Isolation and Characterization of Human Colon Adenocarcinoma Stem-Like Cells Based on the Endogenous Expression of the Stem Markers
Abstract
:1. Introduction
2. Results
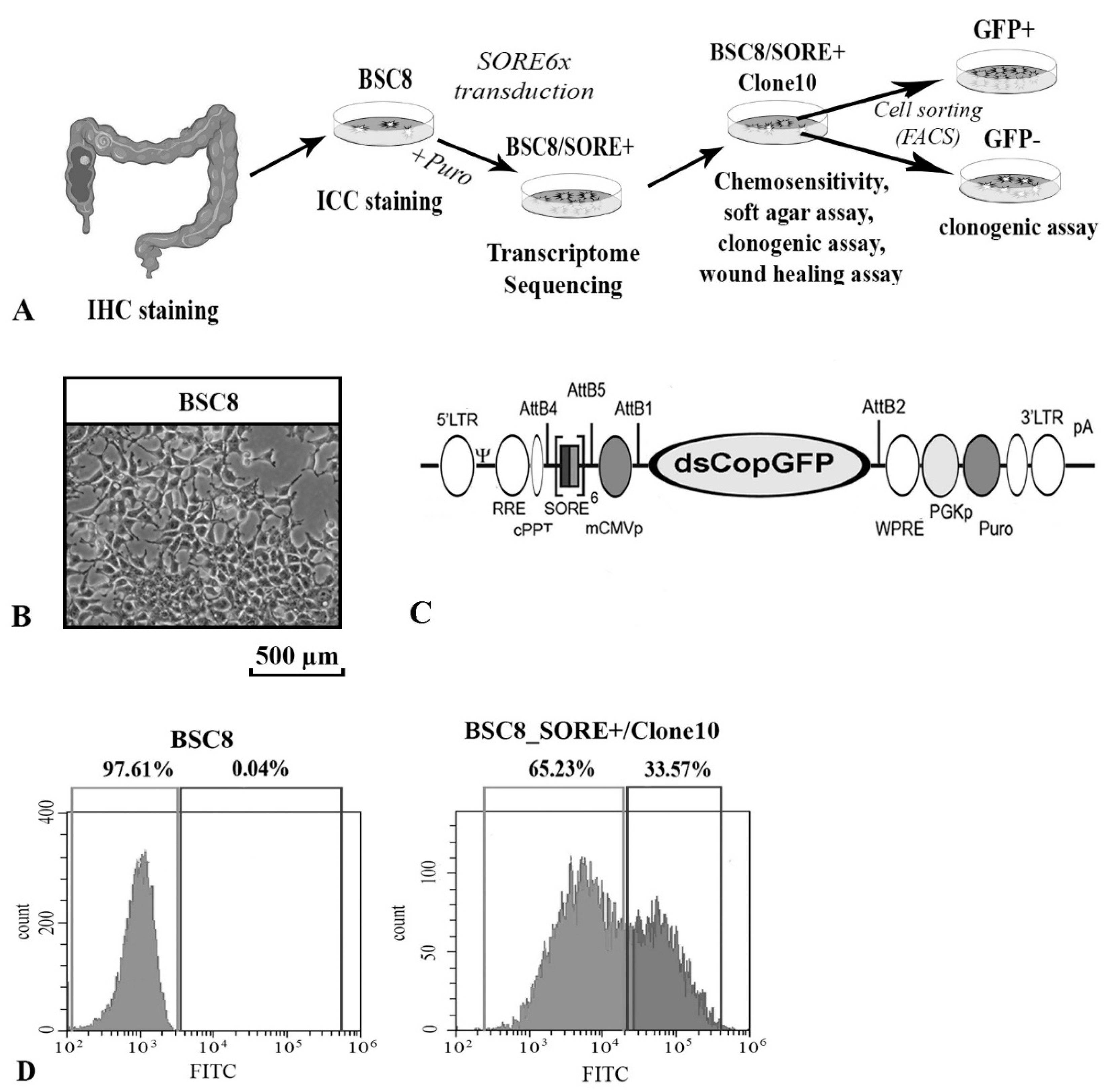
2.1. Enrichment of Cancer Stem-Like Cells from the Primary Culture of the Human Colon Cancer
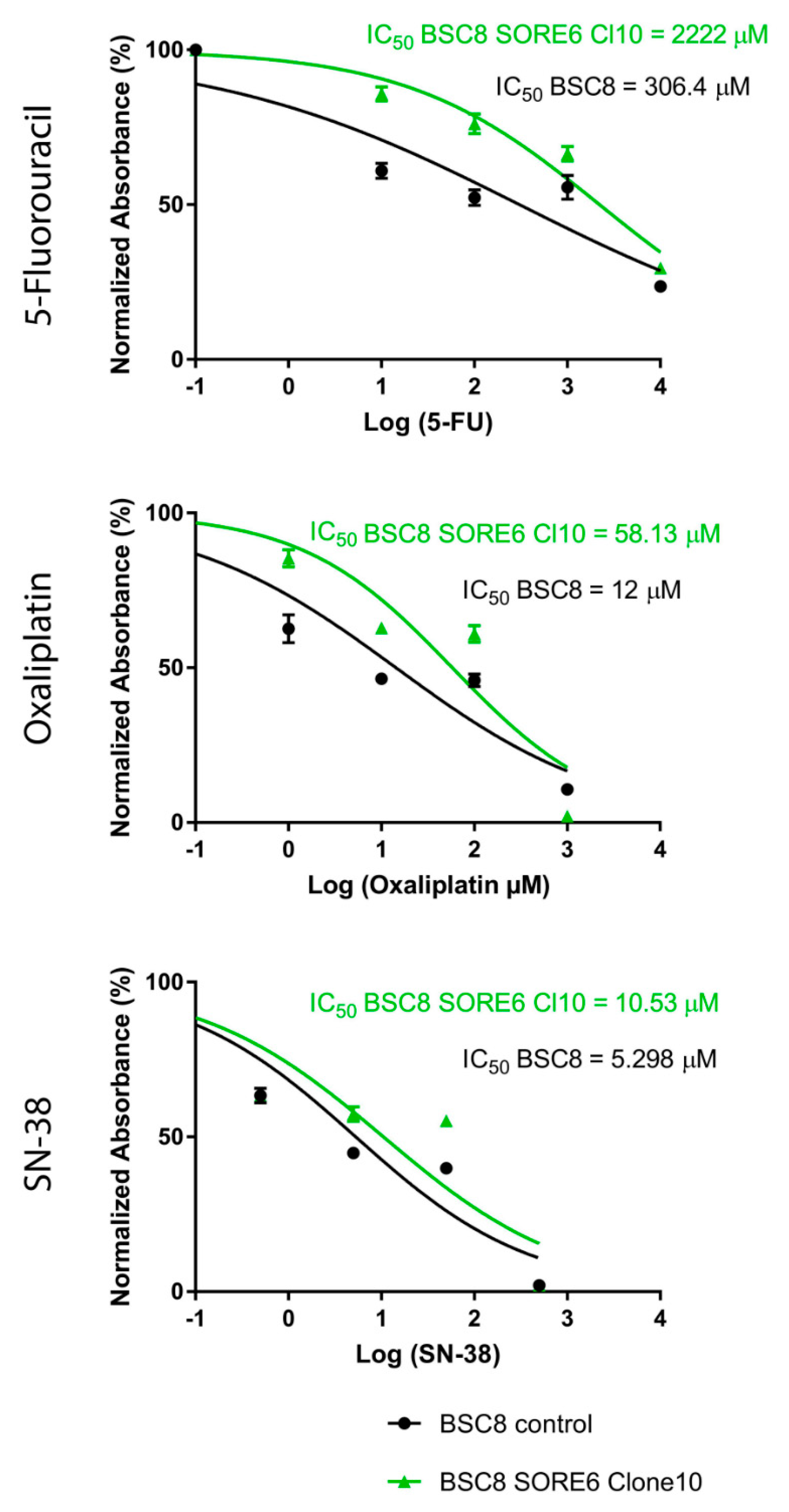
2.2. The Increased Cytostatic Resistance of Cell Clones Selected after Reporter Transduction
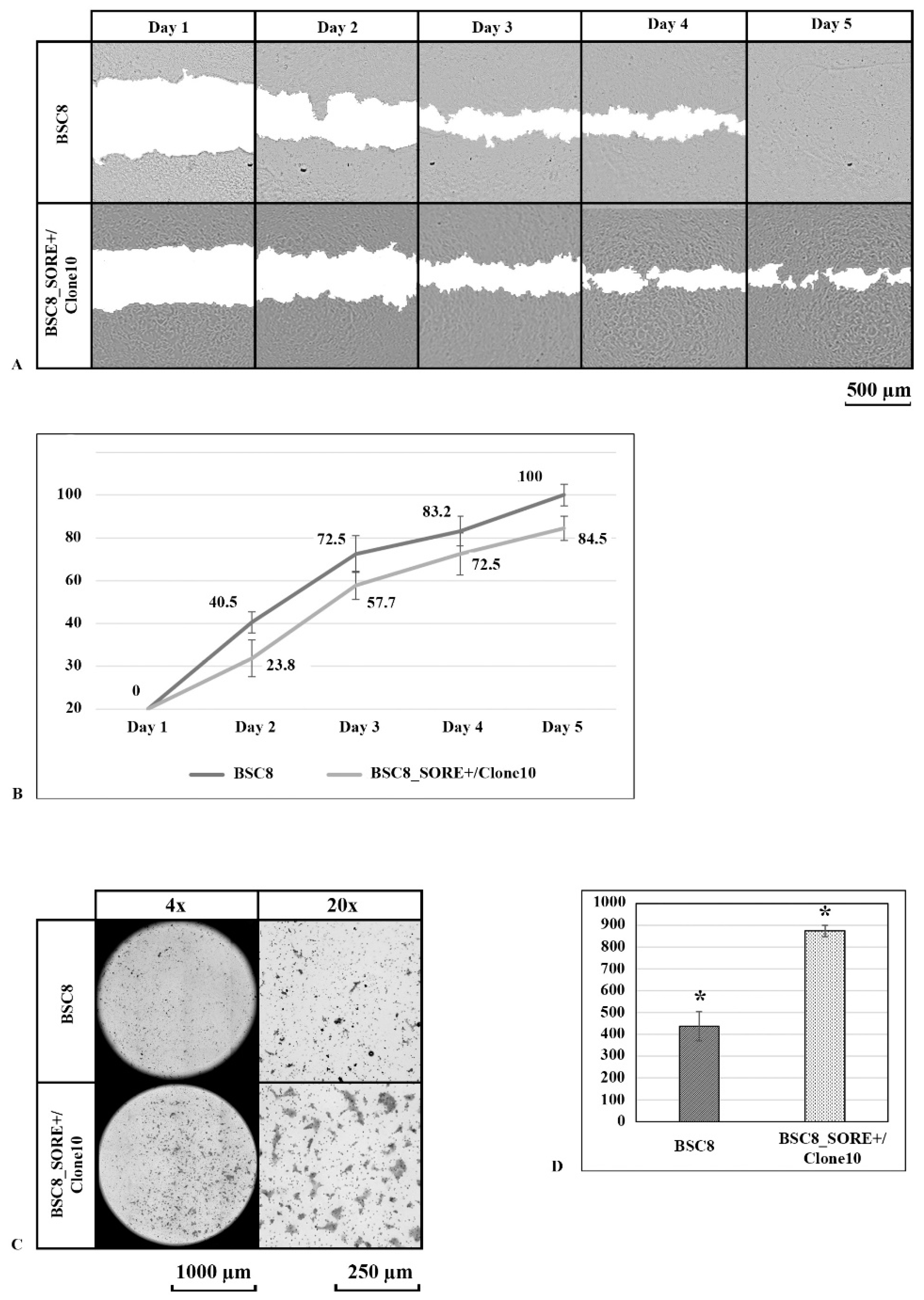
2.3. Clone Formation under Normoxia and Hypoxia and Cell Migratory Activity Analysis
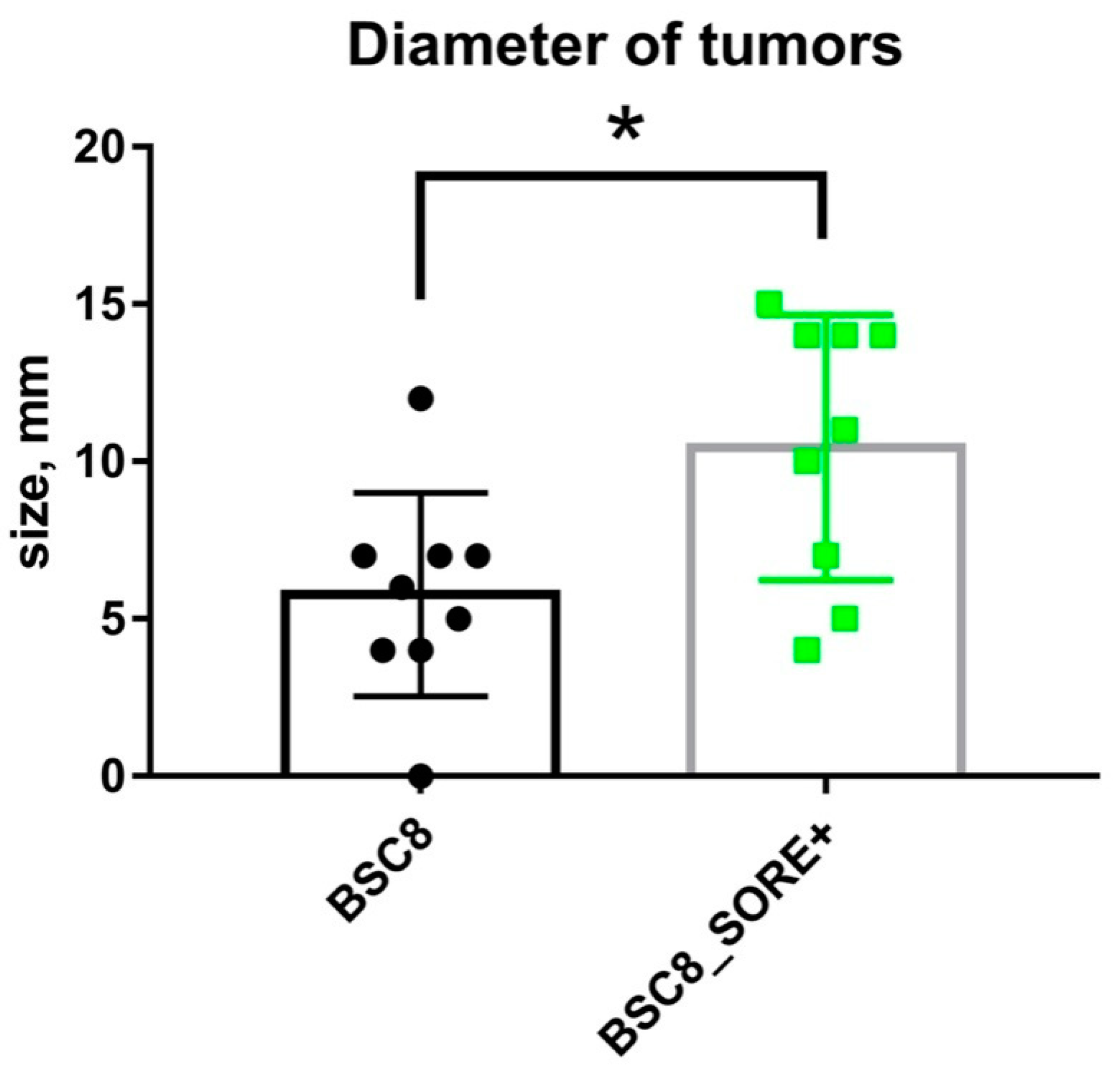
2.4. Tumorogenic Activity of 5-FU Resistant Cells Compared to the Original Cells in the Xenograft Assay
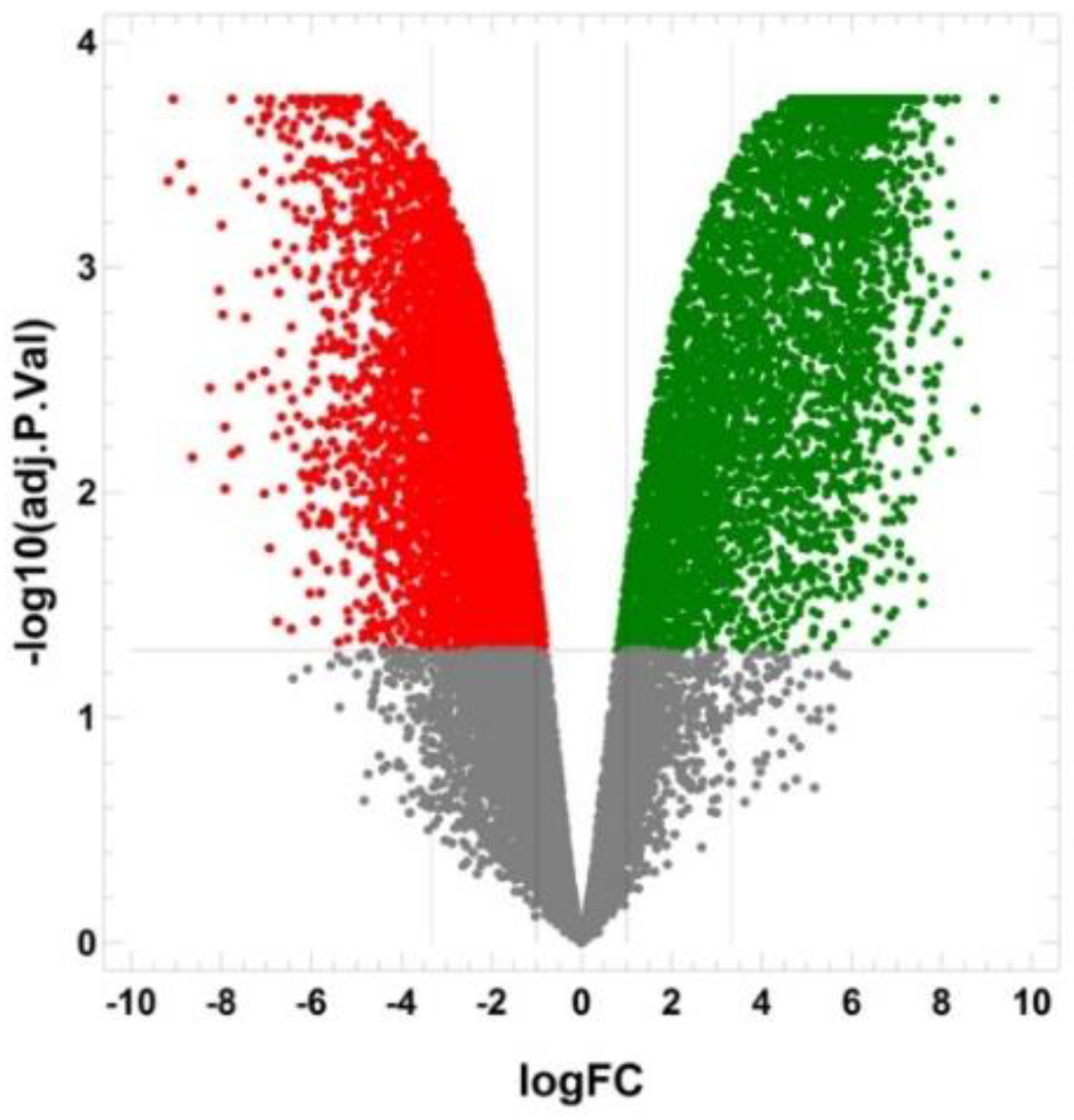
2.5. The Transcriptomic Landscape of the SORE+ Cells
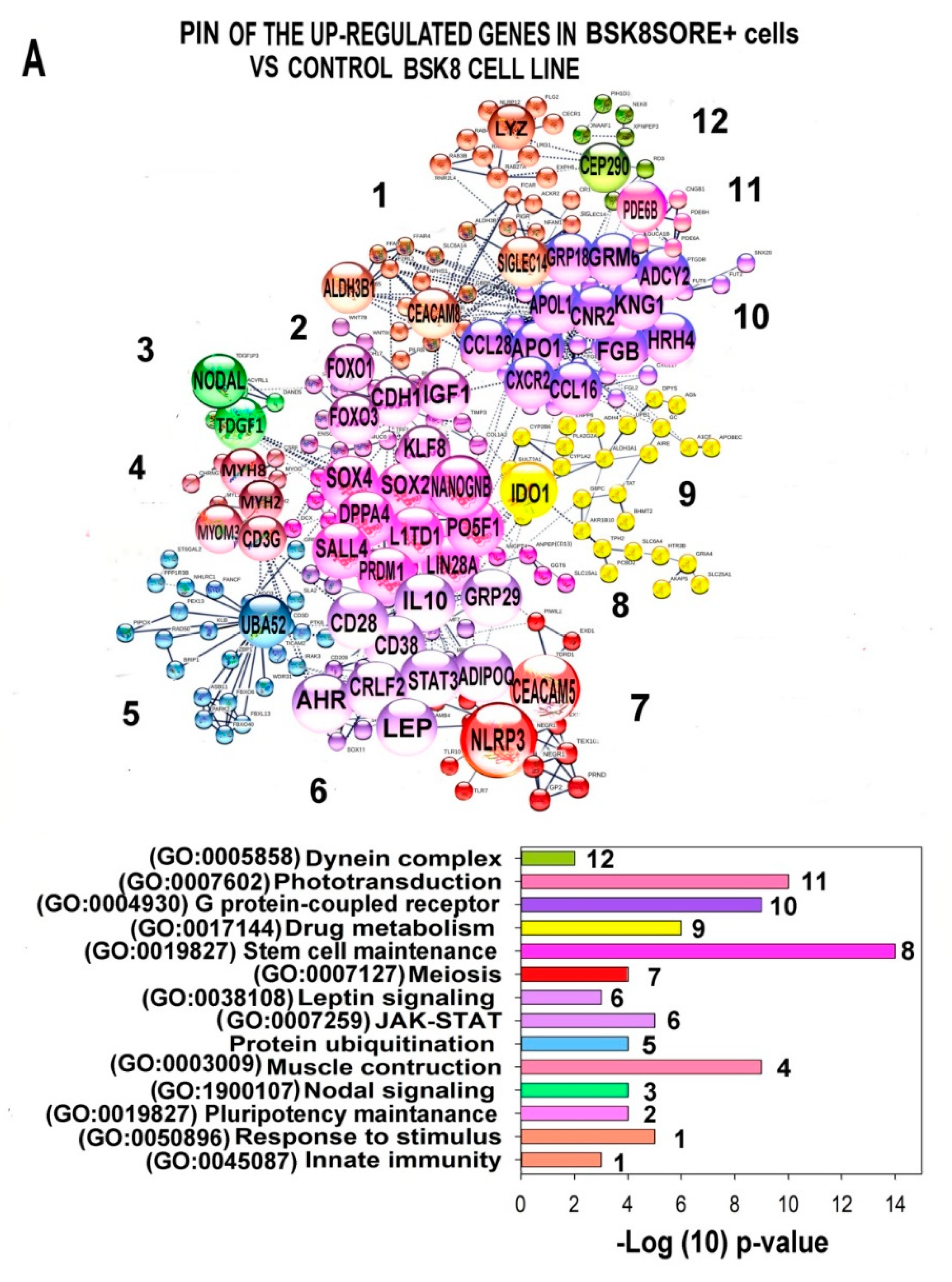
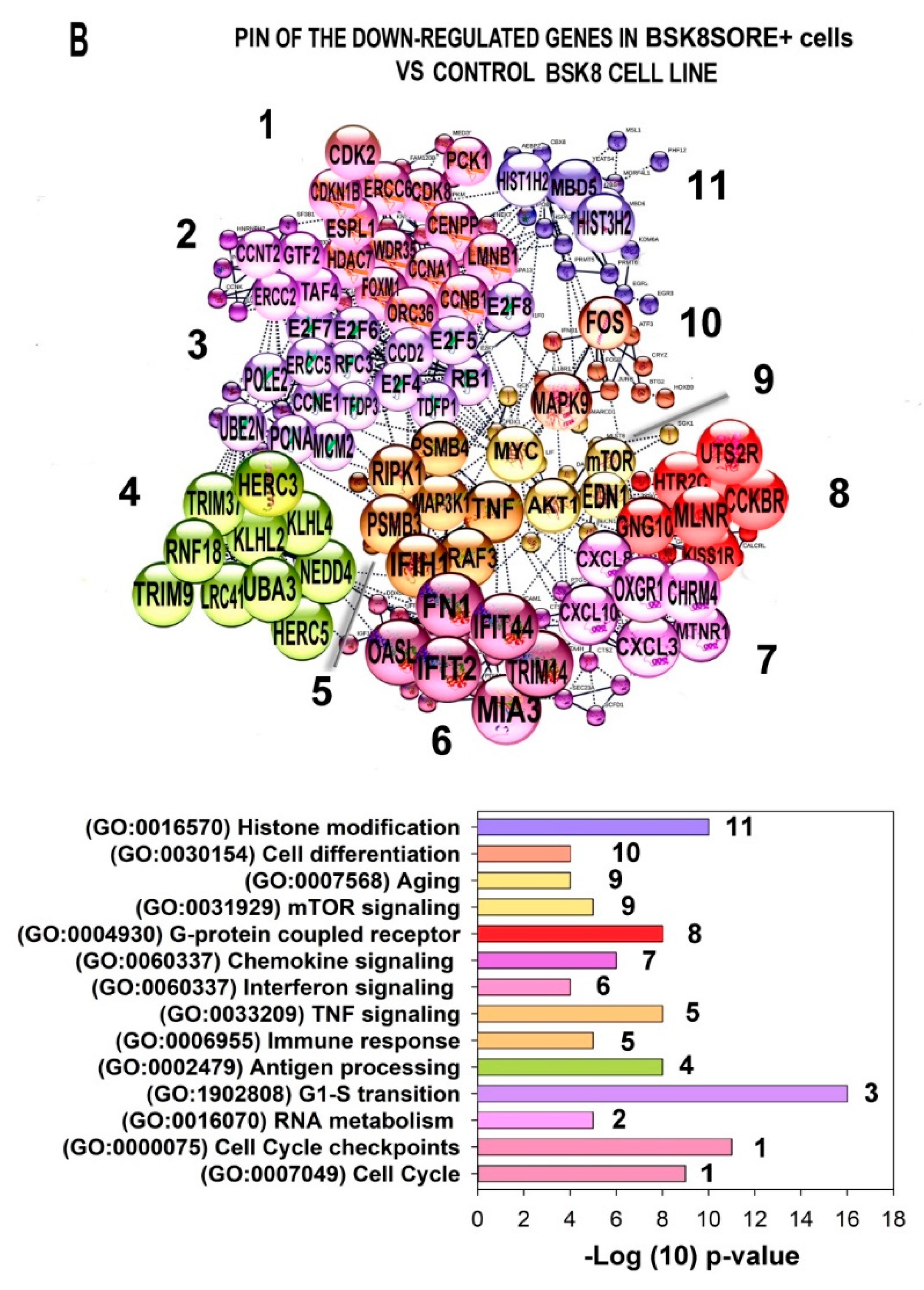
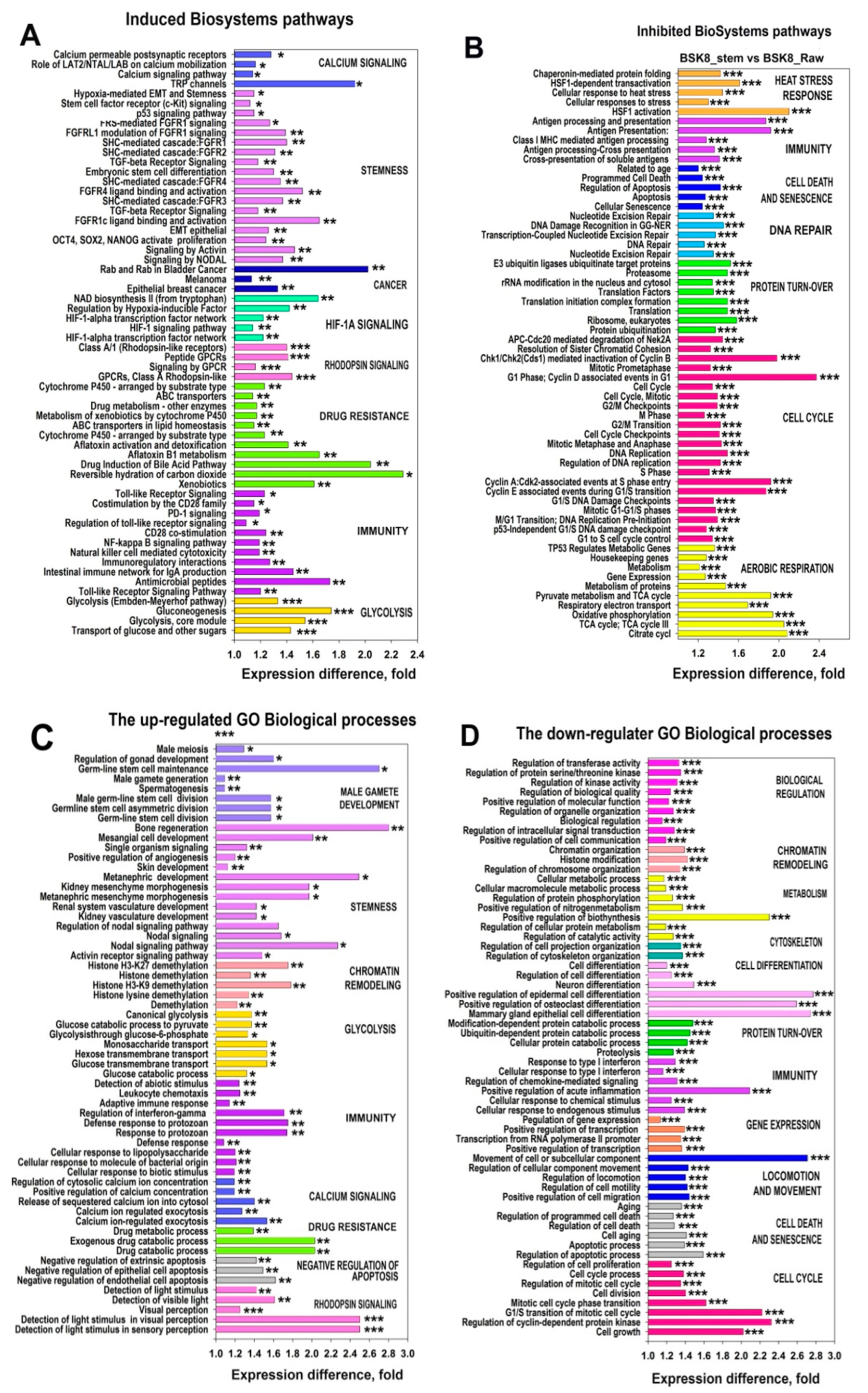
2.6. Characterization of the PINs and Enrichment of Gene Modules for Genes Differently Expressed in BSC8_SORE+ vs. BSC8
2.7. Gene Module Analysis
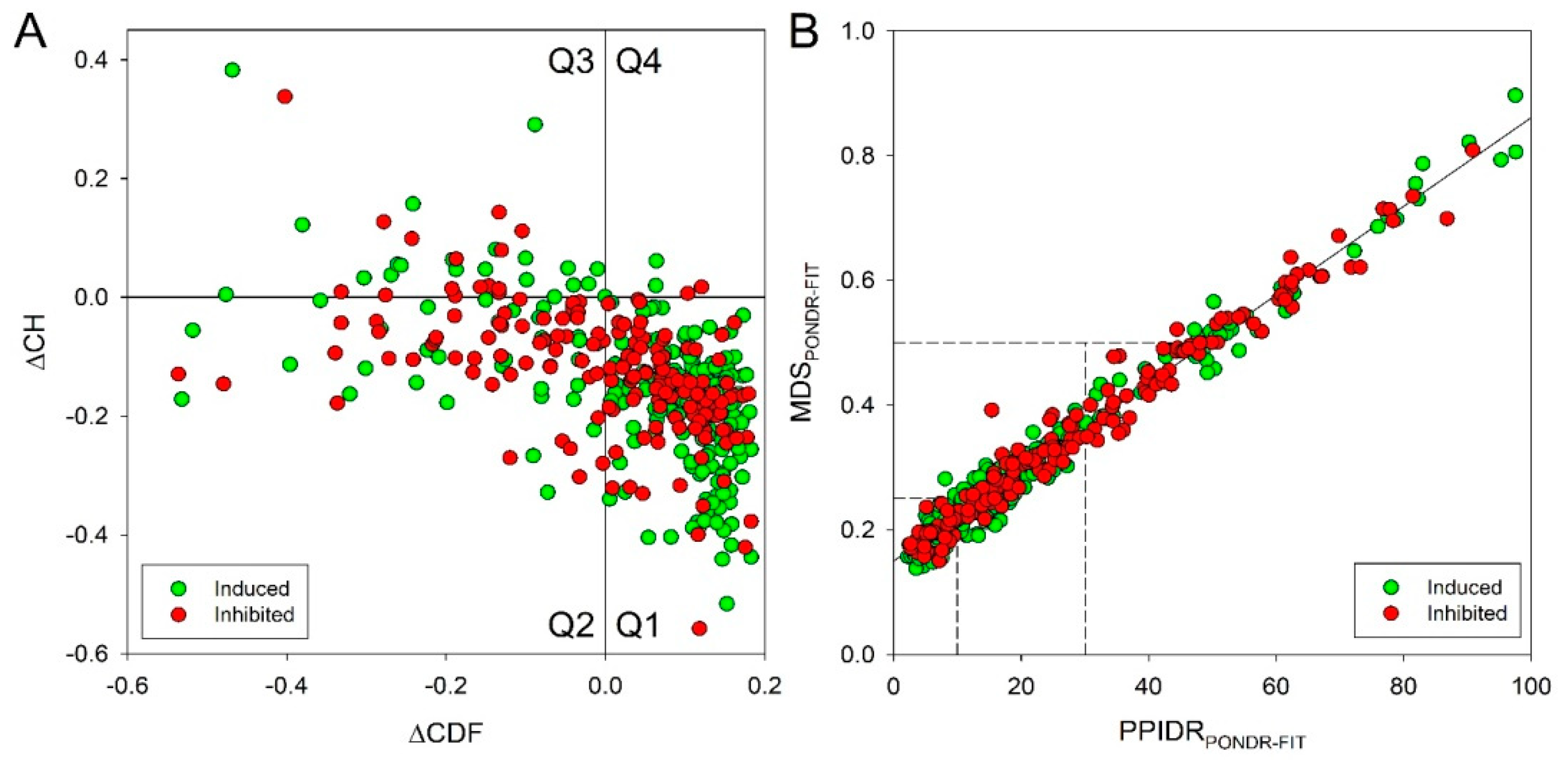
2.8. Intrinsic Disorder Predisposition of Proteins Encoded by Genes Differently Expressed in the BSC8_SORE+ Cells vs. BSC8 Control Cells
3. Discussion
4. Materials and Methods
4.1. Obtaining the Primary Cell Line
4.2. Packaging of VIral Particles, Infecting Cells in Vitro, and Selecting Clones with Integrated SORE6x Reporter
4.3. Immunofluorescence Staining of Formalin-Fixed Paraffin-Embedded Tissue Sections
4.4. Determining the Resistance to Cytostatics by the MTT Method
4.5. Cell Proliferation and Colony Formation Assaying
4.6. Soft Agar Cloning
4.7. Wound Repair Assay
4.8. Migration Assay
4.9. Timelapse Microscopy and Confocal Imaging
4.10. RNA Isolation for Transcriptome Sequencing
4.11. Transcriptome Sequencing and Analysis
4.12. Gene Module Analysis
4.13. Protein-Protein Interaction Network Analysis
4.14. Intrinsic Disorder Predisposition Analysis
4.15. Statistical Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacharakis, M.; Xynos, I.D.; Lazaris, A.; Smaro, T.; Kosmas, C.; Dokou, A.; Felekouras, E.; Antoniou, E.; Polyzos, A.; Sarantonis, J.; et al. Predictors of survival in stage IV metastatic colorectal cancer. Anticancer Res. 2010, 30, 653–660. [Google Scholar] [PubMed]
- Coppedè, F.; Lopomo, A.; Spisni, R.; Migliore, L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J. Gastroenterol. 2014, 20, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.A.; Song, S.; Hochster, H.S.; Steinberg, I.B. Cancer stem cells: The promise and the potential. Semin. Oncol. 2015, 42, S3–S17. [Google Scholar] [CrossRef]
- Wong, J.F. Probing the biology of cancer stem cells. Genet. Eng. Biotechnol. News. 2007, 27, 45–47. [Google Scholar]
- Dalerba, P.; Cho, R.W.; Clarke, M.F. Cancer Stem Cells: Models and Concepts. Annu. Rev. Med. 2007, 58, 267–284. [Google Scholar] [CrossRef] [Green Version]
- Tang, D. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.M.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells—Perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef] [Green Version]
- Baumann, M.; Krause, M.; Hill, R. Clonogens and cancer stem cells. Nat. Rev. Cancer 2008, 8, 990. [Google Scholar] [CrossRef]
- Jen, J.; Tang, Y.A.; Lu, Y.H.; Lin, C.C.; Lai, W.W.; Wang, Y.C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef] [Green Version]
- Shan, J.; Shen, J.; Liu, L.; Xia, F.; Xu, C.; Duan, G.; Xu, Y.; Ma, Q.; Yang, Z.; Zhang, Q.; et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology 2012, 56, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; de Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef]
- Munz, M.; Baeuerle, P.A.; Gires, O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009, 69, 5627–5629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, D.; Lee, H.W.; Hur, K.Y.; Kim, J.J.; Park, G.S.; Jang, S.H.; Song, Y.S.; Jang, K.S.; Paik, S.S. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J. Gastroenterol. 2009, 15, 2258–2264. [Google Scholar]
- Tachezy, M.; Zander, H.; Gebauer, F.; Marx, A.; Kaifi, J.T.; Izbicki, J.R.; Bockhorn, M. Activated leukocyte cell adhesion molecule (CD166)—Its prognostic power for colorectal cancer patients. J. Surg. Res. 2012, 177, e15–e20. [Google Scholar] [CrossRef] [PubMed]
- Corti, S.; Locatelli, F.; Papadimitriou, D.; Donadoni, C.; Salani, S.; del Bo, R.; Strazzer, S.; Bresolin, N.; Comi, G.P. Identification of a Primitive Brain-Derived Neural Stem Cell Population Based on Aldehyde Dehydrogenase Activity. Stem Cells 2006, 24, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Arcaroli, J.; Chen, Y.; Thompson, D.C.; Messersmith, W.; Jimeno, A.; Vasiliou, V. ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/β-catenin, Notch and PI3K/Akt signaling pathways. PLoS ONE 2015, 10, e0121648. [Google Scholar] [CrossRef]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Alman, B.A. Side population cells in human cancers. Cancer Lett. 2008, 268, 1–9. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Storms, R.W.; Trujillo, A.P.; Springer, J.B.; Shah, L.; Colvin, O.M.; Ludeman, S.M.; Smith, C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9118–9123. [Google Scholar] [CrossRef] [Green Version]
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; Clair, R.S.; Baljevic, M.; White, I.; Jin, D.K.; et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J. Clin. Investig. 2008, 118, 2111–2120. [Google Scholar] [CrossRef]
- Masters, J.R.; Foley, C.L.; Bisson, I.; Ahmed, A. Cancer stem cells. BJU Int. 2003, 92, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Saygin, C.; Samour, M.; Chumakova, A.; Jarrar, A.; Lathia, J.D.; Reizes, O. Reporter systems to study cancer stem cells. Methods Mol. Biol. 2016, 1516, 319–333. [Google Scholar] [PubMed]
- Tang, B.; Raviv, A.; Esposito, D.; Flanders, K.C.; Daniel, C.; Nghiem, B.T.; Garfield, S.; Lim, L.; Mannan, P.; Robles, A.I.; et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Rep. 2015, 4, 155–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiechert, A.; Saygin, C.; Thiagarajan, P.S.; Rao, V.S.; Hale, J.S.; Gupta, N.; Hitomi, M.; Nagaraj, A.B.; DiFeo, A.; Lathia, J.D.; et al. Cisplatin induces stemness in ovarian cancer. Oncotarget 2016, 7, 30511–30522. [Google Scholar] [CrossRef] [Green Version]
- Kurtova, A.V.; Xiao, J.; Mo, Q.; Pazhanisamy, S.K.; Krasnow, R.; Lerner, S.P.; Chen, F.; Roh, T.T.; Lay, E.; Ho, P.L.; et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 2015, 517, 209–213. [Google Scholar] [CrossRef]
- Cherepanova, O.A.; Gomez, D.; Shankman, L.S.; Swiatlowska, P.; Williams, J.; Sarmento, O.F.; Alencar, G.F.; Hess, D.L.; Bevard, M.H.; Greene, E.S.; et al. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat. Med. 2016, 22, 657–665. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.W.; Li, C.H.; Peng, Y.J.; Cheng, Y.W.; Chen, H.W.; Liao, P.L.; Kang, J.J.; Yeng, M.H. Snail regulates Nanog status during the epithelial-mesenchymal transition via the Smad1/Akt/GSK3β signaling pathway in non-small-cell lung cancer. Oncotarget 2014, 5, 3880–3894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.Y.; Cai, Y.; Mao, Y.; Lin, Y.; Zheng, H.; Wu, T.; Huang, Y.; Fang, X.; Lin, S.; Feng, Q.; et al. Twist2 promotes self-renewal of liver cancer stem-like cells by regulating CD24. Carcinogenesis 2014, 35, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Yang, R.; Gao, W.Q. Contributions of epithelial-mesenchymal transition and cancer stem cells to the development of castration resistance of prostate cancer. Mol. Cancer 2014, 13, 55. [Google Scholar] [CrossRef] [Green Version]
- Chaffer, C.L.; Marjanovic, N.D.; Lee, T.; Bell, G.; Kleer, C.G.; Reinhardt, F.; D’Alessio, A.C.; Young, R.A.; Weinberg, R.A. XPoised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013, 154, 61. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Cano, A. Tumorigenesis: Twist1 links EMT to self-renewal. Nat. Cell Biol. 2010, 12, 924–925. [Google Scholar] [CrossRef]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, S.; Chanda, P.; Thandavarayan, R.A.; Cooke, J.P. Transflammation: How Innate Immune Activation and Free Radicals Drive Nuclear Reprogramming. Antioxid. Redox Signal. 2018, 29, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Baek, K.H. Cellular functions of stem cell factors mediated by the ubiquitin—Proteasome system. Cell. Mol. Life Sci. 2018, 75, 1947–1957. [Google Scholar] [CrossRef] [PubMed]
- Boutet, A. The evolution of asymmetric photosensitive structures in metazoans and the Nodal connection. Mech. Dev. 2017, 147, 49–60. [Google Scholar] [CrossRef]
- Machida, K. Tumor-initiating stem-like cells and drug resistance: Carcinogenesis through Toll-like receptors, environmental factors, and virus. Drug Deliv. Transl. Res. 2013, 3, 152–164. [Google Scholar] [CrossRef]
- Harbuzariu, A.; Gonzalez-Perez, R.R. Leptin-Notch axis impairs 5-fluorouracil effects on pancreatic cancer. Oncotarget 2018, 9, 18239–18253. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.E.; Kirk, J.; Ji, Y.; Tang, D.G. Tumor Dormancy and Slow-Cycling Cancer Cells. In Advances in Experimental Medicine and Biology; Springer LLC: New York, NY, USA, 2019; Volume 1164, pp. 199–206. [Google Scholar]
- Eggenberger, J.; Blanco-Melo, D.; Panis, M.; Brennand, K.J.; Ten Oever, B.R. Type I interferon response impairs differentiation potential of pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2019, 116, 1384–1393. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.K.; Yoon, B.H.; Kang, S.M.; Yoon, Y.G.; Kim, S.Y.; Kim, W. Cdrgator: An integrative navigator of cancer drug resistance gene signatures. Mol. Cells 2019, 42, 237–244. [Google Scholar]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013, 19, 4191–4213. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Multitude of binding modes attainable by intrinsically disordered proteins: A portrait gallery of disorder-based complexes. Chem. Soc. Rev. 2011, 40, 1623–1634. [Google Scholar] [CrossRef]
- Uversky, V.N. The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett. 2015, 589, 2498–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mammen, M.; Choi, S.K.; Whitesides, G.M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998, 37, 2755–2794. [Google Scholar] [CrossRef]
- Schulz, G.E. Nucleotide Binding Proteins. In Molecular Mechanism of Biological Recognition; Balaban, M., Ed.; Elsevier/North-Holland Biomedical Press: New York, NY, USA, 1979; pp. 79–94. [Google Scholar]
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyson, H.J.; Wright, P.E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002, 12, 54–60. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008, 9, S1–S20. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins and their “mysterious” (meta)physics. Front. Phys. 2019, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [PubMed]
- Patil, A.; Nakamura, H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006, 580, 2041–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekman, D.; Light, S.; Bjorklund, A.K.; Elofsson, A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006, 7, R45. [Google Scholar] [CrossRef] [Green Version]
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006, 2, e100. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Chen, J.; Dunker, A.K.; Simon, I.; Tompa, P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 2006, 11, 2985–2995. [Google Scholar] [CrossRef]
- Dash, D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins 2007, 68, 602–605. [Google Scholar]
- Singh, G.P.; Ganapathi, M.; Dash, D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins 2007, 66, 761–765. [Google Scholar] [CrossRef]
- Xue, B.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. CDF it all: Consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009, 583, 1469–1474. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Oldfield, C.J.; Xue, B.; Hsu, W.L.; Meng, J.; Liu, X.; Shen, L.; Romero, P.; Uversky, V.N.; Dunker, A. Improving protein order-disorder classification using charge-hydropathy plots. BMC Bioinform. 2014, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. BioSyst. 2008, 4, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pas. Symp. Biocomputing 2012, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Santoro, A.; Vlachou, T.; Carminati, M.; Pelicci, P.G.; Mapelli, M. Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep. 2016, 17, 1700–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, G.; Harrington, K.I.; Lovegrove, H.E.; Page, D.J.; Chakravartula, S.; Bentley, K.; Herbert, S.P. Asymmetric division coordinates collective cell migration in angiogenesis. Nat. Cell Biol. 2016, 18, 1292–1301. [Google Scholar] [CrossRef] [Green Version]
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; Di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73. [Google Scholar]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Aiken, C.; Werbowetski-Ogilvie, T. Animal Models of Cancer Stem Cells: What are They Really Telling Us? Curr. Pathobiol. Rep. 2013, 1, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Fujino, S.; Miyoshi, N. OCT4 gene expression in primary colorectal cancer promotes liver metastasis. Stem Cells Int. 2019, 2, 7896524. [Google Scholar] [CrossRef] [Green Version]
- Cai, W.; Wang, Z.; Wei, C.; Wu, M.; Zheng, W.; Zhang, H.; Liu, C.; Liu, L. Prognostic evaluation of NANOG and OCT4 expression for posttransplantation hepatocellular carcinoma recurrence. J. Cell. Biochem. 2019, 120, 8419–8429. [Google Scholar] [CrossRef]
- Basati, G.; Mohammadpour, H.; Emami Razavi, A. Association of High Expression Levels of SOX2, NANOG, and OCT4 in Gastric Cancer Tumor Tissues with Progression and Poor Prognosis. J. Gastrointest. Cancer 2020, 51, 41–47. [Google Scholar] [CrossRef]
- Mohiuddin, I.S.; Wei, S.J.; Kang, M.H. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165432. [Google Scholar] [CrossRef]
- Li, W.; Zhou, Y.; Zhang, X.; Yang, Y.; Dan, S.; Su, T.; She, S.; Dong, W.; Zhao, Q.; Jia, J.; et al. Dual inhibiting OCT4 and AKT potently suppresses the propagation of human cancer cells. Sci. Rep. 2017, 7, 46246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forghanifard, M.M.; Moghbeli, M.; Raeisossadati, R.; Tavassoli, A.; Mallak, A.J.; Boroumand-Noughabi, S.; Abbaszadegan, M.R. Role of SALL4 in the progression and metastasis of colorectal cancer. J. Biomed. Sci. 2013, 20, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolè, L.; Sanavia, T.; Veronese, N.; Cappellesso, R.; Luchini, C.; Dabrilli, P.; Fassina, A. Oncofetal gene SALL4 and prognosis in cancer: A systematic review with meta-analysis. Oncotarget 2017, 8, 22968–22979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatetsu, H.; Kong, N.R.; Chong, G.; Amabile, G.; Tenen, D.G.; Chai, L. SALL4, the missing link between stem cells, development and cancer. Gene 2016, 584, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.H.; Wei, W.; Xu, J.; Guo, Z.X.; Xiao, C.Z.; Zhang, Y.F.; Jian, P.E.; Wu, X.L.; Shi, M.; Guo, R.P. Elevated expression of Cripto-1 correlates with poor prognosis in hepatocellular carcinoma. Oncotarget 2015, 6, 35116–35128. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; Li, R.; Ma, J.; Lei, J.; Xu, Q.; Jiang, Z.; Nan, L.; Li, X.; Wang, Z.; Huo, X.; et al. Overexpression of Nodal induces a metastatic phenotype in pancreatic cancer cells via the Smad2/3 pathway. Oncotarget 2015, 6, 1490–1506. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Chen, W.; Wang, X.; Zhao, J.; Li, Q.; Fu, Z. Cripto-1 Promotes the Epithelial-Mesenchymal Transition in Esophageal Squamous Cell Carcinoma Cells. Evid. Based Complement. Altern. Med. 2015, 2015, 421285. [Google Scholar] [CrossRef]
- Klauzinska, M.; Castro, N.P.; Rangel, M.C.; Spike, B.T.; Gray, P.C.; Bertolette, D.; Cuttitta, F.; Salomon, D. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin. Cancer Biol. 2014, 29, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Lonardo, E.; Hermann, P.C.; Mueller, M.-T.; Huber, S.; Balic, A.; Miranda-Lorenzo, I.; Zagorac, S.; Alcala, S.; Rodriguez-Arabaolaza, I.; Ramirez, J.C.; et al. Nodal/activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell 2011, 9, 433–446. [Google Scholar] [CrossRef] [Green Version]
- Wilson, B.J.; Schatton, T.; Zhan, Q.; Gasser, M.; Jie, M.; Saab, K.R.; Schanche, R.; Waaga-Gasser, A.M.; Gold, J.S.; Huang, Q.; et al. ABCB5 identifies a therapy-refractory tumor cell population in colorectal cancer patients. Cancer Res. 2011, 71, 5307–5316. [Google Scholar] [CrossRef] [Green Version]
- Kugimiya, N.; Nishimoto, A.; Hosoyama, T.; Ueno, K.; Enoki, T.; Li, T.S.; Hamano, K. The c-MYC-ABCB5 axis plays a pivotal role in 5-fluorouracil resistance in human colon cancer cells. J. Cell. Mol. Med. 2015, 19, 1569–1581. [Google Scholar] [CrossRef] [Green Version]
- Grimm, M.; Krimmel, M.; Polligkeit, J.; Alexander, D.; Munz, A.; Kluba, S.; Keutel, C.; Hoffmann, J.; Reinert, S.; Hoefert, S. ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur. J. Cancer 2012, 48, 3186–3197. [Google Scholar] [CrossRef]
- Guo, Q.; Grimmig, T.; Gonzalez, G.; Giobbie-Hurder, A.; Berg, G.; Carr, N.; Wilson, B.J.; Banerjee, P.; Ma, J.; Gold, J.S.; et al. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J. Biol. Chem. 2018, 293, 11166–11178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferdinande, L.; Decaestecker, C.; Verset, L.; Mathieu, A.; Moles Lopez, X.; Negulescu, A.M.; van Maerken, T.; Salmon, I.; Cuvelier, C.A.; Demetter, P.; et al. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br. J. Cancer 2012, 106, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Uyttenhove, C.; Pilotte, L.; Théate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef]
- Moffett, J.R.; Namboodiri, M.A. Tryptophan and the immune response. Immunol. Cell Biol. 2003, 81, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Fallarino, F.; Grohmann, U.; You, S.; McGrath, B.C.; Cavener, D.R.; Vacca, C.; Orabona, C.; Bianchi, R.; Belladonna, M.L.; Volpi, C.; et al. The Combined Effects of Tryptophan Starvation and Tryptophan Catabolites Down-Regulate T Cell Receptor ζ-Chain and Induce a Regulatory Phenotype in Naive T Cells. J. Immunol. 2006, 176, 6752–6761. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, G.J.; Visvader, J.E. Insights into the cell of origin in breast cancer and breast cancer stem cells. Asia Pac. J. Clin. Oncol. 2010, 6, 89–97. [Google Scholar] [CrossRef]
- Li, S.; Kennedy, M.; Payne, S.; Kennedy, K.; Seewaldt, V.L.; Pizzo, S.V.; Bachelder, R.E. Model of tumor dormancy/recurrence after short-term chemotherapy. PLoS ONE 2014, 9, e98021. [Google Scholar] [CrossRef] [Green Version]
- Dembinski, J.L.; Krauss, S. A distinct slow-cycling cancer stem-like subpopulation of pancreatic adenocarcinoma cells is maintained in Vivo. Cancers 2010, 2, 2011–2025. [Google Scholar] [CrossRef]
- Atkins, R.J.; Stylli, S.S.; Kurganovs, N.; Mangiola, S.; Nowell, C.J.; Ware, T.M.; Corcoran, N.M.; Brown, D.V.; Kaye, A.H.; Morokoff, A.; et al. Cell quiescence correlates with enhanced glioblastoma cell invasion and cytotoxic resistance. Exp. Cell Res. 2019, 374, 353–364. [Google Scholar] [CrossRef]
- Ohlsson, L.; Hammarström, M.L.; Lindmark, G.; Hammarström, S.; Sitohy, B. Ectopic expression of the chemokine CXCL17 in colon cancer cells. Br. J. Cancer 2016, 114, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Sjöberg, E.; Meyrath, M.; Milde, L.; Herrera, M.; Lövrot, J.; Hägerstrand, D.; Frings, O.; Bartish, M.; Rolny, C.; Sonnhammer, E.; et al. A novel ACKR2-Dependent role of fibroblast-derived CXCL14 in epithelial-to-mesenchymal transition and metastasis of breast cancer. Clin. Cancer Res. 2019, 25, 3702–3717. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.L.; Liu, K.Y.; Lin, F.J.; Shi, H.M.; Ou, Z.L. CCL28 promotes breast cancer growth and metastasis through MAPK-mediated cellular anti-apoptosis and pro-metastasis. Oncol. Rep. 2017, 38, 1393–1401. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Rosen, D.; Liu, G.; Yang, F.; Guo, X.; Xiao, X.; Xue, F.; Mercado-Uribe, I.; Huang, J.; Lin, S.H.; et al. CXCR2 promotes ovarian cancer growth through dysregulated cell cycle, diminished apoptosis, and enhanced angiogenesis. Clin. Cancer Res. 2010, 16, 3875–3886. [Google Scholar] [CrossRef] [Green Version]
- Cheng, W.L.; Wang, C.S.; Huang, Y.H.; Tsai, M.M.; Liang, Y.; Lin, K.H. Overexpression of CXCL1 and its receptor CXCR2 promote tumor invasion in gastric cancer. Ann. Oncol. 2011, 22, 2267–2276. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, H.; Shen, Z.; Wang, X.; Zhang, H.; Qin, J.; Xu, J.; Sun, Y.; Qin, X. The prognostic value of CXC-chemokine receptor 2 (CXCR2) in gastric cancer patients. BMC Cancer 2015, 15, 766. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Ou, B.; Feng, H.; Wang, P.; Yin, S.; Zhu, C.; Wang, S.; Chen, C.; Zheng, M.; Zong, Y.; et al. Overexpression of CXCR2 predicts poor prognosis in patients with colorectal cancer. Oncotarget 2017, 8, 28442–28454. [Google Scholar] [CrossRef] [Green Version]
- Varney, M.L.; Singh, S.; Li, A.; Mayer-Ezell, R.; Bond, R.; Singh, R.K. Small molecule antagonists for CXCR2 and CXCR1 inhibit human colon cancer liver metastases. Cancer Lett. 2011, 300, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Augustin, H.G.; Young Koh, G.; Thurston, G.; Alitalo, K. Control of vascular morphogenesis and homeostasis through the angiopoietin—Tie system. Nat. Rev. Mol. Cell Biol. 2009, 10, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Gavalas, N.G.; Liontos, M.; Trachana, S.P.; Bagratuni, T.; Arapinis, C.; Liacos, C.; Dimopoulos, M.A.; Bamias, A. Angiogenesis-related pathways in the pathogenesis of ovarian cancer. Int. J. Mol. Sci. 2013, 14, 15885–15909. [Google Scholar] [CrossRef] [Green Version]
- Sfiligoi, C.; de Luca, A.; Cascone, I.; Sorbello, V.; Fuso, L.; Ponzone, R.; Biglia, N.; Audero, E.; Arisio, R.; Bussolino, F.; et al. Angiopoietin-2 expression in breast cancer correlates with lymph node invasion and short survival. Int. J. Cancer 2003, 103, 466–474. [Google Scholar] [CrossRef]
- Mitsuhashi, N.; Shimizu, H.; Ohtsuka, M.; Wakabayashi, Y.; Ito, H.; Kimura, F.; Yoshidome, H.; Kato, A.; Nukui, Y.; Miyazaki, M.; et al. Angiopoietins and Tie-2 expression in angiogenesis and proliferation of human hepatocellular carcinoma. Hepatology 2003, 37, 1105–1113. [Google Scholar] [CrossRef]
- Ramanathan, R.; Olex, A.L.; Dozmorov, M.; Bear, H.D.; Fernandez, L.J.; Takabe, K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res. Treat. 2017, 162, 191–198. [Google Scholar] [CrossRef]
- Huang, H.; Bhat, A.; Woodnutt, G.; Lappe, R. Targeting the ANGPT-TIE2 pathway in malignancy. Nat. Rev. Cancer 2010, 10, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Wan, S.; Zhou, F.; Myers, R.E.; Guo, X.; Li, B.; Fu, X.; Palazzo, J.P.; Dou, K.; Yang, H.; et al. Genetic polymorphism in a vegf-independent angiogenesis gene angpt1 and overall survival of colorectal cancer patients after surgical resection. PLoS ONE 2012, 7, e34758. [Google Scholar] [CrossRef] [Green Version]
- Cortese, M.S.; Uversky, V.N.; Dunker, A. Intrinsic disorder in scaffold proteins: Getting more from less. Prog. Biophys. Mol. Biol. 2008, 98, 85–106. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Obradovic, Z. The protein trinity-linking function and disorder. Nat. Biotechnol. 2001, 19, 805–806. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Uversky, V.N. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008, 4, 229–230. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic Disorder and Functional Proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010, 2010, 568068. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef]
- Vucetic, S.; Xie, H.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J. Proteome Res. 2007, 6, 1899–1916. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007, 6, 1917–1932. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N.; Obradovic, Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007, 6, 1882–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dave, V.; Iakoucheva, L.M.; Malaney, P.; Metallo, S.J.; Pathak, R.R.; Joerger, A.C. Pathological unfoldomics of uncontrolled chaos: Intrinsically disordered proteins and human diseases. Chem. Rev. 2014, 114, 6844–6879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Amyloidogenesis of natively unfolded proteins. Curr. Alzheimer Res. 2008, 5, 260–287. [Google Scholar] [CrossRef]
- Cheng, Y.; LeGall, T.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry 2006, 45, 10448–10460. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Uversky, V.N. A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes. Int. J. Mol. Sci. 2017, 18, 2010. [Google Scholar] [CrossRef]
- Uversky, V.N. The triple power of D3: Protein intrinsic disorder in degenerative diseases. Front. Biosci. 2014, 19, 181–258. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front. Biosci. 2009, 14, 5188–5238. [Google Scholar] [CrossRef]
- Uversky, V.N. Wrecked regulation of intrinsically disordered proteins in diseases: Pathogenicity of deregulated regulators. Front. Mol. Biosci. 2014, 1, 6. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genom. 2009, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Oldfield, C.J.; Van, Y.-Y.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and induced pluripotent stem cells. Mol. BioSyst. 2012, 8, 134–150. [Google Scholar] [CrossRef]
- Yu, C.S.; Huang, A.C.; Lai, K.C.; Huang, Y.P.; Lin, M.W.; Yang, J.S.; Chung, J.G. Diallyl trisulfide induces apoptosis in human primary colorectal cancer cells. Oncol. Rep. 2012, 28, 949–954. [Google Scholar] [CrossRef] [Green Version]
- Koshkin, S.; Danilova, A.; Raskin, G.; Petrov, N.; Bajenova, O.; O’Brien, S.J.; Tomilin, A.; Tolkunova, E. Primary cultures of human colon cancer as a model to study cancer stem cells. Tumour Biol. 2016, 37, 12833–12842. [Google Scholar] [CrossRef]
- Young, L.; Sung, J.; Masters, J.R. Detection of mycoplasma in cell cultures. Nat. Protoc. 2010, 5, 929–934. [Google Scholar] [CrossRef]
- Bajenova, O.; Chaika, N.; Tolkunova, E.; Davydov-Sinitsyn, A.; Gapon, S.; Thomas, P.; O’Brien, S. Carcinoembryonic antigen promotes colorectal cancer progression by targeting adherens junction complexes. Exp. Cell Res. 2014, 324, 115–123. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Shilina, M.A.; Anatskaya, O.V.; Alekseenko, L.L.; Fridlyanskaya, I.I.; Krasnenko, A.; Kim, A.; Korostin, D.; Ilynsky, V.; Elmuratov, A.; et al. Molecular genetic analysis of human endometrial mesenchymal stem cells that survived sublethal heat shock. Stem Cells Int. 2017, 2017, 2362630. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Lindner, R.; Friedel, C.C. A Comprehensive Evaluation of Alignment Algorithms in the Context of RNA-Seq. PLoS ONE 2012, 7, e52403. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Gene Golden Age paradox and its partial solution. Genomics 2019, 111, 115–126. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E.; Vainshelbaum, N.M.; Giuliani, A.; Erenpreisa, J. Phylostratic Shift of Whole-Genome Duplications in Normal Mammalian Tissues towards Unicellularity Is Driven by Developmental Bivalent Genes and Reveals a Link to Cancer. Int. J. Mol. Sci. 2020, 21, 8759. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Cell-cycle dependence of transcriptome gene modules: Comparison of regression lines. FEBS J. 2020, 287, 4427–4439. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [Green Version]
- Sayers, E.W.; Beck, J.; Brister, J.R.; Bolton, E.E.; Canese, K.; Comeau, D.C.; Funk, K.; Ketter, A.; Kim, S.; Kimchi, A.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 48, D9–D16. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein—Protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Vazquez-Martin, A.; Anatskaya, O.V.; Giuliani, A.; Erenpreisa, J.; Huang, S.; Salmina, K.; Inashkina, I.; Huna, A.; Nikolsky, N.N.; Vinogradov, A.E. Somatic polyploidy is associated with the upregulation of c-MYC interacting genes and EMT-like signature. Oncotarget 2016, 7, 75235–75260. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Salmina, K.; Anatskaya, O.; Cragg, M.S. Paradoxes of cancer: Survival at the brink. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar] [CrossRef]
- Shilina, M.A.; Grinchuk, T.M.; Anatskaya, O.V.; Vinogradov, A.E.; Alekseenko, L.L.; Elmuratov, A.U.; Nikolsky, N.N. Cytogenetic and Transcriptomic Analysis of Human Endometrial MSC Retaining Proliferative Activity after Sublethal Heat Shock. Cells 2018, 7, 184. [Google Scholar] [CrossRef] [Green Version]
- Vinogradov, A.E.; Anatskaya, O.V. Systemic evolutionary changes in mammalian gene expression. Biosystems 2020, 198, 104256. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Evolutionary framework of the human interactome: Unicellular and multicellular giant clusters. Biosystems 2019, 181, 82–87. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. DNA helix: The importance of being AT-rich. Mamm. Genome 2017, 28, 455–464. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Somatic polyploidy promotes cell function under stress and energy depletion: Evidence from tissue-specific mammal transcriptome. Funct. Integr. Genom. 2010, 10, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [Green Version]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [Green Version]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshkin, S.A.; Anatskaya, O.V.; Vinogradov, A.E.; Uversky, V.N.; Dayhoff, G.W., II; Bystriakova, M.A.; Pospelov, V.A.; Tolkunova, E.N. Isolation and Characterization of Human Colon Adenocarcinoma Stem-Like Cells Based on the Endogenous Expression of the Stem Markers. Int. J. Mol. Sci. 2021, 22, 4682. https://doi.org/10.3390/ijms22094682
Koshkin SA, Anatskaya OV, Vinogradov AE, Uversky VN, Dayhoff GW II, Bystriakova MA, Pospelov VA, Tolkunova EN. Isolation and Characterization of Human Colon Adenocarcinoma Stem-Like Cells Based on the Endogenous Expression of the Stem Markers. International Journal of Molecular Sciences. 2021; 22(9):4682. https://doi.org/10.3390/ijms22094682
Chicago/Turabian StyleKoshkin, Sergei A., Olga V. Anatskaya, Alexander E. Vinogradov, Vladimir N. Uversky, Guy W. Dayhoff, II, Margarita A. Bystriakova, Valery A. Pospelov, and Elena N. Tolkunova. 2021. "Isolation and Characterization of Human Colon Adenocarcinoma Stem-Like Cells Based on the Endogenous Expression of the Stem Markers" International Journal of Molecular Sciences 22, no. 9: 4682. https://doi.org/10.3390/ijms22094682
APA StyleKoshkin, S. A., Anatskaya, O. V., Vinogradov, A. E., Uversky, V. N., Dayhoff, G. W., II, Bystriakova, M. A., Pospelov, V. A., & Tolkunova, E. N. (2021). Isolation and Characterization of Human Colon Adenocarcinoma Stem-Like Cells Based on the Endogenous Expression of the Stem Markers. International Journal of Molecular Sciences, 22(9), 4682. https://doi.org/10.3390/ijms22094682






