NUP-98 Rearrangements Led to the Identification of Candidate Biomarkers for Primary Induction Failure in Pediatric Acute Myeloid Leukemia
Abstract
1. Introduction
2. Results
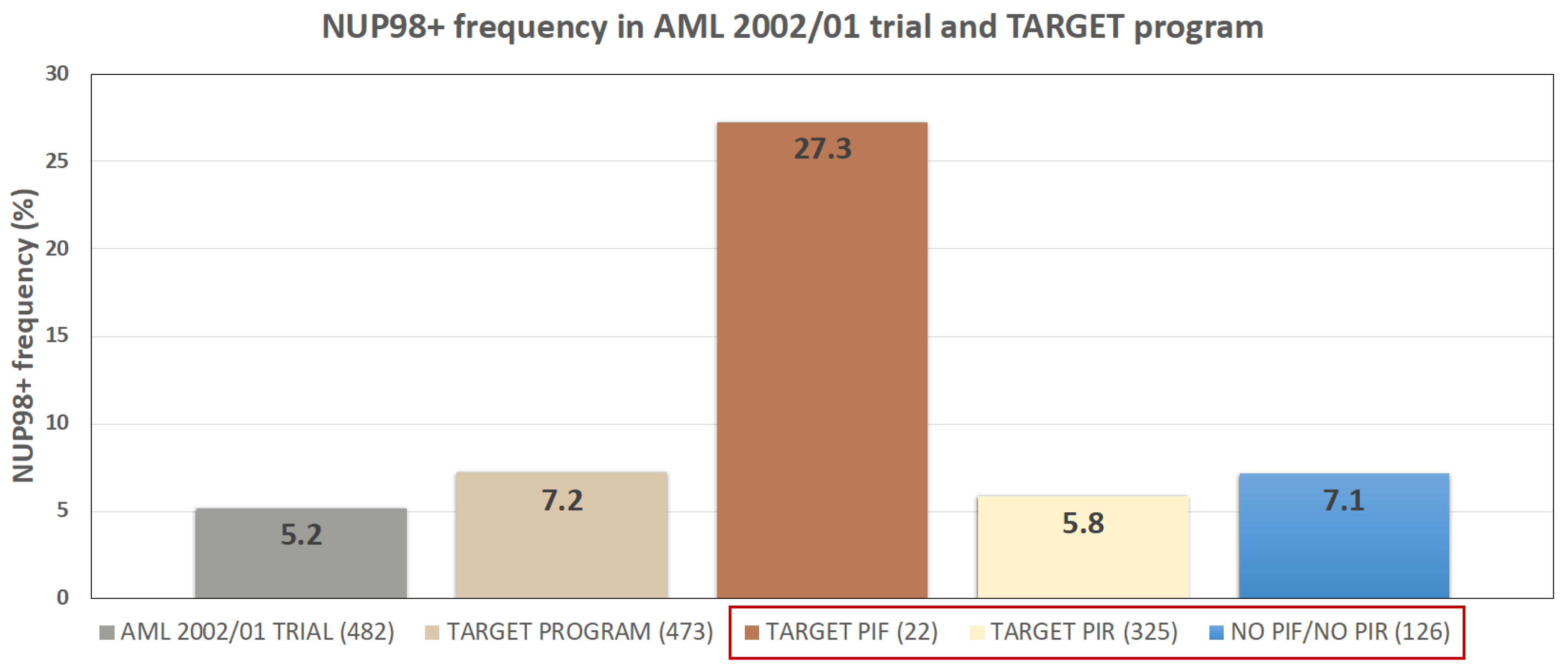
2.1. NUP98 Rearrangements Frequency Is Comparable in Cohorts Belonging to Three Different Programs (AIEOP-AML Group; TARGET; Local Cohort)
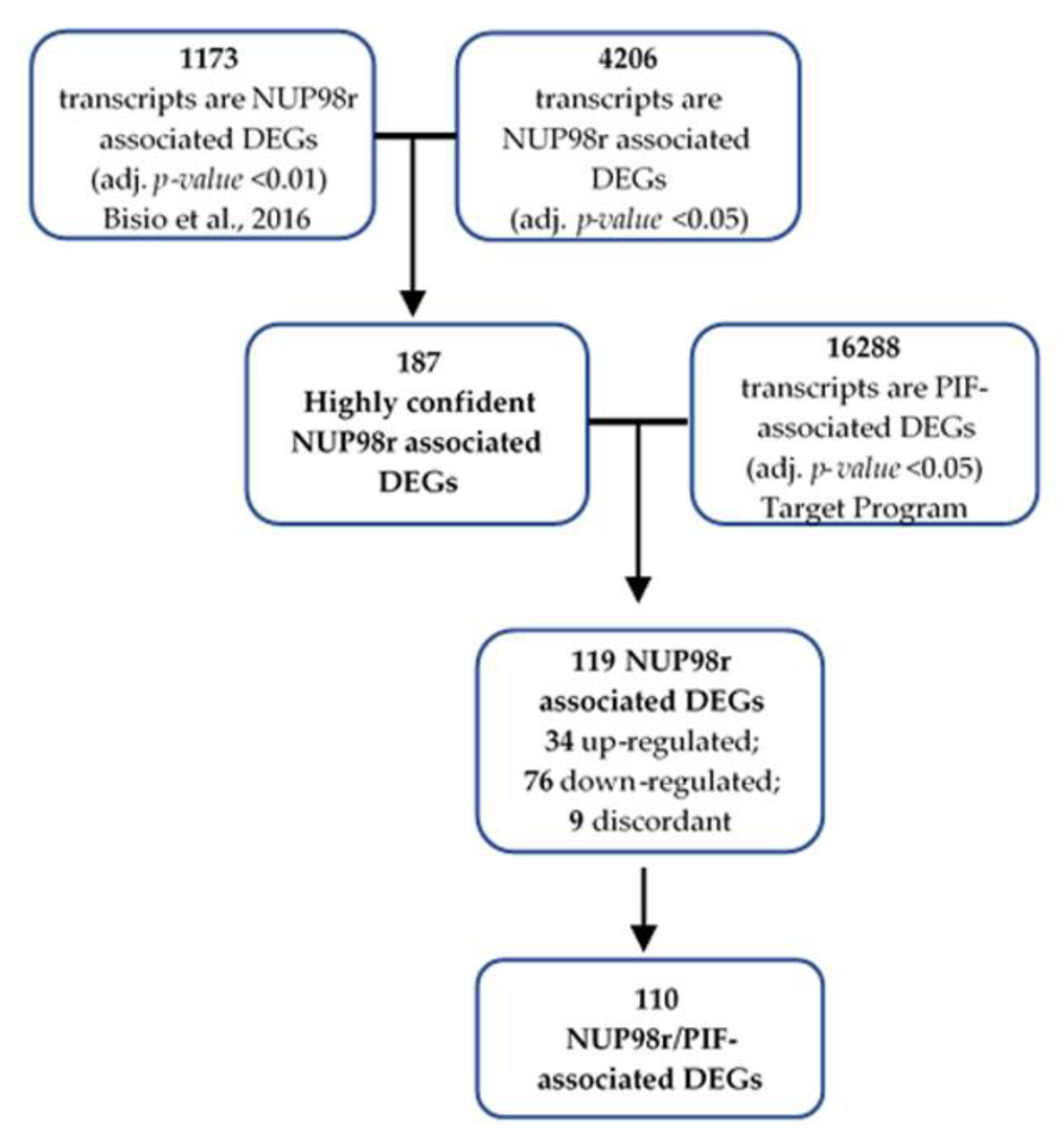
2.2. Differentially Expressed Genes between NUP98r+ and NUP98r− Samples
2.3. Primary Induction Failure-Associated DEGs Are Enriched among Highly Confident NUP98r Associated DEGs
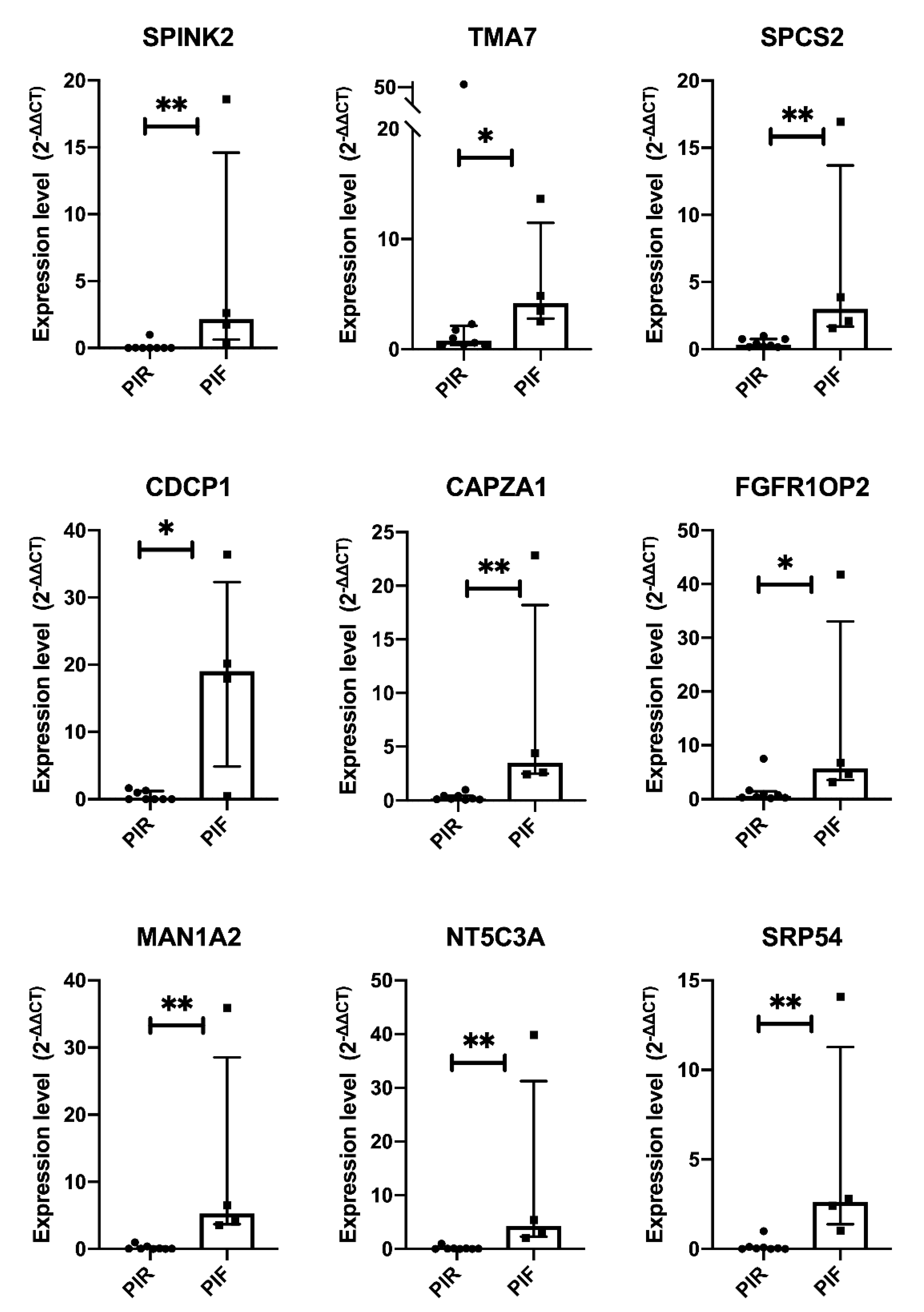
2.4. Validation by Quantitative Real-Time PCR in Local Cohort
2.5. NUP98-NSD1 and NUP98-KMD5A Comparison
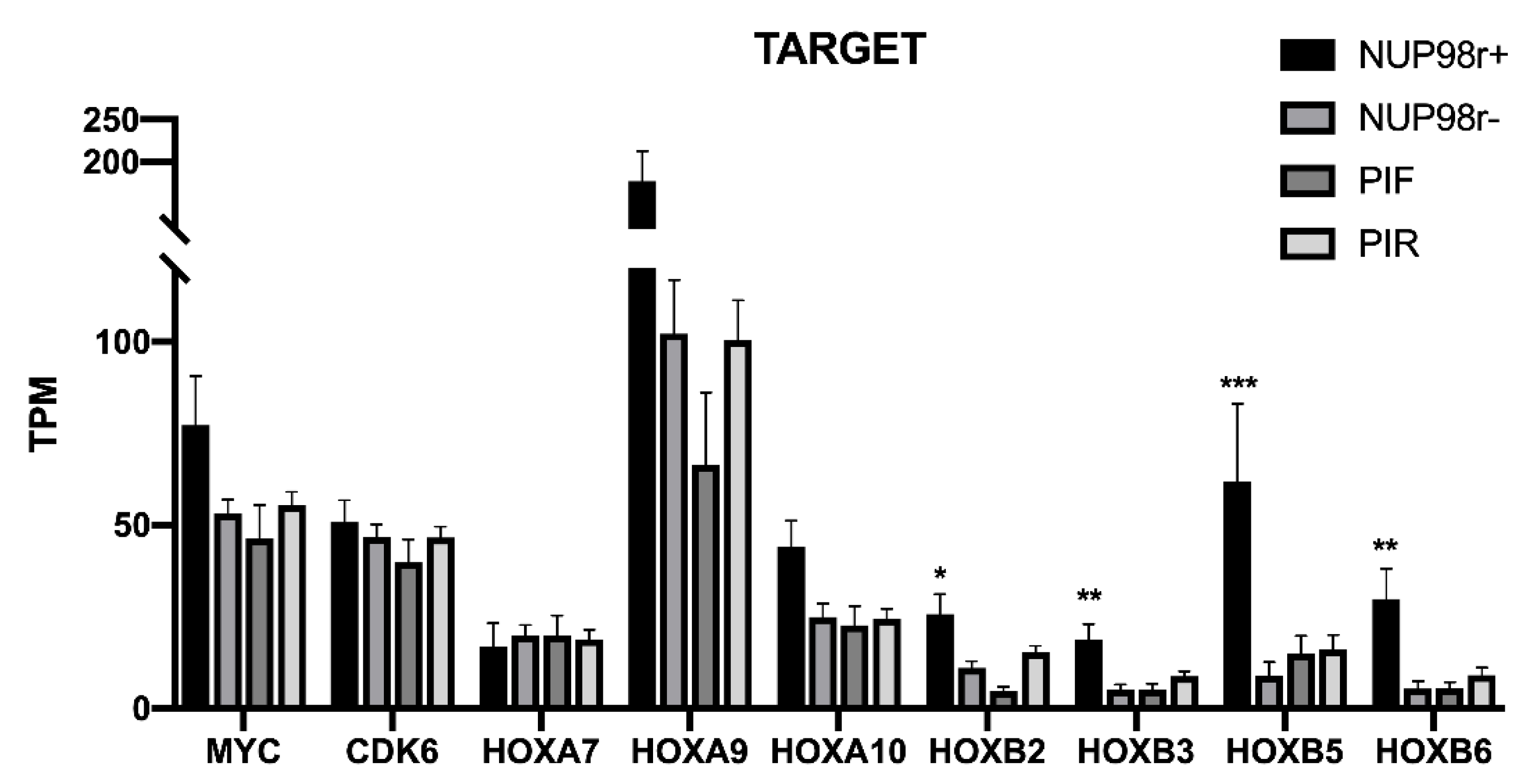
2.6. HOXA and HOXB Clusters, MYC and CDK6 Expression
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Human Transcriptome Array 2.0
4.3. Gene Expression Profiling Dataset
4.4. RNA Extraction and Quantitative PCR
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gibson, B.E.S.; Sauer, M.G.; Amrolia, P. Acute Myeloid Leukemia in Children. In The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies; Carreras, E., Dufour, C., Mohty, M., Kröger, N., Eds.; Springer: Cham, Switzerland, 2019; ISBN 978-3-030-02277-8. [Google Scholar]
- Tsukimoto, I.; Tawa, A.; Horibe, K.; Tabuchi, K.; Kigasawa, H.; Tsuchida, M.; Yabe, H.; Nakayama, H.; Kudo, K.; Kobayashi, R.; et al. Risk-Stratified Therapy and the Intensive Use of Cytarabine Improves the Outcome in Childhood Acute Myeloid Leukemia: The AML99 Trial From the Japanese Childhood AML Cooperative Study Group. JCO 2009, 27, 4007–4013. [Google Scholar] [CrossRef]
- Pession, A.; Masetti, R.; Rizzari, C.; Putti, M.C.; Casale, F.; Fagioli, F.; Luciani, M.; Lo Nigro, L.; Menna, G.; Micalizzi, C.; et al. Results of the AIEOP AML 2002/01 Multicenter Prospective Trial for the Treatment of Children with Acute Myeloid Leukemia. Blood 2013, 122, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional Genomic Landscape of Acute Myeloid Leukaemia. Nature 2018, 562, 526–531. [Google Scholar] [CrossRef]
- Stanisic, S.; Kalaycio, M. Treatment of Refractory and Relapsed Acute Myelogenous Leukemia. Expert Rev. Anticancer Ther. 2002, 2, 287–295. [Google Scholar] [CrossRef]
- Quarello, P.; Fagioli, F.; Basso, G.; Putti, M.C.; Berger, M.; Luciani, M.; Rizzari, C.; Menna, G.; Masetti, R.; Locatelli, F. Outcome of Children with Acute Myeloid Leukaemia (AML) Experiencing Primary Induction Failure in the AIEOP AML 2002/01 Clinical Trial. Br. J. Haematol. 2015, 171, 566–573. [Google Scholar] [CrossRef]
- Brown, F.C.; Cifani, P.; Drill, E.; He, J.; Still, E.; Zhong, S.; Balasubramanian, S.; Pavlick, D.; Yilmazel, B.; Knapp, K.M.; et al. Genomics of Primary Chemoresistance and Remission Induction Failure in Paediatric and Adult Acute Myeloid Leukaemia. Br. J. Haematol. 2017, 176, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Meggendorfer, M.; Bacher, U.; Alpermann, T.; Haferlach, C.; Kern, W.; Gambacorti-Passerini, C.; Haferlach, T.; Schnittger, S. SETBP1 Mutations Occur in 9% of MDS/MPN and in 4% of MPN Cases and Are Strongly Associated with Atypical CML, Monosomy 7, Isochromosome i(17)(Q10), ASXL1 and CBL Mutations. Leukemia 2013, 27, 1852–1860. [Google Scholar] [CrossRef]
- Bolouri, H.; Farrar, J.E.; Triche, T.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The Molecular Landscape of Pediatric Acute Myeloid Leukemia Reveals Recurrent Structural Alterations and Age-Specific Mutational Interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Ostronoff, F.; Othus, M.; Gerbing, R.B.; Loken, M.R.; Raimondi, S.C.; Hirsch, B.A.; Lange, B.J.; Petersdorf, S.; Radich, J.; Appelbaum, F.R.; et al. NUP98/NSD1 and FLT3/ITD Coexpression Is More Prevalent in Younger AML Patients and Leads to Induction Failure: A COG and SWOG Report. Blood 2014, 124, 2400–2407. [Google Scholar] [CrossRef]
- McNeer, N.A.; Philip, J.; Geiger, H.; Ries, R.E.; Lavallée, V.-P.; Walsh, M.; Shah, M.; Arora, K.; Emde, A.-K.; Robine, N.; et al. Genetic Mechanisms of Primary Chemotherapy Resistance in Pediatric Acute Myeloid Leukemia. Leukemia 2019, 33, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [CrossRef]
- Chou, W.-C.; Chen, C.-Y.; Hou, H.-A.; Lin, L.-I.; Tang, J.-L.; Yao, M.; Tsay, W.; Ko, B.-S.; Wu, S.-J.; Huang, S.-Y.; et al. Acute Myeloid Leukemia Bearing t(7;11)(P15;P15) Is a Distinct Cytogenetic Entity with Poor Outcome and a Distinct Mutation Profile: Comparative Analysis of 493 Adult Patients. Leukemia 2009, 23, 1303–1310. [Google Scholar] [CrossRef]
- Hollink, I.H.I.M.; van den Heuvel-Eibrink, M.M.; Arentsen-Peters, S.T.C.J.M.; Pratcorona, M.; Abbas, S.; Kuipers, J.E.; van Galen, J.F.; Beverloo, H.B.; Sonneveld, E.; Kaspers, G.-J.J.L.; et al. NUP98/NSD1 Characterizes a Novel Poor Prognostic Group in Acute Myeloid Leukemia with a Distinct HOX Gene Expression Pattern. Blood 2011, 118, 3645–3656. [Google Scholar] [CrossRef]
- Struski, S.; Lagarde, S.; Bories, P.; Puiseux, C.; Prade, N.; Cuccuini, W.; Pages, M.-P.; Bidet, A.; Gervais, C.; Lafage-Pochitaloff, M.; et al. NUP98 Is Rearranged in 3.8% of Pediatric AML Forming a Clinical and Molecular Homogenous Group with a Poor Prognosis. Leukemia 2017, 31, 565–572. [Google Scholar] [CrossRef]
- Niktoreh, N.; Walter, C.; Zimmermann, M.; von Neuhoff, C.; von Neuhoff, N.; Rasche, M.; Waack, K.; Creutzig, U.; Hanenberg, H.; Reinhardt, D. Mutated WT1, FLT3-ITD, and NUP98-NSD1 Fusion in Various Combinations Define a Poor Prognostic Group in Pediatric Acute Myeloid Leukemia. J. Oncol. 2019, 2019. [Google Scholar] [CrossRef]
- Pineault, N.; Abramovich, C.; Humphries, R.K. Transplantable Cell Lines Generated with NUP98—Hox Fusion Genes Undergo Leukemic Progression by Meis1 Independent of Its Binding to DNA. Leukemia 2005, 19, 636–643. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Michmerhuizen, N.L.; Klco, J.M.; Mullighan, C.G. Mechanistic Insights and Potential Therapeutic Approaches for NUP98-Rearranged Hematologic Malignancies. Blood 2020, 136, 2275–2289. [Google Scholar] [CrossRef]
- Schmoellerl, J.; Barbosa, I.A.M.; Eder, T.; Brandstoetter, T.; Schmidt, L.; Maurer, B.; Troester, S.; Pham, H.T.T.; Sagarajit, M.; Ebner, J.; et al. CDK6 Is an Essential Direct Target of NUP98-Fusion Proteins in Acute Myeloid Leukemia. Blood 2020, 136, 387–400. [Google Scholar] [CrossRef]
- Bisio, V.; Zampini, M.; Tregnago, C.; Manara, E.; Salsi, V.; Di Meglio, A.; Masetti, R.; Togni, M.; Di Giacomo, D.; Minuzzo, S.; et al. NUP98-Fusion Transcripts Characterize Different Biological Entities within Acute Myeloid Leukemia: A Report from the AIEOP-AML Group. Leukemia 2017, 31, 974–977. [Google Scholar] [CrossRef]
- Conze, T.; Lammers, R.; Kuçi, S.; Scherl-Mostageer, M.; Schweifer, N.; Kanz, L.; Bühring, H.-J. CDCP1 Is a Novel Marker for Hematopoietic Stem Cells. Ann. N. Y. Acad. Sci. 2003, 996, 222–226. [Google Scholar] [CrossRef]
- Bühring, H.-J.; Kuçi, S.; Conze, T.; Rathke, G.; Bartolović, K.; Grünebach, F.; Scherl-Mostageer, M.; Brümmendorf, T.H.; Schweifer, N.; Lammers, R. CDCP1 Identifies a Broad Spectrum of Normal and Malignant Stem/Progenitor Cell Subsets of Hematopoietic and Nonhematopoietic Origin: CDCP1 Identifies a Broad Spectrum of Normal and Malignant Stem/Progenitor Cell Subsets of Hematopoietic and Nonhematopoietic Origin. Stem Cells 2004, 22, 334–343. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Noort, S.; Wander, P.; Alonzo, T.A.; Smith, J.; Ries, R.E.; Gerbing, R.B.; Dolman, M.E.M.; Locatelli, F.; Reinhardt, D.; Baruchel, A.; et al. The Clinical and Biological Characteristics of NUP98-KDM5A in Pediatric Acute Myeloid Leukemia. Haematologica 2021, 106, 630–634. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, J.D.E.; Hollink, I.H.I.M.; Arentsen-Peters, S.T.C.J.M.; van Galen, J.F.; Berna Beverloo, H.; Baruchel, A.; Trka, J.; Reinhardt, D.; Sonneveld, E.; Zimmermann, M.; et al. NUP98/JARID1A Is a Novel Recurrent Abnormality in Pediatric Acute Megakaryoblastic Leukemia with a Distinct HOX Gene Expression Pattern. Leukemia 2013, 27, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.G.; Cai, L.; Pasillas, M.P.; Kamps, M.P. NUP98-NSD1 Links H3K36 Methylation to Hox-A Gene Activation and Leukaemogenesis. Nat. Cell Biol. 2007, 9, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Heitmann, J.S.; Hagelstein, I.; Hinterleitner, C.; Roerden, M.; Jung, G.; Salih, H.R.; Märklin, M.; Kauer, J. Identification of CD318 (CDCP1) as Novel Prognostic Marker in AML. Ann. Hematol. 2020, 99, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Möritz, A.; Grzeschik, K.H.; Wingender, E.; Fink, E. Organization and Sequence of the Gene Encoding the Human Acrosin-Trypsin Inhibitor (HUSI-II). Gene 1993, 123, 277–281. [Google Scholar] [CrossRef][Green Version]
- Chen, T.; Lee, T.-R.; Liang, W.-G.; Chang, W.-S.W.; Lyu, P.-C. Identification of Trypsin-Inhibitory Site and Structure Determination of Human SPINK2 Serine Proteinase Inhibitor. Proteins 2009, 77, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Zhang, J.; Zhang, G.; Xue, Y.; Zhang, G.; Wu, X. Elevated SPINK2 Gene Expression Is a Predictor of Poor Prognosis in Acute Myeloid Leukemia. Oncol. Lett. 2019, 18, 2877–2884. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.; Tan, B.; Rosenwald, A.; Hurt, E.H.; Wiestner, A.; Staudt, L.M. A Gene Expression-Based Method to Diagnose Clinically Distinct Subgroups of Diffuse Large B Cell Lymphoma. Proc. Natl. Acad. Sci. USA 2003, 100, 9991–9996. [Google Scholar] [CrossRef]
- Hoefnagel, J.J.; Dijkman, R.; Basso, K.; Jansen, P.M.; Hallermann, C.; Willemze, R.; Tensen, C.P.; Vermeer, M.H. Distinct Types of Primary Cutaneous Large B-Cell Lymphoma Identified by Gene Expression Profiling. Blood 2005, 105, 3671–3678. [Google Scholar] [CrossRef]
- Scherl-Mostageer, M.; Sommergruber, W.; Abseher, R.; Hauptmann, R.; Ambros, P.; Schweifer, N. Identification of a Novel Gene, CDCP1, Overexpressed in Human Colorectal Cancer. Oncogene 2001, 20, 4402–4408. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Morii, E.; Ikeda, J.; Ezoe, S.; Xu, J.-X.; Nakamichi, N.; Tomita, Y.; Shibayama, H.; Kanakura, Y.; Aozasa, K. Role of DNA Methylation for Expression of Novel Stem Cell Marker CDCP1 in Hematopoietic Cells. Leukemia 2006, 20, 1551–1556. [Google Scholar] [CrossRef] [PubMed]
- Gioia, R.; Leroy, C.; Drullion, C.; Lagarde, V.; Etienne, G.; Dulucq, S.; Lippert, E.; Roche, S.; Mahon, F.-X.; Pasquet, J.-M. Quantitative Phosphoproteomics Revealed Interplay between Syk and Lyn in the Resistance to Nilotinib in Chronic Myeloid Leukemia Cells. Blood 2011, 118, 2211–2221. [Google Scholar] [CrossRef]
- Huang, D.; Cao, L.; Zheng, S. CAPZA1 Modulates EMT by Regulating Actin Cytoskeleton Remodelling in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2017, 36. [Google Scholar] [CrossRef]
- Hug, C.; Miller, T.M.; Torres, M.A.; Casella, J.F.; Cooper, J.A. Identification and Characterization of an Actin-Binding Site of CapZ. J. Cell Biol. 1992, 116, 923–931. [Google Scholar] [CrossRef] [PubMed]
- LEE, Y.-J.; JEONG, S.-H.; HONG, S.-C.; CHO, B.-I.; HA, W.-S.; PARK, S.-T.; CHOI, S.-K.; JUNG, E.-J.; JU, Y.-T.; JEONG, C.-Y.; et al. Prognostic Value of CAPZA1 Overexpression in Gastric Cancer. Int. J. Oncol. 2013, 42, 1569–1577. [Google Scholar] [CrossRef]
- Lin, A.; Hokugo, A.; Choi, J.; Nishimura, I. Small Cytoskeleton-Associated Molecule, Fibroblast Growth Factor Receptor 1 Oncogene Partner 2/Wound Inducible Transcript-3.0 (FGFR1OP2/Wit3.0), Facilitates Fibroblast-Driven Wound Closure. Am. J. Pathol. 2010, 176, 108–121. [Google Scholar] [CrossRef]
- Jin, Y.; Zhen, Y.; Haugsten, E.M.; Wiedlocha, A. The Driver of Malignancy in KG-1a Leukemic Cells, FGFR1OP2–FGFR1, Encodes an HSP90 Addicted Oncoprotein. Cell. Signal. 2011, 23, 1758–1766. [Google Scholar] [CrossRef]
- Herscovics, A. Structure and Function of Class I Alpha 1,2-Mannosidases Involved in Glycoprotein Synthesis and Endoplasmic Reticulum Quality Control. Biochimie 2001, 83, 757–762. [Google Scholar] [CrossRef]
- Kim, S.J.; Sohn, I.; Do, I.-G.; Jung, S.H.; Ko, Y.H.; Yoo, H.Y.; Paik, S.; Kim, W.S. Gene Expression Profiles for the Prediction of Progression-Free Survival in Diffuse Large B Cell Lymphoma: Results of a DASL Assay. Ann. Hematol. 2014, 93, 437–447. [Google Scholar] [CrossRef]
- Li, L.; Fridley, B.; Kalari, K.; Jenkins, G.; Batzler, A.; Safgren, S.; Hildebrandt, M.; Ames, M.; Schaid, D.; Wang, L. Gemcitabine and Cytosine Arabinoside Cytotoxicity: Association with Lymphoblastoid Cell Expression. Cancer Res. 2008, 68, 7050–7058. [Google Scholar] [CrossRef] [PubMed]
- Cheong, H.S.; Koh, Y.; Ahn, K.-S.; Lee, C.; Shin, H.D.; Yoon, S.-S. NT5C3 Polymorphisms and Outcome of First Induction Chemotherapy in Acute Myeloid Leukemia. Pharm. Genom. 2014, 24, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, L.; Simon, A.J.; Rechavi, G.; Lev, A.; Barel, O.; Kunik, V.; Toren, A.; Schiby, G.; Tamary, H.; Steinberg-Shemer, O.; et al. Congenital Neutropenia with Variable Clinical Presentation in Novel Mutation of the SRP54 Gene. Pediatric Blood Cancer 2020, 67, e28237. [Google Scholar] [CrossRef] [PubMed]
- Bellanné-Chantelot, C.; Schmaltz-Panneau, B.; Marty, C.; Fenneteau, O.; Callebaut, I.; Clauin, S.; Docet, A.; Damaj, G.-L.; Leblanc, T.; Pellier, I.; et al. Mutations in the SRP54 Gene Cause Severe Congenital Neutropenia as Well as Shwachman-Diamond-like Syndrome. Blood 2018, 132, 1318–1331. [Google Scholar] [CrossRef]
- Carden, M.A.; Connelly, J.A.; Weinzierl, E.P.; Kobrynski, L.J.; Chandrakasan, S. Severe Congenital Neutropenia Associated with SRP54 Mutation in 22q11.2 Deletion Syndrome: Hematopoietic Stem Cell Transplantation Results in Correction of Neutropenia with Adequate Immune Reconstitution. J. Clin. Immunol. 2018, 38, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Barresi, V.; Trovato-Salinaro, A.; Spampinato, G.; Musso, N.; Castorina, S.; Rizzarelli, E.; Condorelli, D.F. Transcriptome Analysis of Copper Homeostasis Genes Reveals Coordinated Upregulation of SLC31A1,SCO1, and COX11 in Colorectal Cancer. FEBS Open Bio 2016, 6, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Condorelli, D.F.; Spampinato, G.; Valenti, G.; Musso, N.; Castorina, S.; Barresi, V. Positive Caricature Transcriptomic Effects Associated with Broad Genomic Aberrations in Colorectal Cancer. Sci. Rep. 2018, 8, 14826. [Google Scholar] [CrossRef]
- Tregnago, C.; Manara, E.; Zampini, M.; Bisio, V.; Borga, C.; Bresolin, S.; Aveic, S.; Germano, G.; Basso, G.; Pigazzi, M. CREB Engages C/EBPδ to Initiate Leukemogenesis. Leukemia 2016, 30, 1887–1896. [Google Scholar] [CrossRef]
- Condorelli, D.F.; Privitera, A.P.; Barresi, V. Chromosomal Density of Cancer Up-Regulated Genes, Aberrant Enhancer Activity and Cancer Fitness Genes Are Associated with Transcriptional Cis-Effects of Broad Copy Number Gains in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4652. [Google Scholar] [CrossRef] [PubMed]
- Barresi, V.; Valenti, G.; Spampinato, G.; Musso, N.; Castorina, S.; Rizzarelli, E.; Condorelli, D.F. Transcriptome Analysis Reveals an Altered Expression Profile of Zinc Transporters in Colorectal Cancer. J. Cell. Biochem. 2018, 119, 9707–9719. [Google Scholar] [CrossRef] [PubMed]
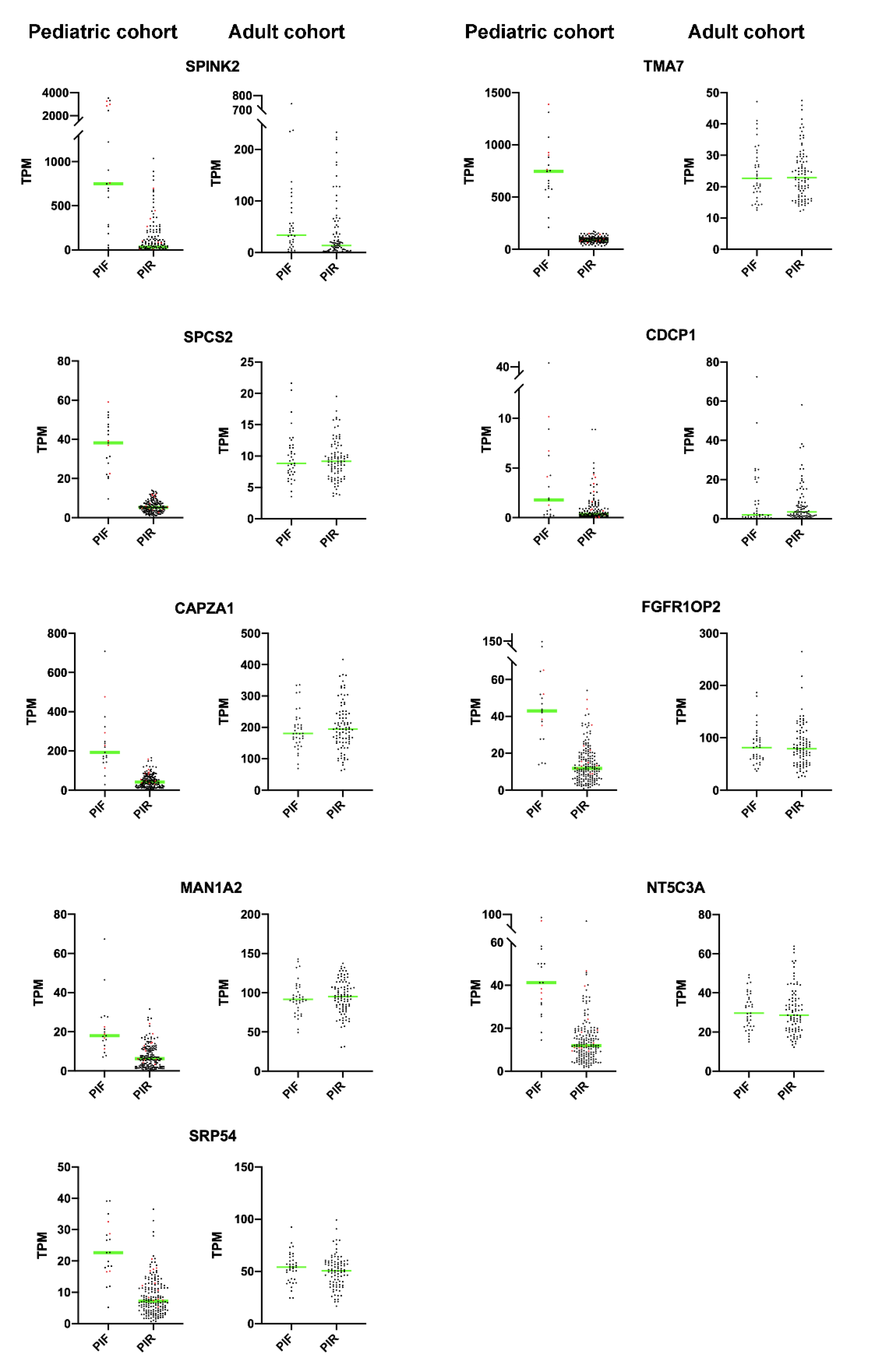

| Gene Symbol | Linear Fold Change (PIF vs. PIR) | AUC | p-Value AUC |
|---|---|---|---|
| SPINK2 | 1.96 | 0.89 | <0.0001 |
| TMA7 | 1.51 | 1.00 | <0.0001 |
| CAPZA1 | 16.13 | 0.95 | <0.0001 |
| TRDMT1 | 1.48 | 0.94 | <0.0001 |
| FGFR1OP2 | 2.86 | 0.92 | <0.0001 |
| NDUFA5 | 1.39 | 0.93 | <0.0001 |
| MAN1A2 | 3.08 | 0.89 | <0.0001 |
| NT5C3A | 2.09 | 0.93 | <0.0001 |
| SRP54 | 2.73 | 0.91 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barresi, V.; Di Bella, V.; Andriano, N.; Privitera, A.P.; Bonaccorso, P.; La Rosa, M.; Iachelli, V.; Spampinato, G.; Pulvirenti, G.; Scuderi, C.; et al. NUP-98 Rearrangements Led to the Identification of Candidate Biomarkers for Primary Induction Failure in Pediatric Acute Myeloid Leukemia. Int. J. Mol. Sci. 2021, 22, 4575. https://doi.org/10.3390/ijms22094575
Barresi V, Di Bella V, Andriano N, Privitera AP, Bonaccorso P, La Rosa M, Iachelli V, Spampinato G, Pulvirenti G, Scuderi C, et al. NUP-98 Rearrangements Led to the Identification of Candidate Biomarkers for Primary Induction Failure in Pediatric Acute Myeloid Leukemia. International Journal of Molecular Sciences. 2021; 22(9):4575. https://doi.org/10.3390/ijms22094575
Chicago/Turabian StyleBarresi, Vincenza, Virginia Di Bella, Nellina Andriano, Anna Provvidenza Privitera, Paola Bonaccorso, Manuela La Rosa, Valeria Iachelli, Giorgia Spampinato, Giulio Pulvirenti, Chiara Scuderi, and et al. 2021. "NUP-98 Rearrangements Led to the Identification of Candidate Biomarkers for Primary Induction Failure in Pediatric Acute Myeloid Leukemia" International Journal of Molecular Sciences 22, no. 9: 4575. https://doi.org/10.3390/ijms22094575
APA StyleBarresi, V., Di Bella, V., Andriano, N., Privitera, A. P., Bonaccorso, P., La Rosa, M., Iachelli, V., Spampinato, G., Pulvirenti, G., Scuderi, C., Condorelli, D. F., & Lo Nigro, L. (2021). NUP-98 Rearrangements Led to the Identification of Candidate Biomarkers for Primary Induction Failure in Pediatric Acute Myeloid Leukemia. International Journal of Molecular Sciences, 22(9), 4575. https://doi.org/10.3390/ijms22094575







