Target Prediction Model for Natural Products Using Transfer Learning
Abstract
1. Introduction
2. Results and Discussion
2.1. Hyperparameters Optimization of the Pre-Trained Model
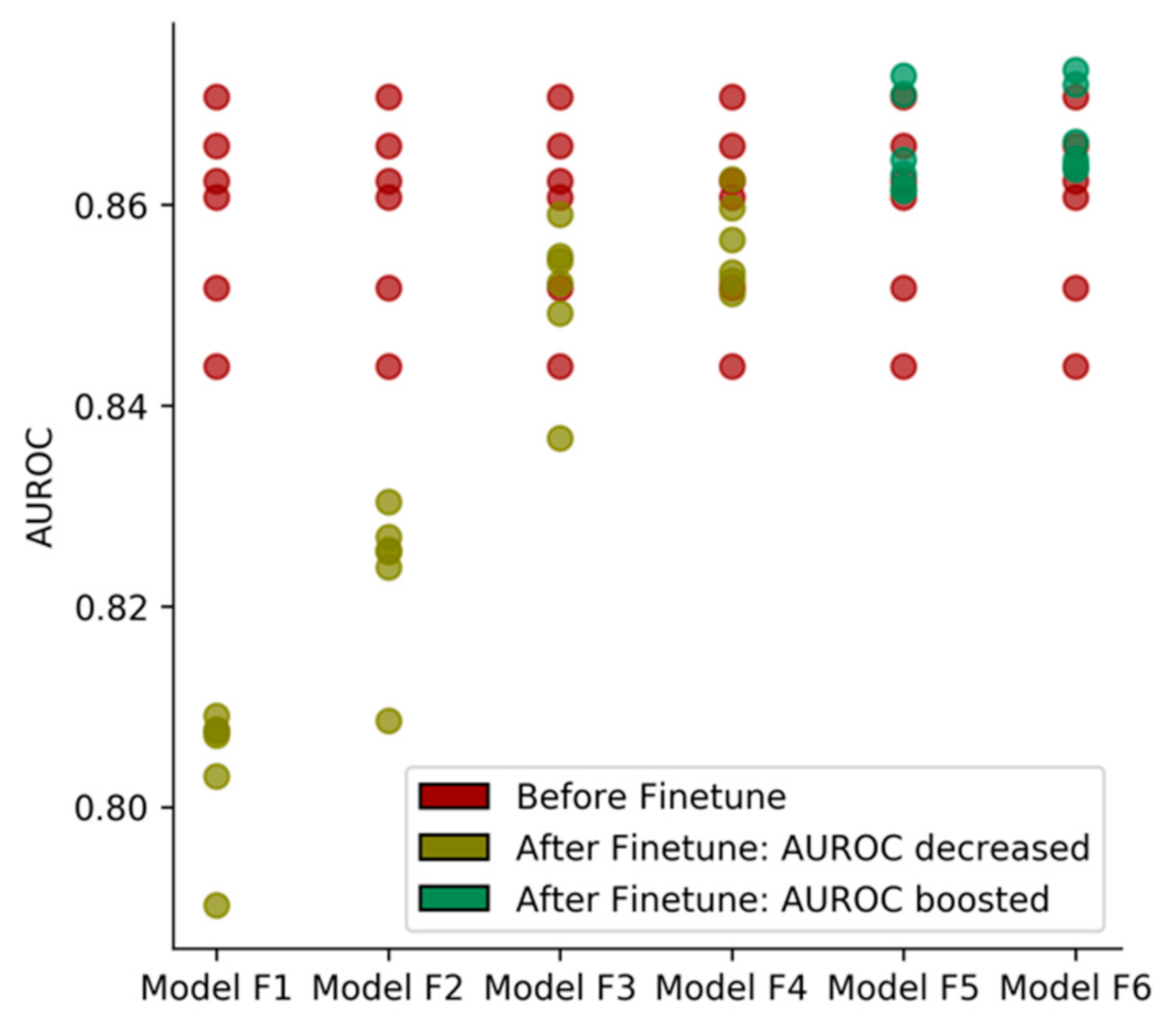
2.2. Effectiveness of Transfer Learning
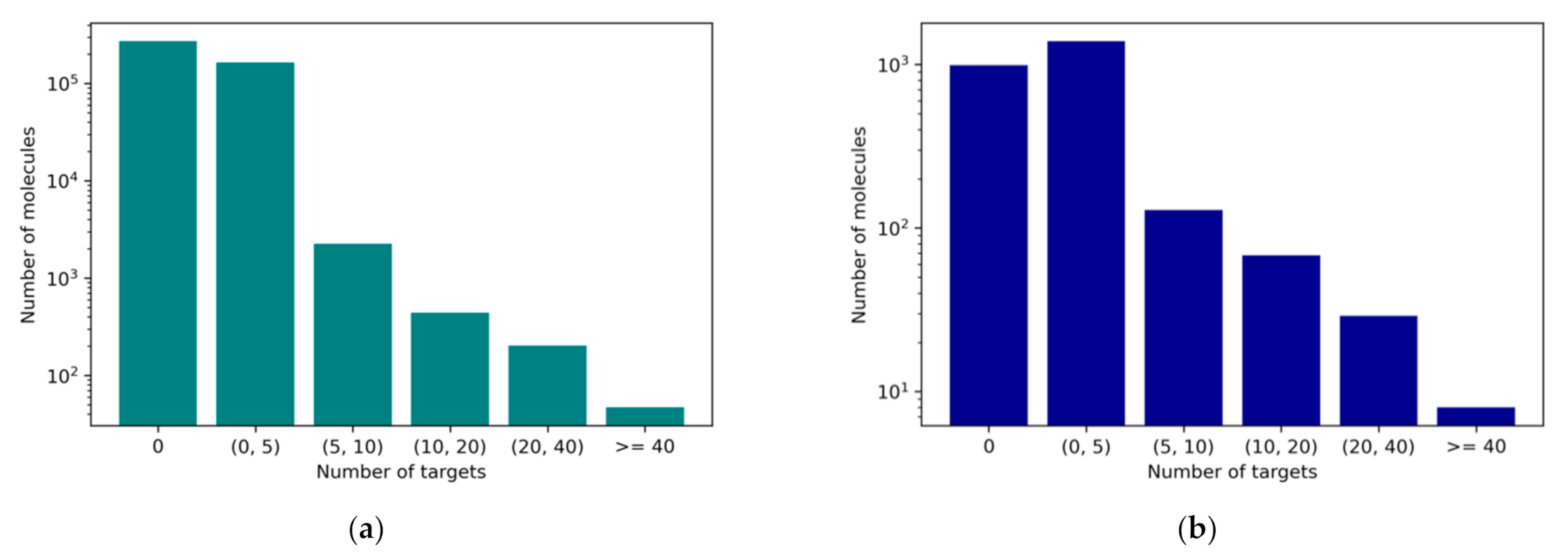
2.3. Data Balance
2.4. Comparison with References
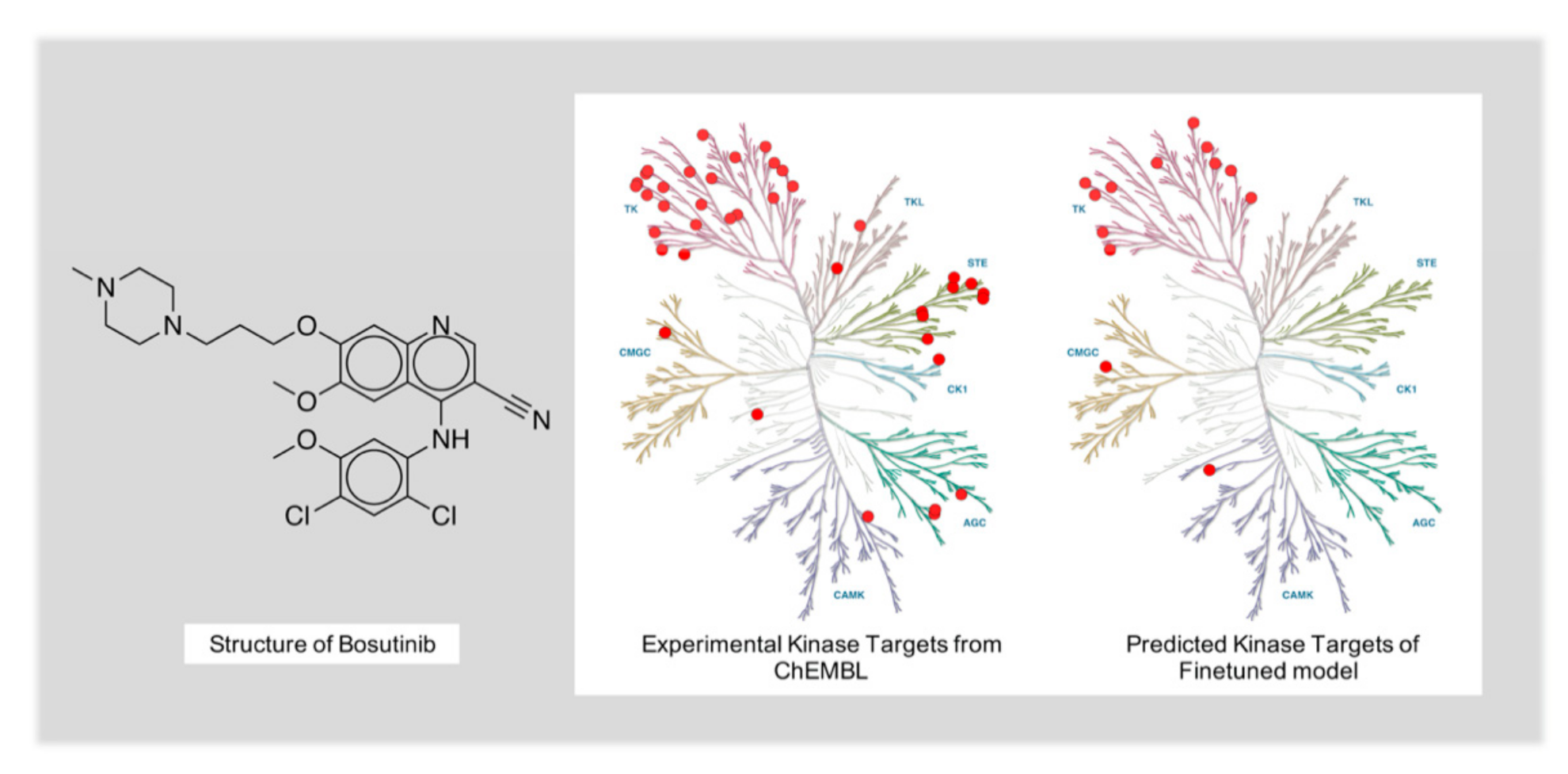
2.5. Case Study
3. Methods
3.1. Preparation of the Dataset
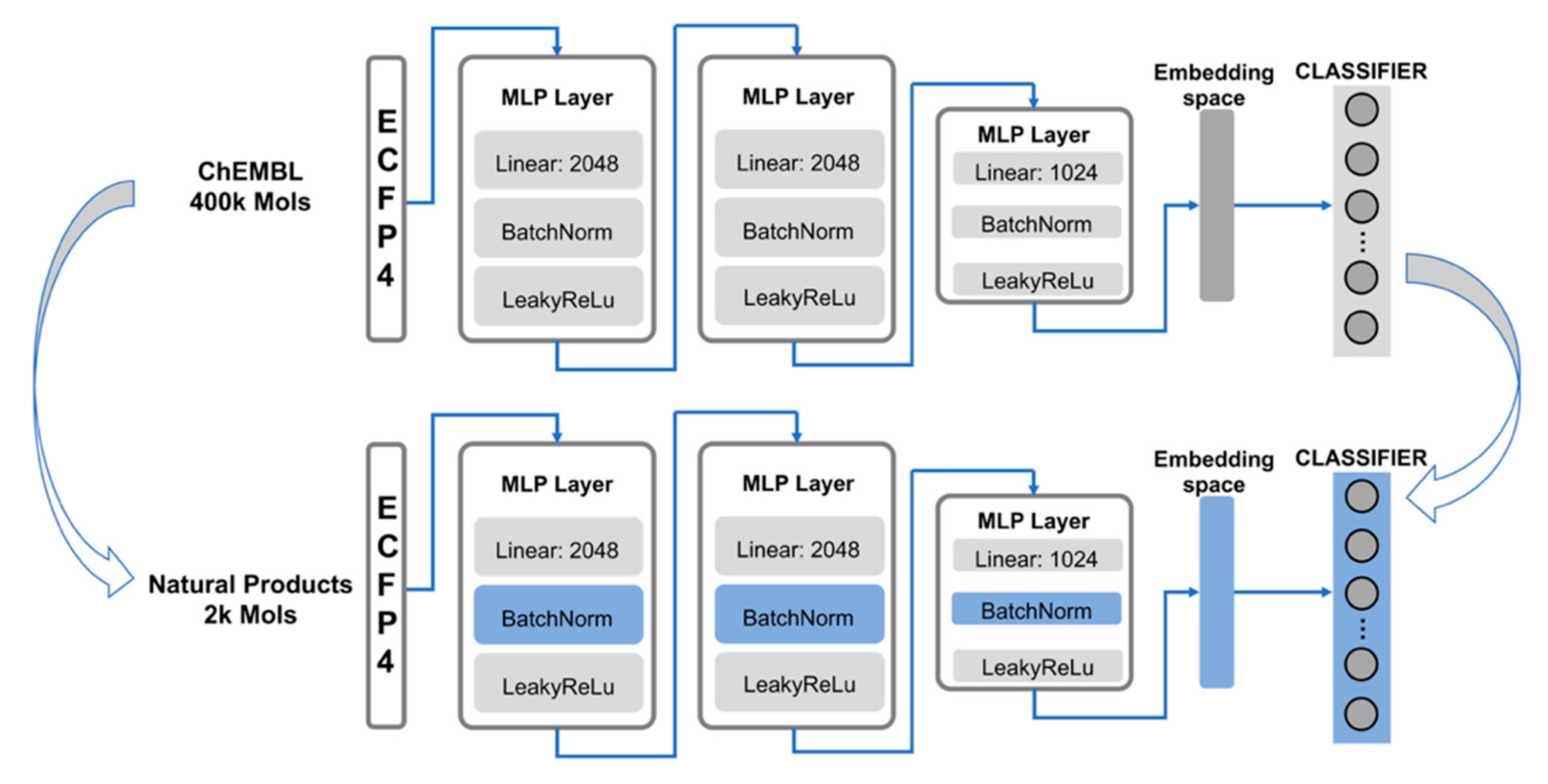
3.2. Model Structure
3.3. Pre-Training and Transfer Learning
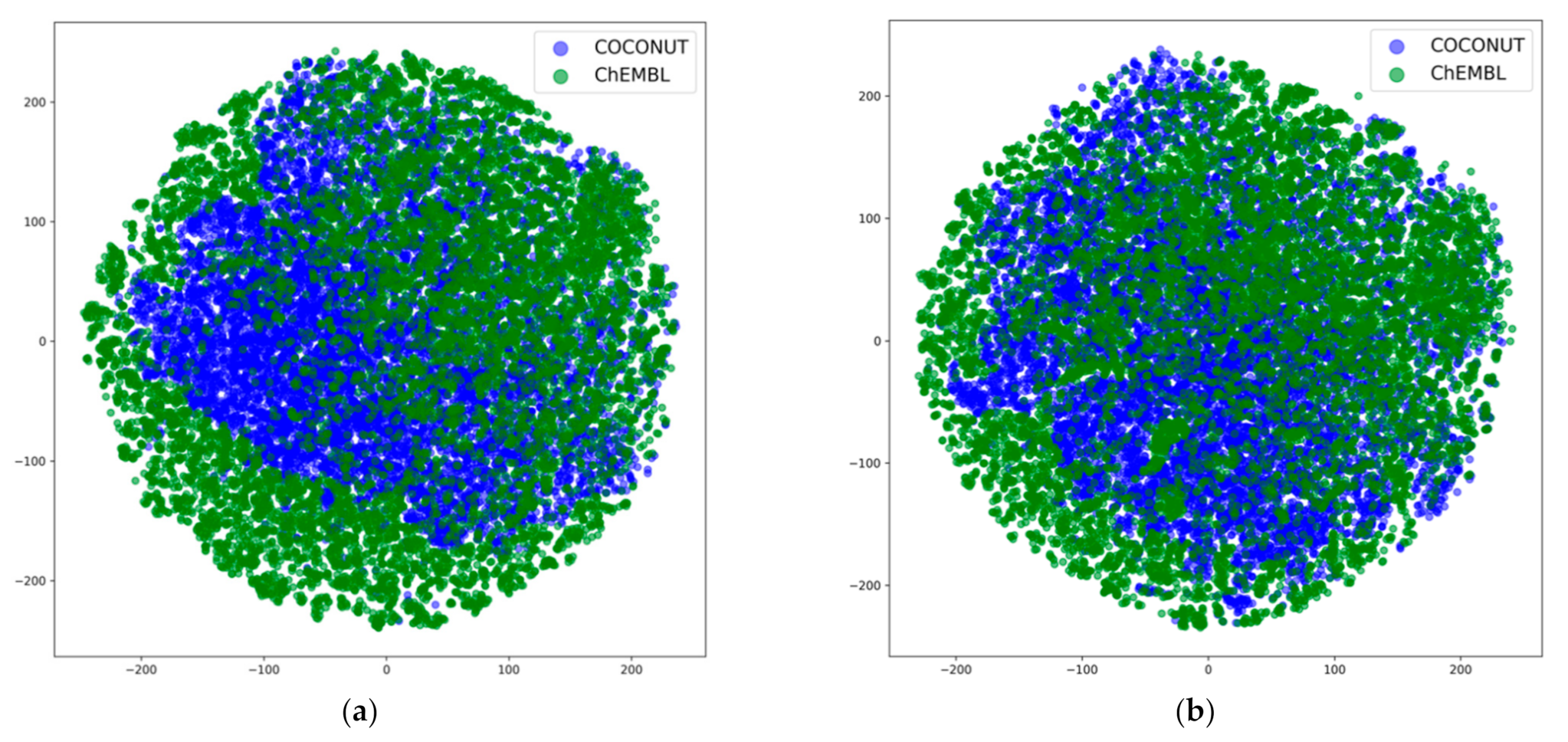
3.4. Dimensionality Reduction
3.5. Data Balance
3.6. Validation Methods
3.7. Case Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.; Schuhmann, T. Cheminformatics Analysis of Natural Product Scaffolds: Comparison of Scaffolds Produced by Animals, Plants, Fungi and Bacteria. Mol. Inform. 2020, 39, 2000017. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Reker, D.; Schneider, P.; Schneider, G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Ian, P.; Anderson, E.A. The Renaissance of Natural Products as Drug Candidates. Science 2005, 310, 451. [Google Scholar]
- Gordon, C.; David, M.; Newman, J. Natural Products: A Continuing Source of Novel Drug Leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar]
- Byrne, R.; Gisbert, S. In Silico Target Prediction for Small Molecules. In Systems Chemical Biology: Methods and Protocols; Ziegler, S., Waldmann, H., Eds.; Springer: New York, NY, USA, 2019; pp. 273–309. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Liu, X.; Fu, Z.; Xiong, Z.; Wu, X.; Tan, X.; Zhao, J.; Zhong, F.; Wan, X.; et al. KinomeX: A web application for predicting kinome-wide polypharmacology effect of small molecules. Bioinformation 2019, 35, 5354–5356. [Google Scholar] [CrossRef]
- Reker, D.; Rodrigues, T.; Schneider, P.; Schneider, G. Identifying the Macromolecular Targets of De Novo-Designed Chemical Entities through Self-Organizing Map Consensus. Proc. Natl. Acad. Sci. USA 2014, 111, 4067–4072. [Google Scholar] [CrossRef]
- Gawehn, E.; Hiss, J.A.; Brown, J.B.; Schneider, G. Advancing drug discovery via GPU-based deep learning. Expert Opin. Drug Discov. 2018, 13, 579–582. [Google Scholar] [CrossRef]
- Mendez, D.; Anna, G.; Bento, P.A.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. Chembl: Towards Direct Deposition of Bioassay Data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Steijaert, M.; Wegner, J.K.; Ceulemans, H.; Clevert, D.A.; Hochreiter, S. Large-Scale Comparison of Machine Learning Methods for Drug Target Prediction on Chembl. Chem. Sci. 2018, 9, 5441–5451. [Google Scholar] [CrossRef]
- Matus, T. Benefits of Depth in Neural Networks. Proc. Mach. Learn. Res. 2016, 49, 1517–1539. [Google Scholar]
- Ntie-Kang, F.; Zofou, D.; Babiaka, S.B.; Meudom, R.; Scharfe, M.; Lifongo, L.L.; Mbah, J.A.; Mbaze, L.M.; Sippl, W.; Efange, S.M.N. AfroDb: A Select Highly Potent and Diverse Natural Product Library from African Medicinal Plants. PLoS ONE 2013, 8, e78085. [Google Scholar] [CrossRef]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef]
- Pilon, A.C.; Marilia, V.; Dametto, A.C.; Pinto, M.E.F.; Freire, R.T.; Castro-Gamboa, I.; Adriano, D.A.; Bolzani, V.S. Nubbedb: An Updated Database to Uncover Chemical and Biological Information from Brazilian Biodiversity. Sci. Rep. 2017, 7, 7215. [Google Scholar] [CrossRef]
- Cai, C.; Wang, S.; Xu, Y.; Zhang, W.; Tang, K.; Ouyang, Q.; Lai, L.; Pei, J. Transfer Learning for Drug Discovery. J. Med. Chem. 2020, 63, 8683–8694. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Lu, J.; Wang, C.; Zhang, Y. Predicting Molecular Energy Using Force-Field Optimized Geometries and Atomic Vector Representations Learned from an Improved Deep Tensor Neural Network. J. Chem. Theory Comput. 2019, 15, 4113–4121. [Google Scholar] [CrossRef]
- Girschick, T.; Rückert, U.; Kramer, S. Adapted Transfer of Distance Measures for Quantitative Structure-Activity Relationships and Data-Driven Selection of Source Datasets. Comput. J. 2012, 56, 274–288. [Google Scholar] [CrossRef]
- Li, X.; Fourches, D. Inductive Transfer Learning for Molecular Activity Prediction: Next-Gen Qsar Models with Molpmofit. J. Cheminform. 2020, 12, 1–15. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Wang, X.; Zhang, C.; Ge, J.; Tang, J.; Su, A.; Duan, H. Data augmentation and transfer learning strategies for reaction prediction in low chemical data regimes. Org. Chem. Front. 2021. [Google Scholar] [CrossRef]
- Zheng, S.; Yan, X.; Gu, Q.; Yang, Y.; Du, Y.; Lu, Y.; Xu, J. QBMG: Quasi-biogenic molecule generator with deep recurrent neural network. J. Chemin. 2019, 11, 5. [Google Scholar] [CrossRef]
- Lai, J.; Hu, J.; Wang, Y.; Zhou, X.; Li, Y.; Zhang, L.; Liu, Z. Privileged Scaffold Analysis of Natural Products with Deep Learning-Based Indication Prediction Model. Mol. Inf. 2020, 39, 2000057. [Google Scholar] [CrossRef]
- Keum, J.; Yoo, S.; Lee, D.; Nam, H. Prediction of compound-target interactions of natural products using large-scale drug and protein information. BMC Bioinform. 2016, 17, 417–425. [Google Scholar] [CrossRef]
- Cockroft, N.T.; Cheng, X.; Fuchs, J.R. STarFish: A Stacked Ensemble Target Fishing Approach and its Application to Natural Products. J. Chem. Inf. Model. 2019, 59, 4906–4920. [Google Scholar] [CrossRef]
- Chen, Y.; Mathai, N.; Kirchmair, J. Scope of 3D Shape-Based Approaches in Predicting the Macromolecular Targets of Structurally Complex Small Molecules Including Natural Products and Macrocyclic Ligands. J. Chem. Inf. Model. 2020, 60, 2858–2875. [Google Scholar] [CrossRef]
- Mathai, N.; Chen, Y.; Kirchmair, J. Validation strategies for target prediction methods. Briefings Bioinform. 2019, 21, 791–802. [Google Scholar] [CrossRef]
- Li, Y.; Wang, N.; Shi, J.; Hou, X.; Liu, J. Adaptive Batch Normalization for practical domain adaptation. Pattern Recognit. 2018, 80, 109–117. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. Coconut online: Collection of Open Natural Products database. J. Chem. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Landrum, G. Rdkit: Open-Source Cheminformatics. Available online: http://www.rdkit.org (accessed on 1 March 2021).
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Korkmaz, S. Deep Learning-Based Imbalanced Data Classification for Drug Discovery. J. Chem. Inf. Model. 2020, 60, 4180–4190. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; Wu, X.; Xiong, Z.; Yang, T.; Fu, Z.; Liu, X.; Tan, X.; Zhong, F.; Wan, X.; et al. Deep Learning Enhancing Kinome-Wide Polypharmacology Profiling: Model Construction and Experiment Validation. J. Med. Chem. 2020, 63, 8723–8737. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Assempour, N. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Eid, S.; Turk, S.; Volkamer, A.; Rippmann, F.; Fulle, S. KinMap: A web-based tool for interactive navigation through human kinome data. BMC Bioinform. 2017, 18, 16. [Google Scholar] [CrossRef]
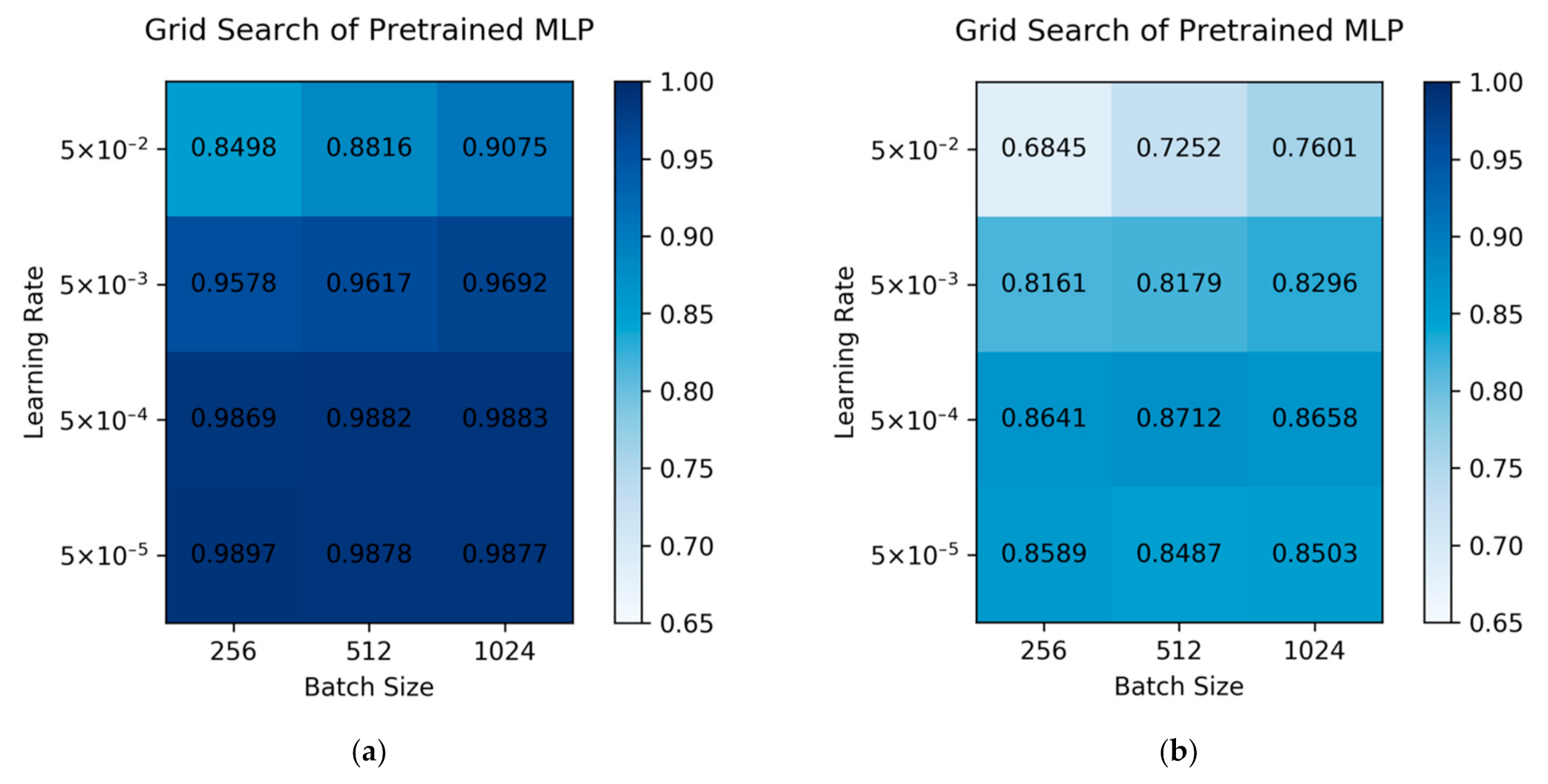

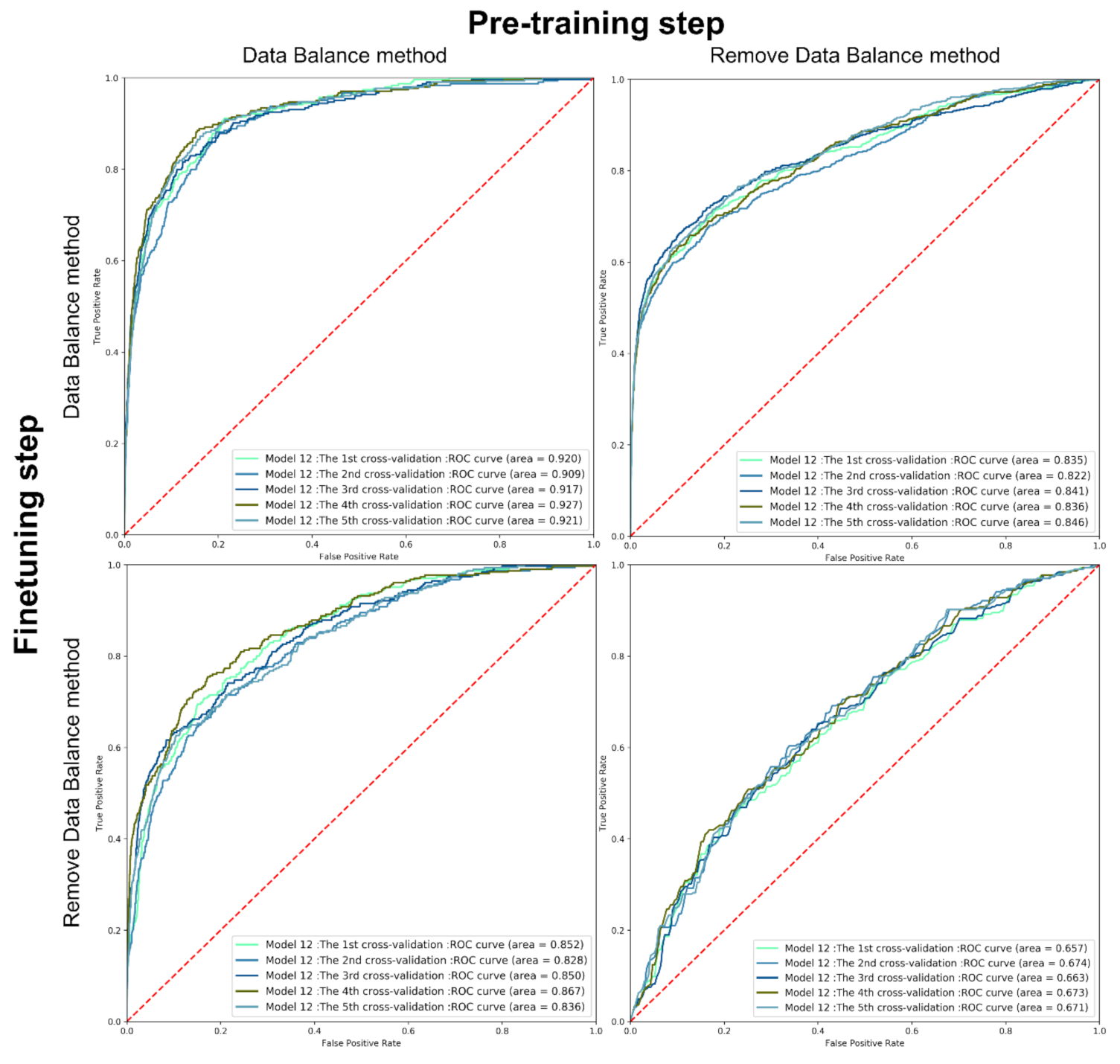


| Grid Search Space | |
|---|---|
| Model 1 | Learning Rate: 5 × 10−2 Batch size: 256 |
| Model 2 | Learning Rate: 5 × 10−2 Batch size: 512 |
| Model 3 | Learning Rate: 5 × 10−2 Batch size: 1024 |
| Model 4 | Learning Rate: 5 × 10−3 Batch size: 256 |
| Model 5 | Learning Rate: 5 × 10−3 Batch size: 512 |
| Model 6 | Learning Rate: 5 × 10−3 Batch size: 1024 |
| Model 7 | Learning Rate: 5 × 10−4 Batch size: 256 |
| Model 8 | Learning Rate: 5 × 10−4 Batch size: 512 |
| Model 9 | Learning Rate: 5 × 10−4 Batch size: 1024 |
| Model 10 | Learning Rate: 5 × 10−5 Batch size: 256 |
| Model 11 | Learning Rate: 5 × 10−5 Batch size: 512 |
| Model 12 | Learning Rate: 5 × 10−5 Batch size: 1024 |
| AUROC | Sensitivity (SE) | Specificity (SP) | Precision (PR) | Accuracy (ACC) | Matthews Correlation Coefficient (MCC) | |
|---|---|---|---|---|---|---|
| Model: Pre-trained model Test: Natural Products (drug removed) | 0.8461 | 0.3842 | 0.9932 | 0.2884 | 0.9889 | 0.3274 |
| Model: Pre-trained model Test: Natural Products (drug set) | 0.8548 | 0.4167 | 0.9935 | 0.4158 | 0.9872 | 0.4098 |
| Model: Fine-tuned model Test: Natural Products (random split 10% set) | 0.8849 ± 0.00097 | 0.6394 ± 0.005909 | 0.9401 ± 0.000012 | 0.06617 ± 0.000113 | 0.9380 ± 0.000008 | 0.1908 ± 0.000319 |
| Model: Fine-tuned model Test: Natural Products (drug set) | 0.7646 | 0.5321 | 0.9159 | 0.0654 | 0.9117 | 0.1636 |
| Probability of Top15 | Probability of Top20 | AUROC | |
|---|---|---|---|
| Fine-tuned Model | 0.817 | 0.860 | 0.910 |
| STarFish | 0.621 | 0.653 | 0.899 |
| Structure of Fluphenazine | Predicted Experimental High-Frequency Targets | Recommended High-Frequency Targets |
|---|---|---|
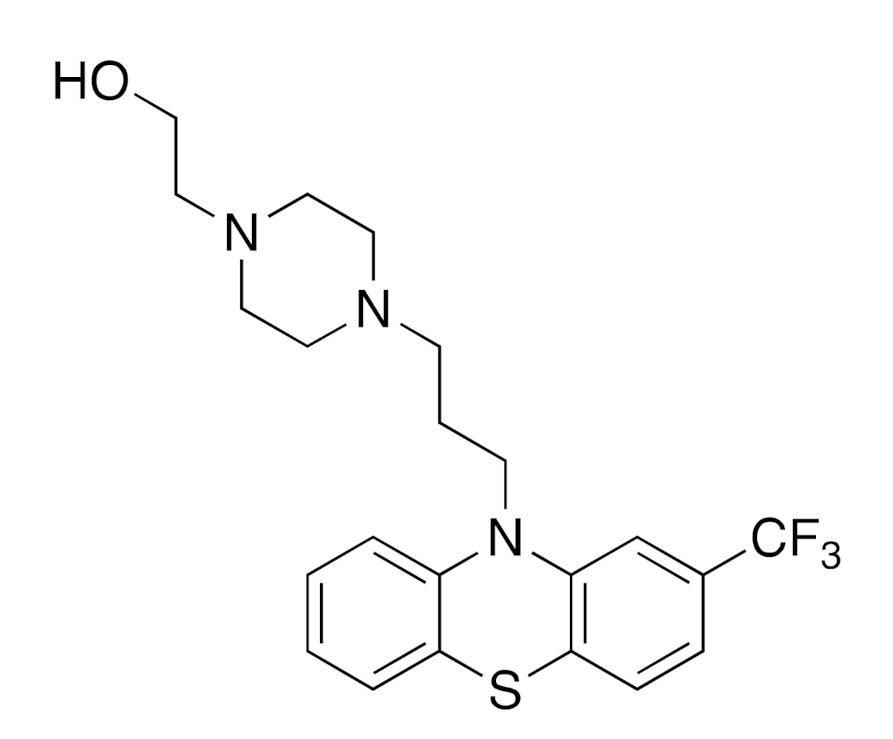 | ChEMBL 217, ChEMBL 234, ChEMBL 224, ChEMBL 3371, ChEMBL 225, ChEMBL 287, ChEMBL 1833, ChEMBL 223, ChEMBL 231, ChEMBL 2056, ChEMBL 1867, ChEMBL 319, ChEMBL 1916, ChEMBL 1942, ChEMBL 315 | ChEMBL 214, ChEMBL 273 *, ChEMBL 228 *, ChEMBL 339, ChEMBL 313, ChEMBL 222 *, ChEMBL 219, ChEMBL3155, ChEMBL 229, ChEMBL 322, ChEMBL 245 *, ChEMBL 216 *, ChEMBL 211 *, ChEMBL 3602, ChEMBL 265, ChEMBL 3943, ChEMBL 2035 *, ChEMBL 6007, ChEMBL 4081 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiang, B.; Lai, J.; Jin, H.; Zhang, L.; Liu, Z. Target Prediction Model for Natural Products Using Transfer Learning. Int. J. Mol. Sci. 2021, 22, 4632. https://doi.org/10.3390/ijms22094632
Qiang B, Lai J, Jin H, Zhang L, Liu Z. Target Prediction Model for Natural Products Using Transfer Learning. International Journal of Molecular Sciences. 2021; 22(9):4632. https://doi.org/10.3390/ijms22094632
Chicago/Turabian StyleQiang, Bo, Junyong Lai, Hongwei Jin, Liangren Zhang, and Zhenming Liu. 2021. "Target Prediction Model for Natural Products Using Transfer Learning" International Journal of Molecular Sciences 22, no. 9: 4632. https://doi.org/10.3390/ijms22094632
APA StyleQiang, B., Lai, J., Jin, H., Zhang, L., & Liu, Z. (2021). Target Prediction Model for Natural Products Using Transfer Learning. International Journal of Molecular Sciences, 22(9), 4632. https://doi.org/10.3390/ijms22094632








