Targeting Mitochondrial Metabolism in Clear Cell Carcinoma of the Ovaries
Abstract
1. Introduction
2. Results
2.1. ARID1A-Deficient Cells Have Increased Expression of OXPHOS Genes and Pathways
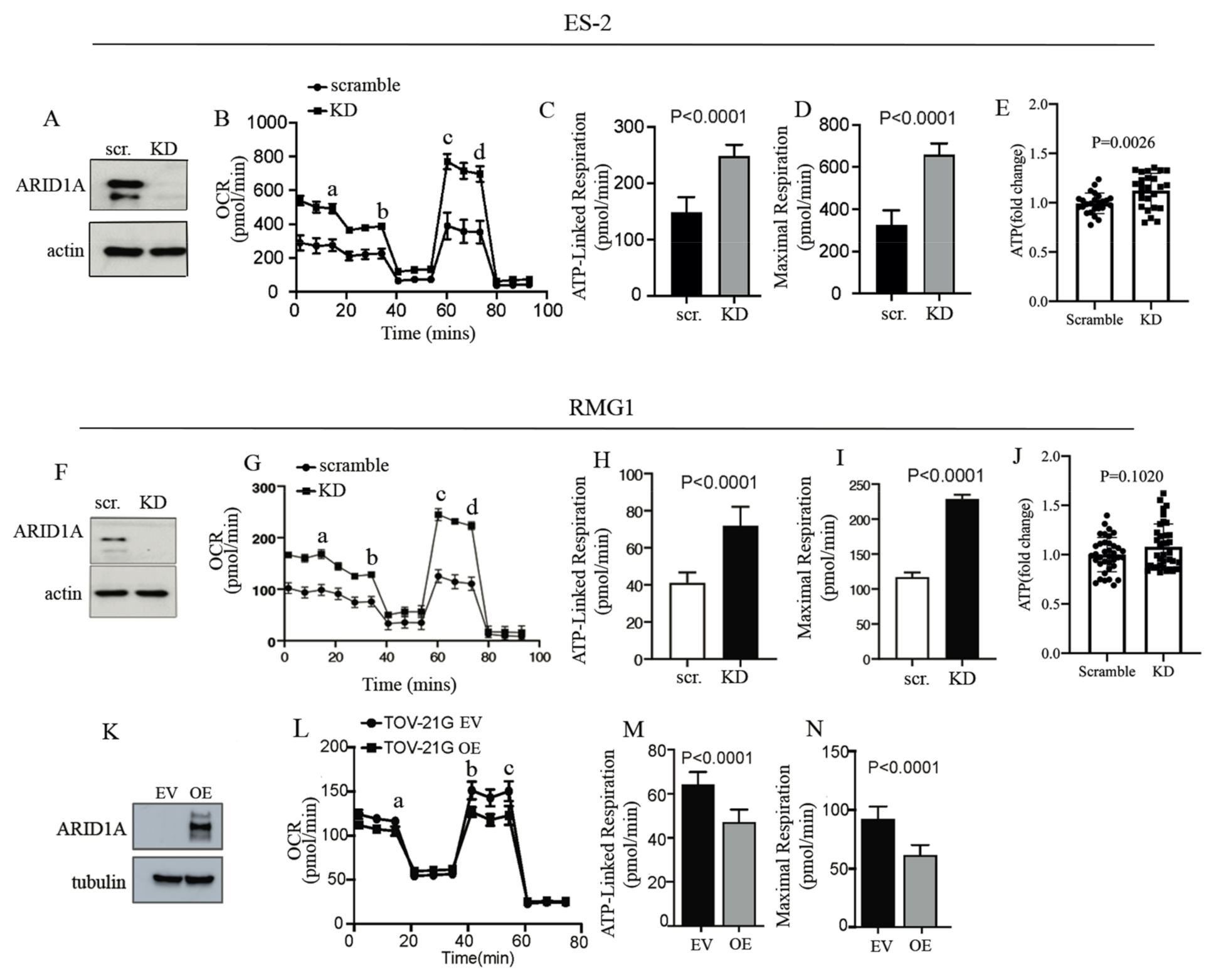
2.2. ARID1A Regulates Mitochondrial Respiration in OCCC Cells
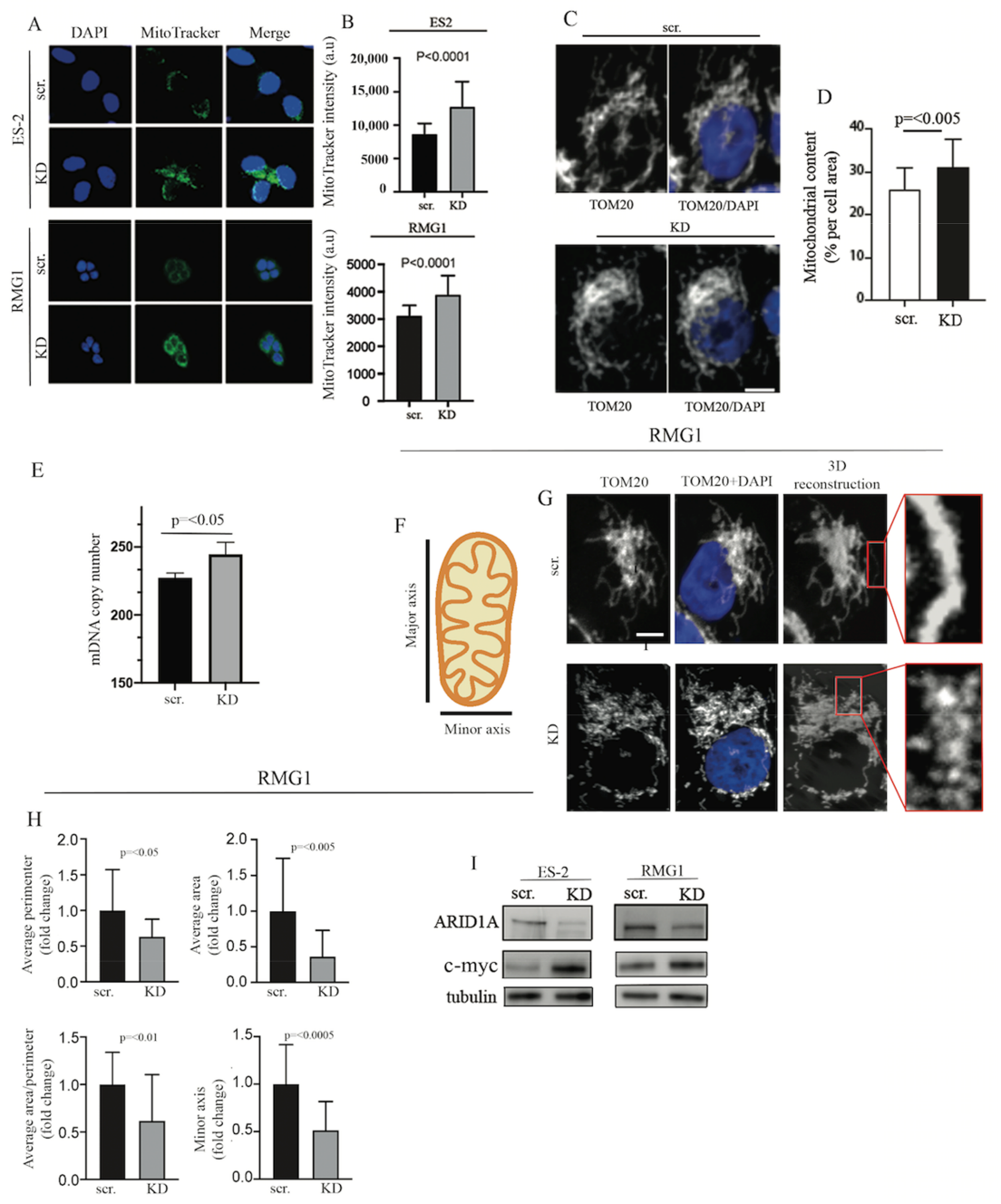
2.3. ARID1A Controls Mitochondrial Membrane Potential, Mitochondrial Number, and Size
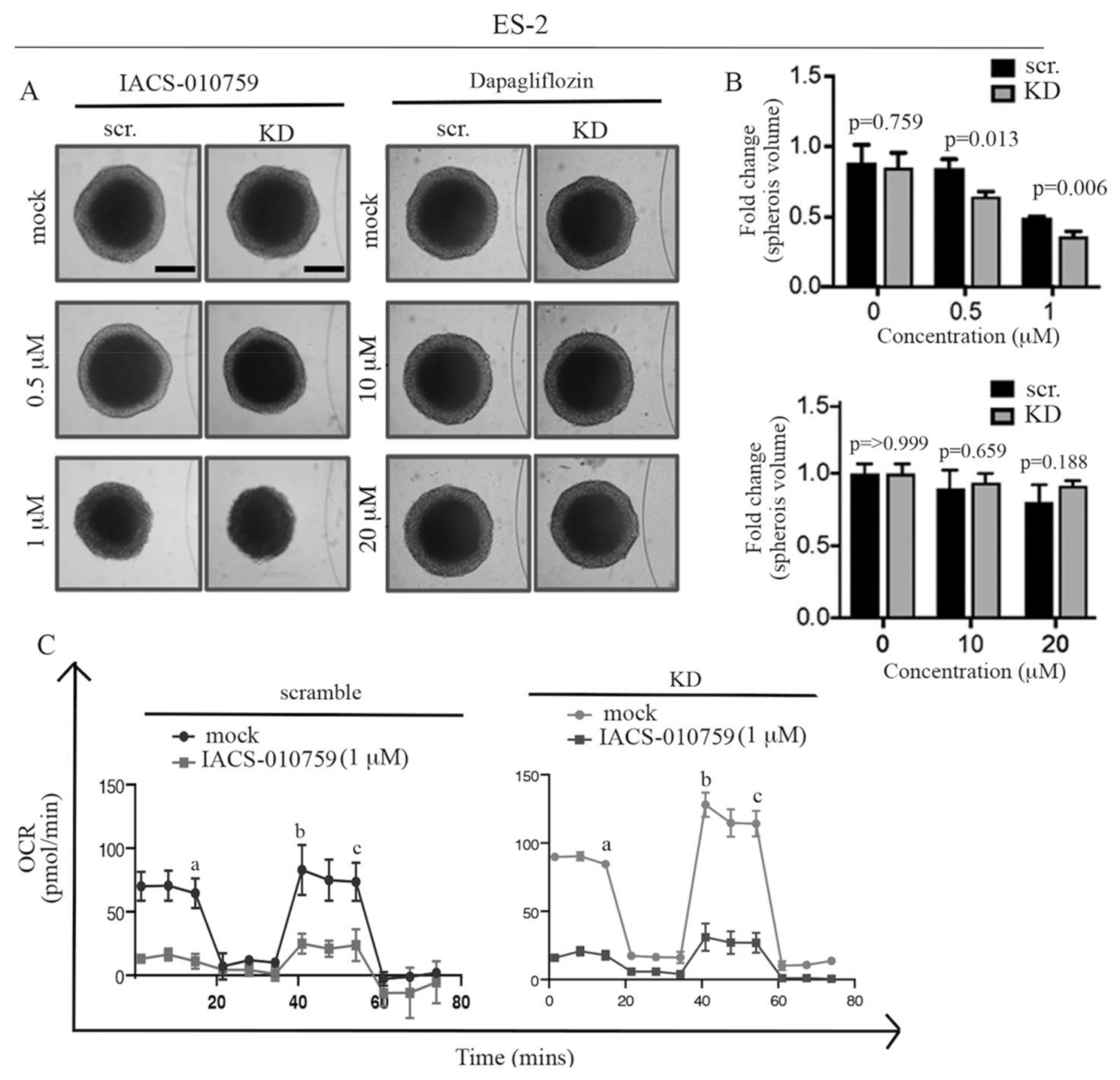
2.4. ARID1A Loss Results in Selective Sensitivity to the Mitochondrial Inhibitor IACS-010759 in 2D
2.5. ARID1A Loss Results in Selective Sensitivity to the Mitochondrial Inhibitor IACS-010759 in Three-Dimensional Culture
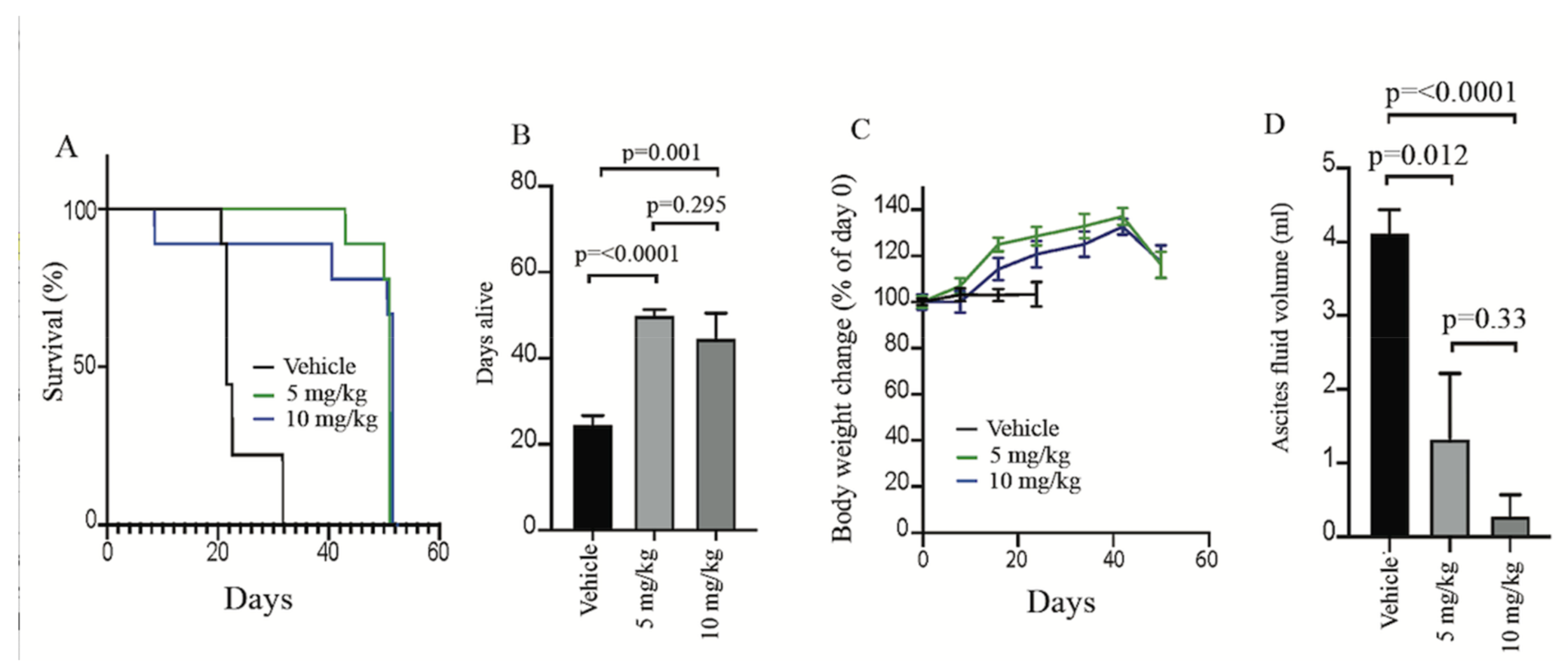
2.6. IACS-010759 Treatment Prolongs Survival in a Model of ARID1A-Mutated OCCCs
3. Discussion
4. Materials and Methods
4.1. Chemicals and Antibodies
4.2. Cell Culture
4.3. siRNA
4.4. Western Blot
4.5. RNA-SEQ Experiments
4.6. Isolation of Mitochondria
4.7. Native Gel
4.8. Measurements of Oxygen Consumption
4.9. ATP Quantification Assay
4.10. Confocal Microscopy
4.11. Analysis of Mitochondrial Morphology
4.12. Measurement of mDNA Content
4.13. Plasmid Transfection
4.14. Generation of Spheroids
4.15. Crystal Violet Staining
4.16. Colony Formation Assay
4.17. Preclinical Testing of IACS-010759
4.18. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Fukumoto, T.; Magno, E.; Zhang, R. SWI/SNF Complexes in Ovarian Cancer: Mechanistic Insights and Therapeutic Implications. Mol. Cancer Res. 2018, 16, 1819–1825. [Google Scholar] [CrossRef] [PubMed]
- Jelinic, W.; Mueller, J.J.; Olvera, N.; Dao, F.; Scott, S.N.; Shah, R.; Gao, J.J.; Schultz, N.; Gonen, M.; Soslow, R.A.; et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Gen. 2014, 46, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Dier, U.; Shin, D.H.; Hemachandra, L.P.; Uusitalo, L.M.; Hempel, N. Bioenergetic analysis of ovarian cancer cell lines: Profiling of histological subtypes and identification of a mitochondria-defective cell line. PLoS ONE 2014, 9, e98479. [Google Scholar] [CrossRef]
- Kwan, S.Y.; Cheng, X.; Tsang, Y.T.; Choi, J.S.; Kwan, S.Y.; Izaguirre, D.I.; Kwan, H.S.; Gershenson, D.M.; Wong, K.K. Loss of ARID1A expression leads to sensitivity to ROS-inducing agent elesclomol in gynecologic cancer cells. Oncotarget 2016, 7, 56933–56943. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, H.; Takahashi, K.; Sasaki, M.; Kuroda, T.; Yoshida, H.; Watanabe, R.; Maruyama, A.; Makinoshima, H.; Chiwaki, F.; Sasaki, H.; et al. Targeting the Vulnerability of Glutathione Metabolism in ARID1A-Deficient Cancers. Cancer Cell 2019, 35, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.R.; Wardell, S.E.; Cakir, M.; Yip, C.; Ahn, Y.R.; Ali, M.; Yllanes, A.P.; Chao, C.A.; McDonnell, D.P.; Wood, K.C. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat. Commun. 2018, 9, 1677. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.L. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013, 369, 2236–2251. [Google Scholar] [CrossRef] [PubMed]
- Maycotte, P.; Marin-Hernandez, A.; Goyri-Aguirre, M.; Anaya-Ruiz, M.; Reyes-Leyva, J.; Cortes-Hernandez, P. Mitochondrial dynamics and cancer. Tumour Biol. 2017, 39, 1010428317698391. [Google Scholar] [CrossRef]
- Samant, S.A.; Zhang, H.J.; Hong, Z.; Pillai, V.B.; Sundaresan, N.R.; Wolfgeher, D.; Archer, S.L.; Chan, D.C.; Gupta, M.P. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol. Cell. Biol. 2014, 34, 807–819. [Google Scholar] [CrossRef]
- Trotta, A.P.; Chipuk, J.E. Mitochondrial dynamics as regulators of cancer biology. Cell. Mol. Life Sci. 2017, 74, 1999–2017. [Google Scholar] [CrossRef]
- Kashatus, J.A.; Nascimento, A.; Myers, L.J.; Sher, A.; Byrne, F.L.; Hoehn, K.L.; Counter, C.M.; Kashatus, D.F. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 2015, 57, 537–551. [Google Scholar] [CrossRef]
- Malhotra, A.; Dey, A.; Prasad, N.; Kenney, A.M. Sonic Hedgehog Signaling Drives Mitochondrial Fragmentation by Suppressing Mitofusins in Cerebellar Granule Neuron Precursors and Medulloblastoma. Mol. Cancer Res. MCR 2016, 14, 114–124. [Google Scholar] [CrossRef]
- Wieder, S.Y.; Serasinghe, M.N.; Sung, J.C.; Choi, D.C.; Birge, M.B.; Yao, J.L.; Bernstein, E.; Celebi, J.T.; Chipuk, J.E. Activation of the Mitochondrial Fragmentation Protein DRP1 Correlates with BRAF(V600E) Melanoma. J. Investig. Derm. 2015, 135, 2544–2547. [Google Scholar] [CrossRef]
- Zhang, G.E.; Jin, H.L.; Lin, X.K.; Chen, C.; Liu, X.S.; Zhang, Q.; Yu, J.R. Anti-tumor effects of Mfn2 in gastric cancer. Int. J. Mol. Sci. 2013, 14, 13005–13021. [Google Scholar] [CrossRef]
- Zhao, X.; Tian, C.; Puszyk, W.M.; Ogunwobi, O.O.; Cao, M.; Wang, T.; Cabrera, R.; Nelson, D.R.; Liu, C. OPA1 downregulation is involved in sorafenib-induced apoptosis in hepatocellular carcinoma. Lab. Investig. 2013, 93, 8–19. [Google Scholar] [CrossRef]
- Peiris-Pages, M.; Bonuccelli, G.; Sotgia, F.; Lisanti, M.P. Mitochondrial fission as a driver of stemness in tumor cells: mDIVI1 inhibits mitochondrial function, cell migration and cancer stem cell (CSC) signalling. Oncotarget 2018, 9, 13254–13275. [Google Scholar] [CrossRef]
- Rao, V.A. Targeting Mitochondrial Fission to Trigger Cancer Cell Death. Cancer Res. 2019, 79, 6074–6075. [Google Scholar] [CrossRef]
- Devine, M.J.; Kittler, J.T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 2018, 19, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.A.; Renken, C.W.; Frey, T.G.; Ellisman, M.H. Membrane architecture of mitochondria in neurons of the central nervous system. J. Neurosci. Res. 2001, 66, 857–865. [Google Scholar] [CrossRef]
- Perkins, G.A.; Tjong, J.; Brown, J.M.; Poquiz, P.H.; Scott, R.T.; Kolson, D.R.; Ellisman, M.H.; Spirou, G.A. The micro-architecture of mitochondria at active zones: Electron tomography reveals novel anchoring scaffolds and cristae structured for high-rate metabolism. J. Neurosci. 2010, 30, 1015–1026. [Google Scholar] [CrossRef]
- Picard, M.; Shirihai, O.S.; Gentil, B.J.; Burelle, Y. Mitochondrial morphology transitions and functions: Implications for retrograde signaling? Am. J. Physiol. R. Integr. Comp Physiol. 2013, 304, R393–R406. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Zeller, K.I.; Potter, J.J.; Wonsey, D.R.; O’Donnell, K.A.; Kim, J.W.; Yustein, J.T.; Lee, L.A.; Dang, C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005, 25, 6225–6234. [Google Scholar] [CrossRef] [PubMed]
- Morrish, F.; Hockenbery, D. MYC and mitochondrial biogenesis. Cold Spring Harb Perspect. Med. 2014, 4, a014225. [Google Scholar] [CrossRef]
- Agarwal, E.; Altman, B.J.; Ho Seo, J.; Bertolini, I.; Ghosh, J.C.; Kaur, A.; Kossenkov, A.V.; Languino, L.R.; Gabrilovich, D.I.; Speicher, D.W.; et al. Myc Regulation of a Mitochondrial Trafficking Network Mediates Tumor Cell Invasion and Metastasis. Mol. Cell. Biol. 2019, 39, 109–119. [Google Scholar] [CrossRef]
- Seo, J.H.; Agarwal, E.; Chae, Y.C.; Lee, Y.G.; Garlick, D.S.; Storaci, A.M.; Ferrero, S.; Gaudioso, G.; Gianelli, U.; Vaira, V.; et al. Mitochondrial fission factor is a novel Myc-dependent regulator of mitochondrial permeability in cancer. EBioMedicine 2019, 48, 353–363. [Google Scholar] [CrossRef]
- Casinelli, G.; LaRosa, J.; Sharma, M.; Cherok, E.; Banerjee, S.; Branca, M.; Edmunds, L.; Wang, Y.; Sims-Lucas, S.; Churley, L.; et al. n-Myc overexpression increases cisplatin resistance in neuroblastoma via deregulation of mitochondrial dynamics. Cell Death Discov. 2016, 2, 16082. [Google Scholar] [CrossRef]
- Zeng, M.; Kwiatkowski, N.P.; Zhang, T.; Nabet, B.; Xu, M.; Liang, Y.; Quan, C.; Wang, J.; Hao, M.; Palakurthi, S.; et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. eLife 2018, 7, e39030. [Google Scholar] [CrossRef]
- Dimova, I.; Raitcheva, S.; Dimitrov, R.; Doganov, N.; Toncheva, D. Correlations between c-myc gene copy-number and clinicopathological parameters of ovarian tumours. Eur. J. Cancer 2006, 42, 674–679. [Google Scholar] [CrossRef]
- Nagl, N.G., Jr.; Zweitzig, D.R.; Thimmapaya, B.; Beck, G.R., Jr.; Moran, E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006, 66, 1289–1293. [Google Scholar] [CrossRef]
- Trizzino, M.; Barbieri, E.; Petracovici, A.; Wu, S.; Welsh, S.A.; Owens, T.A.; Licciulli, S.; Zhang, R.; Gardini, A. The Tumor Suppressor ARID1A Controls Global Transcription via Pausing of RNA Polymerase II. Cell Rep. 2018, 23, 3933–3945. [Google Scholar] [CrossRef]
- Emmings, E.; Mullany, S.; Chang, Z.; Landen, C.N., Jr.; Linder, S.; Bazzaro, M. Targeting Mitochondria for Treatment of Chemoresistant Ovarian Cancer. Int. J. Mol. Sci. 2019, 20, 229. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.R.; Sun, Y.; Protopopova, M.; Gera, S.; Bandi, M.; Bristow, C.; McAfoos, T.; Morlacchi, P.; Ackroyd, J.; Agip, A.A.; et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018, 24, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Sica, V.; Bravo-San Pedro, J.M.; Stoll, G.; Kroemer, G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int. J. Cancer 2020, 146, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; De Milito, A.; Demiroglu-Zergeroglu, A.; Gullbo, J.; D’Arcy, P.; Linder, S. Eradicating Quiescent Tumor Cells by Targeting Mitochondrial Bioenergetics. Trends Cancer 2016, 2, 657–663. [Google Scholar] [CrossRef]
- Frattaruolo, L.; Brindisi, M.; Curcio, R.; Marra, F.; Dolce, V.; Cappello, A.R. Targeting the Mitochondrial Metabolic Network: A Promising Strategy in Cancer Treatment. Int. J. Mol. Sci. 2020, 21, 6014. [Google Scholar] [CrossRef]
- Oliveira, G.L.; Coelho, A.R.; Marques, R.; Oliveira, P.J. Cancer cell metabolism: Rewiring the mitochondrial hub. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166016. [Google Scholar] [CrossRef]
- Bajpai, R.; Sharma, A.; Achreja, A.; Edgar, C.L.; Wei, C.; Siddiqa, A.A.; Gupta, V.A.; Matulis, S.M.; McBrayer, S.K.; Mittal, A.; et al. Electron transport chain activity is a predictor and target for venetoclax sensitivity in multiple myeloma. Nat. Commun. 2020, 11, 1228. [Google Scholar] [CrossRef]
- Fischer, G.M.; Jalali, A.; Kircher, D.A.; Lee, W.C.; McQuade, J.L.; Haydu, L.E.; Joon, A.Y.; Reuben, A.; de Macedo, M.P.; Carapeto, F.C.L.; et al. Molecular Profiling Reveals Unique Immune and Metabolic Features of Melanoma Brain Metastases. Cancer Discov. 2019, 9, 628–645. [Google Scholar] [CrossRef]
- Lissanu Deribe, Y.; Sun, Y.; Terranova, C.; Khan, F.; Martinez-Ledesma, J.; Gay, J.; Gao, G.; Mullinax, R.A.; Khor, T.; Feng, N.; et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nat. Med. 2018, 24, 1047–1057. [Google Scholar] [CrossRef]
- Panina, S.B.; Pei, J.; Baran, N.; Konopleva, M.; Kirienko, N.V. Utilizing Synergistic Potential of Mitochondria-Targeting Drugs for Leukemia Therapy. Front. Oncol. 2020, 10, 435. [Google Scholar] [CrossRef]
- Sun, Y.; Bandi, M.; Lofton, T.; Smith, M.; Bristow, C.A.; Carugo, A.; Rogers, N.; Leonard, P.; Chang, Q.; Mullinax, R.; et al. Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation. Cell Rep. 2019, 26, 469–482.e465. [Google Scholar] [CrossRef]
- Teh, J.L.F.; Purwin, T.J.; Han, A.; Chua, V.; Patel, P.; Baqai, U.; Liao, C.; Bechtel, N.; Sato, T.; Davies, M.A.; et al. Metabolic Adaptations to MEK and CDK4/6 Cotargeting in Uveal Melanoma. Mol. Cancer 2020, 19, 1719–1726. [Google Scholar] [CrossRef]
- Tsuji, A.; Akao, T.; Masuya, T.; Murai, M.; Miyoshi, H. IACS-010759, a potent inhibitor of glycolysis-deficient hypoxic tumor cells, inhibits mitochondrial respiratory complex I through a unique mechanism. J. Biol. Chem. 2020, 295, 7481–7491. [Google Scholar] [CrossRef]
- Vashisht Gopal, Y.N.; Gammon, S.; Prasad, R.; Knighton, B.; Pisaneschi, F.; Roszik, J.; Feng, N.; Johnson, S.; Pramanik, S.; Sudderth, J.; et al. A Novel Mitochondrial Inhibitor Blocks MAPK Pathway and Overcomes MAPK Inhibitor Resistance in Melanoma. Clin. Cancer Res. 2019, 25, 6429–6442. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, Y.; Zhang, S.; Liu, Y.; Guo, H.; Ahmed, M.; Bell, T.; Zhang, H.; Han, G.; Lorence, E.; et al. Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci. Transl. Med. 2019, 11, 1167. [Google Scholar] [CrossRef]
- Konovalova, S. Analysis of Mitochondrial Respiratory Chain Complexes in Cultured Human Cells using Blue Native Polyacrylamide Gel Electrophoresis and Immunoblotting. J. Vis. Exp. Jove 2019, 144, e59269. [Google Scholar] [CrossRef]
- McKenzie, M.; Lazarou, M.; Thorburn, D.R.; Ryan, M.T. Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal. Biochem. 2007, 364, 128–137. [Google Scholar] [CrossRef]
- Van Coster, R.; Smet, J.; George, E.; De Meirleir, L.; Seneca, S.; Van Hove, J.; Sebire, G.; Verhelst, H.; De Bleecker, J.; Van Vlem, B.; et al. Blue native polyacrylamide gel electrophoresis: A powerful tool in diagnosis of oxidative phosphorylation defects. Pediatr. Res. 2001, 50, 658–665. [Google Scholar] [CrossRef]
- Lenaz, G.; Genova, M.L. Structure and organization of mitochondrial respiratory complexes: A new understanding of an old subject. Antioxid Redox Signal 2010, 12, 961–1008. [Google Scholar] [CrossRef]
- Hirst, J. Open questions: Respiratory chain supercomplexes-why are they there and what do they do? BMC Biol. 2018, 16, 111. [Google Scholar] [CrossRef]
- Letts, J.A.; Sazanov, L.A. Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 2017, 24, 800–808. [Google Scholar] [CrossRef]
- Cichocki, F.; Wu, C.Y.; Zhang, B.; Felices, M.; Tesi, B.; Tuininga, K.; Dougherty, P.; Taras, E.; Hinderlie, P.; Blazar, B.R.; et al. ARID5B regulates metabolic programming in human adaptive NK cells. J. Exp. Med. 2018, 215, 2379–2395. [Google Scholar] [CrossRef]
- Xiao, B.; Deng, X.; Zhou, W.; Tan, E.K. Flow Cytometry-Based Assessment of Mitophagy Using MitoTracker. Front. Cell Neurosci. 2016, 10, 76. [Google Scholar] [CrossRef]
- Lee, J.E.; Westrate, L.M.; Wu, H.; Page, C.; Voeltz, G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 2016, 540, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Nguyen, N.D.; Huang, Y.; Lin, D.; Fujimoto, T.N.; Molkentine, J.M.; Deorukhkar, A.; Kang, Y.; San Lucas, F.A.; Fernandes, C.J.; et al. Mitochondrial fusion exploits a therapeutic vulnerability of pancreatic cancer. Jci Insight 2019, 5, e126215. [Google Scholar] [CrossRef] [PubMed]
- Romero, O.A.; Setien, F.; John, S.; Gimenez-Xavier, P.; Gomez-Lopez, G.; Pisano, D.; Condom, E.; Villanueva, A.; Hager, G.L.; Sanchez-Cespedes, M. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. Embo Mol. Med. 2012, 4, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.clinicaltrials.gov (accessed on 1 November 2018).
- Nasiri, A.R.; Rodrigues, M.R.; Li, Z.; Leitner, B.P.; Perry, R.J. SGLT2 inhibition slows tumor growth in mice by reversing hyperinsulinemia. Cancer Metab. 2019, 7, 10. [Google Scholar] [CrossRef]
- Zhang, X.; Fryknäs, M.; Hernlund, E.; Fayad, W.; De Milito, A.; Olofsson, M.H.; Gogvadze, V.; Dang, L.; Påhlman, S.; Schughart, L.A.; et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat. Commun. 2014, 5, 3295. [Google Scholar] [CrossRef]
- Penet, M.F.; Krishnamachary, B.; Wildes, F.B.; Mironchik, Y.; Hung, C.F.; Wu, T.C.; Bhujwalla, Z.M. Ascites Volumes and the Ovarian Cancer Microenvironment. Front. Oncol. 2018, 8, 595. [Google Scholar] [CrossRef]
- Ahmed, N.; Stenvers, K.L. Getting to know ovarian cancer ascites: Opportunities for targeted therapy-based translational research. Front. Oncol. 2013, 3, 256. [Google Scholar] [CrossRef]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef]
- Hsieh, A.L.; Walton, Z.E.; Altman, B.J.; Stine, Z.E.; Dang, C.V. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015, 43, 11–21. [Google Scholar] [CrossRef]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef]
- Yaromina, A.; Quennet, V.; Zips, D.; Meyer, S.; Shakirin, G.; Walenta, S.; Mueller-Klieser, W.; Baumann, M. Co-localisation of hypoxia and perfusion markers with parameters of glucose metabolism in human squamous cell carcinoma (hSCC) xenografts. Int. J. Radiat Biol. 2009, 85, 972–980. [Google Scholar] [CrossRef]
- Weinberg, S.E.; Chandel, N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015, 11, 9–15. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Zong, W.X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef]
- Bernal, S.D.; Lampidis, T.J.; McIsaac, R.M.; Chen, L.B. Anticarcinoma activity in vivo of rhodamine 123, a mitochondrial-specific dye. Science 1983, 222, 169–172. [Google Scholar] [CrossRef]
- Lampidis, T.J.; Bernal, S.D.; Summerhayes, I.C.; Chen, L.B. Selective toxicity of rhodamine 123 in carcinoma cells in vitro. Cancer Res. 1983, 43, 716–720. [Google Scholar]
- Wenzel, C.; Riefke, B.; Grundemann, S.; Krebs, A.; Christian, S.; Prinz, F.; Osterland, M.; Golfier, S.; Rase, S.; Ansari, N.; et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp. Cell Res. 2014, 323, 131–143. [Google Scholar] [CrossRef]
- Jha, P.; Wang, X.; Auwerx, J. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE). Curr. Protoc. Mouse Biol. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- ImageJ Documentation Wiki. Available online: https://imagejdocu.tudor.lu/plugin/morphology/mitochondrial_morphology_macro_plug-in/start (accessed on 1 April 2019).
- Fayad, W.; Rickardson, L.; Haglund, C.; Olofsson, M.H.; D’Arcy, P.; Larsson, R.; Linder, S.; Fryknas, M. Identification of agents that induce apoptosis of multicellular tumour spheroids: Enrichment for mitotic inhibitors with hydrophobic properties. Chem. Biol. Drug Des. 2011, 78, 547–557. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Shetty, M.; Clemente, V.; Linder, S.; Bazzaro, M. Targeting Mitochondrial Metabolism in Clear Cell Carcinoma of the Ovaries. Int. J. Mol. Sci. 2021, 22, 4750. https://doi.org/10.3390/ijms22094750
Zhang X, Shetty M, Clemente V, Linder S, Bazzaro M. Targeting Mitochondrial Metabolism in Clear Cell Carcinoma of the Ovaries. International Journal of Molecular Sciences. 2021; 22(9):4750. https://doi.org/10.3390/ijms22094750
Chicago/Turabian StyleZhang, Xiaonan, Mihir Shetty, Valentino Clemente, Stig Linder, and Martina Bazzaro. 2021. "Targeting Mitochondrial Metabolism in Clear Cell Carcinoma of the Ovaries" International Journal of Molecular Sciences 22, no. 9: 4750. https://doi.org/10.3390/ijms22094750
APA StyleZhang, X., Shetty, M., Clemente, V., Linder, S., & Bazzaro, M. (2021). Targeting Mitochondrial Metabolism in Clear Cell Carcinoma of the Ovaries. International Journal of Molecular Sciences, 22(9), 4750. https://doi.org/10.3390/ijms22094750






