role of NADH:Fe(III)EDTA oxidoreductase in ethylene formation from 2-keto-4-methylthiobutyrate. J. Ferment. Bioeng. 1990, 69, 287–291. [Google Scholar] [CrossRef]
- Nagahama, K.; Ogawa, T.; Fujii, T.; Fukuda, H. Classification of ethylene-producing bacteria in terms of biosynthetic pathways to ethylene. J. Ferment. Bioeng. 1992, 73, 1–5. [Google Scholar] [CrossRef]
- Fukuda, H.; Ogawa, T.; Tazaki, M.; Nagahama, K.; Fujiil, T.; Tanase, S.; Morino, Y. Two reactions are simultaneously catalyzed by a single enzyme: The arginine-dependent simultaneous formation of two products, ethylene and succinate, from 2-oxoglutarate by an enzyme from Pseudomonas syringae. Biochem. Biophys. Res. Commun. 1992, 188, 483–489. [Google Scholar] [CrossRef]
- Hausinger, R.P. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 21–68. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, G.H.; Koh, Y.J.; Zhuang, Q.; Jung, J.S. Development of specific markers for identification of biovars 1 and 2 strains of Pseudomonas syringae pv. actinidiae. Plant Pathol. J. 2016, 32, 162–167. [Google Scholar] [CrossRef] [Green Version]
- Ciarroni, S.; Gallipoli, L.; Taratufolo, M.C.; Butler, M.I.; Poulter, R.T.M.; Pourcel, C.; Vergnaud, G.; Balestra, G.M.; Mazzaglia, A. Development of a multiple loci variable number of tandem repeats analysis (MLVA) to unravel the intra-pathovar structure of Pseudomonas syringae pv. actinidiae populations worldwide. PLoS ONE 2015, 10, e0135310. [Google Scholar] [CrossRef] [Green Version]
- Vanneste, J.L.; Yu, J.; Cornish, D.A.; Tanner, D.J.; Windner, R.; Chapman, J.R.; Taylor, R.K.; Mackay, J.F.; Dowlut, S. Identification, virulence, and distribution of two biovars of Pseudomonas syringae pv. actinidiae in New Zealand. Plant Dis. 2013, 97, 708–719. [Google Scholar] [CrossRef] [Green Version]
- Balestra, G.M.; Buriani, G.; Cellini, A.; Donati, I.; Mazzaglia, A.; Spinelli, F. First report of Pseudomonas syringae pv. actinidiae on kiwifruit pollen from Argentina. Plant Dis. 2018, 102, 237. [Google Scholar] [CrossRef]
- Weingart, H.; Volksch, B. Ethylene production by Pseudomonas syringae pathovars in vitro and in planta. Appl. Environ. Microbiol. 1997, 63, 156–161. [Google Scholar] [CrossRef] [Green Version]
- Valls, M.; Genin, S.; Boucher, C. Integrated Regulation of the Type III Secretion System and Other Virulence Determinants in Ralstonia solanacearum. PLoS Pathog. 2006, 2, e82. [Google Scholar] [CrossRef] [Green Version]
- De Pedro-Jové, R.; Puigvert, M.; Sebastià, P.; Macho, A.P.; Monteiro, J.S.; Coll, N.S.; Setúbal, J.C.; Valls, M. Dynamic expression of Ralstonia solanacearum virulence factors and metabolism-controlling genes during plant infection. BMC Genom. 2021, 22, 170. [Google Scholar] [CrossRef]
- Lu, H.; McClung, C.R.; Zhang, C. Tick Tock: Circadian regulation of plant innate immunity. Annu. Rev. Phytopathol. 2017, 55, 287–311. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Ye, N.; Yang, J.; Peng, X.; Zhang, J. Regulation of expression of starch synthesis genes by ethylene and ABA in relation to the development of rice inferior and superior spikelets. J. Exp. Bot. 2011, 62, 3907–3916. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lu, Y.; Li, L.; Chu, Z.; Zhang, H.; Li, H.; Fernie, A.R.; Ouyang, B. Impairment of hormone pathways results in a general disturbance of fruit primary metabolism in tomato. Food Chem. 2019, 274, 170–179. [Google Scholar] [CrossRef]
- Lucena, C.; Waters, B.M.; Romera, F.J.; García, M.J.; Morales, M.; Alcántara, E.; Pérez-Vicente, R. Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J. Exp. Bot. 2006, 57, 4145–4154. [Google Scholar] [CrossRef] [Green Version]
- Aznar, A.; Chen, N.W.; Thomine, S.; Dellagi, A. Immunity to plant pathogens and iron homeostasis. Plant Sci. 2015, 240, 90–97. [Google Scholar] [CrossRef]
- Kim, H.-E.; Shitashiro, M.; Kuroda, A.; Takiguchi, N.; Kato, J. Ethylene chemotaxis in Pseudomonas aeruginosa and other Pseudomonas species. Microbes Environ. 2007, 22, 186–189. [Google Scholar] [CrossRef] [Green Version]
- Augimeri, R.V.; Strap, J.L. The Phytohormone Ethylene Enhances Cellulose Production, Regulates CRP/FNRKx Transcription and Causes Differential Gene Expression within the Bacterial Cellulose Synthesis Operon of Komagataeibacter (Gluconacetobacter) xylinus ATCC 53582. Front. Microbiol. 2015, 6, 1459. [Google Scholar] [CrossRef] [Green Version]
- Nonaka, S.; Yuhashi, K.-I.; Takada, K.; Sugaware, M.; Minamisawa, K.; Ezura, H. Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol. 2008, 178, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit. 2. The most suitable times and environments for infection on new canes. Jpn. J. Phytopathol. 1993, 59, 460–468. [Google Scholar] [CrossRef] [Green Version]
- Young, J.M. Pseudomonas syringae pv. actinidiae in New Zealand. J. Plant Pathol. 2012, 94, S1.5–S1.10. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van Der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [Green Version]
- Mur, L.A.J.; Prats, E.; Pierre, S.; Hall, M.A.; Hebelstrup, K.H. Integrating nitric oxide into salicylic acid and jasmonic acid/ethylene plant defense pathways. Front. Plant Sci. 2013, 4, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, M.N.; Vasconcelos, M.; Pinto, V.; Balestra, G.; Mazzaglia, A.; Gomez-Cadenas, A.; Carvalho, S. Role of methyl jasmonate and salicylic acid in kiwifruit plants further subjected to Psa infection: Biochemical and genetic responses. Plant Physiol. Biochem. 2021, 162, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Biondi, E.; Buriani, G.; Farneti, B.; Rodriguez-Estrada, M.T.; Braschi, I.; Savioli, S.; Blasioli, S.; Rocchi, L.; Biasioli, F.; et al. Characterization of volatile organic compounds emitted by kiwifruit plants infected with Pseudomonas syringae pv. actinidiae and their effects on host defences. Trees-Struct. Funct. 2015, 30, 795–806. [Google Scholar] [CrossRef]
- Michelotti, V.; Lamontanara, A.; Buriani, G.; Orrù, L.; Cellini, A.; Donati, I.; Vanneste, J.L.; Cattivelli, L.; Tacconi, G.; Spinelli, F. Comparative transcriptome analysis of the interaction between Actinidia chinensis var. chinensis and Pseudomonas syringae pv. actinidiae in absence and presence of acibenzolar-S-methyl. BMC Genom. 2018, 19, 585. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Cui, X.; Sun, Y.; Dong, C.-H. Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep. 2013, 32, 1099–1109. [Google Scholar] [CrossRef]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U.; Beckers, G.J.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.A.; Pieterse, C.M.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant-Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, S.M.; Mandon, J.; Arslanov, D.; De Pessemier, J.; Hermans, C.; Harren, F.J.M. Current methods for detecting ethylene in plants. Ann. Bot. 2012, 111, 347–360. [Google Scholar] [CrossRef] [Green Version]
- Cappellin, L.; Makhoul, S.; Schuhfried, E.; Romano, A.; Del Pulgar, J.S.; Aprea, E.; Farneti, B.; Costa, F.; Gasperi, F.; Biasioli, F. Ethylene: Absolute real-time high-sensitivity detection with PTR/SRI-MS. The example of fruits, leaves and bacteria. Int. J. Mass Spectrom. 2014, 365-366, 33–41. [Google Scholar] [CrossRef]
- Cristescu, S.; Persijn, S.; Hekkert, S.T.L.; Harren, F. Laser-based systems for trace gas detection in life sciences. Appl. Phys. A 2008, 92, 343–349. [Google Scholar] [CrossRef]
- Alexeyev, M.F. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. BioTechniques 1999, 26, 824–828. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.-E.; Wang, N.; Wang, J.; Chen, Z.J.; Gross, D.C. Oligonucleotide microarray analysis of the SalA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 2005, 18, 324–333. [Google Scholar] [CrossRef]
- Narusaka, M.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. rpoD gene expression as an indicator of bacterial pathogens in host plants. J. Gen. Plant Pathol. 2011, 77, 75–80. [Google Scholar] [CrossRef]
- Greenwald, J.W.; Greenwald, C.J.; Philmus, B.J.; Begley, T.P.; Gross, D.C. RNA-seq analysis reveals that an ECF σ factor, AcsS, regulates achromobactin biosynthesis in Pseudomonas syringae pv. syringae B728a. PLoS ONE 2012, 7, e34804. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef] [Green Version]



| Strain | Biovar | Origin | Source or Reference |
|---|---|---|---|
| Pseudomonas syringae pv. actinidiae | |||
| SUPP319 | 1 | Japan | [35] |
| SUPP320 | 1 | Japan | [35] |
| NIAS302143 | 1 | Japan | NIAS, National Agriculture and Food Research Organization (Japan) |
| NIAS302145 | 1 | Japan | NIAS, National Agriculture and Food Research Organization (Japan) |
| KACC10594 | 2 | Korea | KACC, National Institute of Agricultural Sciences (South Korea) |
| Psa-K2 | 2 | Korea | [36] |
| CFBP7286 | 3 | Italy | CIRM-CFBP, INRA (France) |
| CFBP7286-GFPuv | 3 | [25] | |
| CFBP7286-Δbep | 3 | This work | |
| 10787 | 3 | New Zealand | [37] |
| Arg2.1 | 3 | Argentina | [38] |
| Pseudomonas syringae pv. syringae | |||
| ICMP3523 | Australia | ICMP, Landcare Research (New Zealand) | |
| LT23 | Italy | JL Vanneste, direct isolation from A. deliciosa | |
| Pseudomonas syringae pv. glycinea | |||
| NCPPB1883 | USA | NCPPB, FERA (UK) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cellini, A.; Donati, I.; Farneti, B.; Khomenko, I.; Buriani, G.; Biasioli, F.; Cristescu, S.M.; Spinelli, F. A Breach in Plant Defences: Pseudomonas syringae pv. actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses. Int. J. Mol. Sci. 2021, 22, 4375. https://doi.org/10.3390/ijms22094375
Cellini A, Donati I, Farneti B, Khomenko I, Buriani G, Biasioli F, Cristescu SM, Spinelli F. A Breach in Plant Defences: Pseudomonas syringae pv. actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses. International Journal of Molecular Sciences. 2021; 22(9):4375. https://doi.org/10.3390/ijms22094375
Chicago/Turabian StyleCellini, Antonio, Irene Donati, Brian Farneti, Iuliia Khomenko, Giampaolo Buriani, Franco Biasioli, Simona M. Cristescu, and Francesco Spinelli. 2021. "A Breach in Plant Defences: Pseudomonas syringae pv. actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses" International Journal of Molecular Sciences 22, no. 9: 4375. https://doi.org/10.3390/ijms22094375
APA StyleCellini, A., Donati, I., Farneti, B., Khomenko, I., Buriani, G., Biasioli, F., Cristescu, S. M., & Spinelli, F. (2021). A Breach in Plant Defences: Pseudomonas syringae pv. actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses. International Journal of Molecular Sciences, 22(9), 4375. https://doi.org/10.3390/ijms22094375











