Epigenetic Biomarkers as Diagnostic Tools for Neurodegenerative Disorders
Abstract
:1. Introduction
2. Results
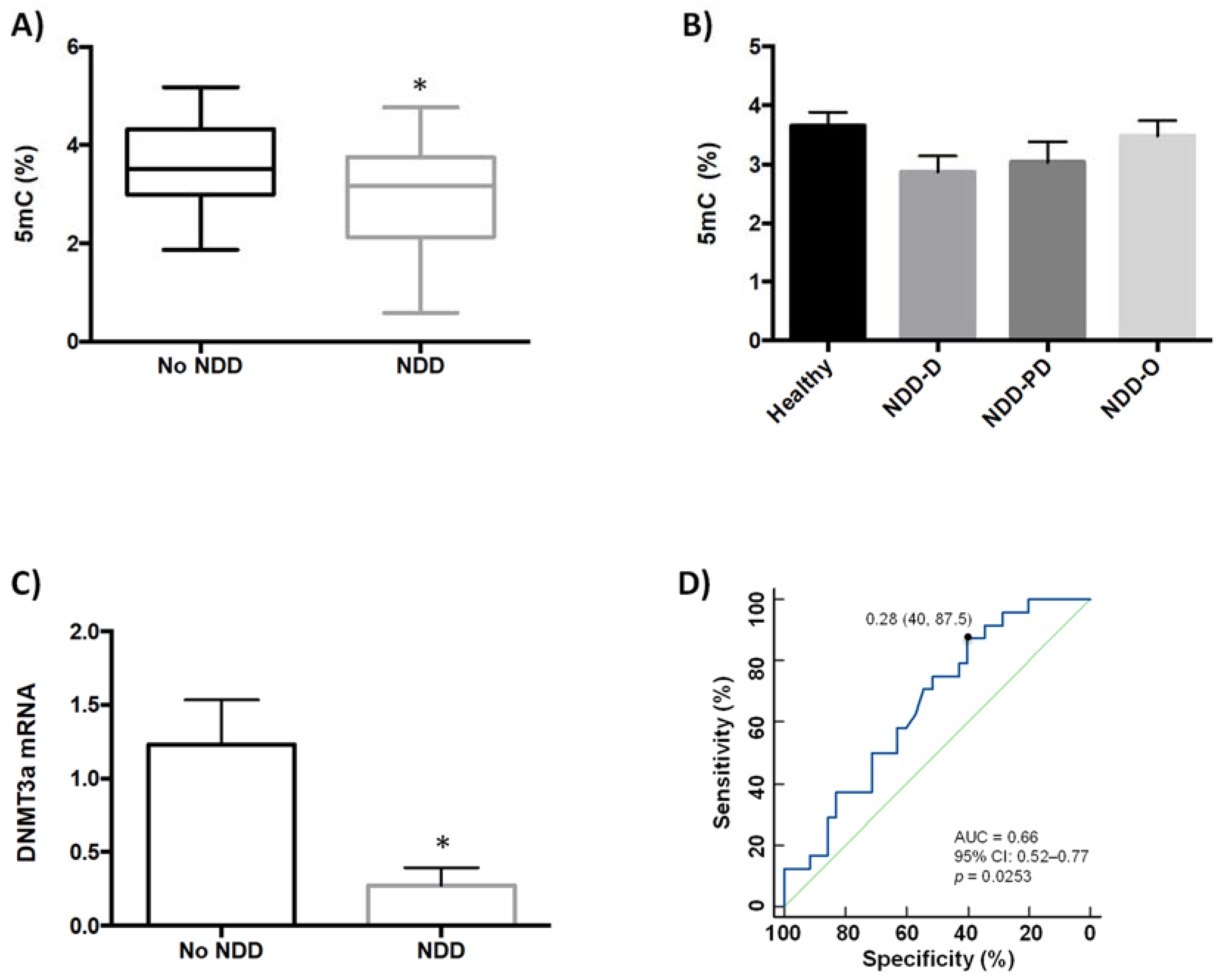
2.1. Global DNA Methylation Is Reduced in Patients with Neurodegenerative Disorders
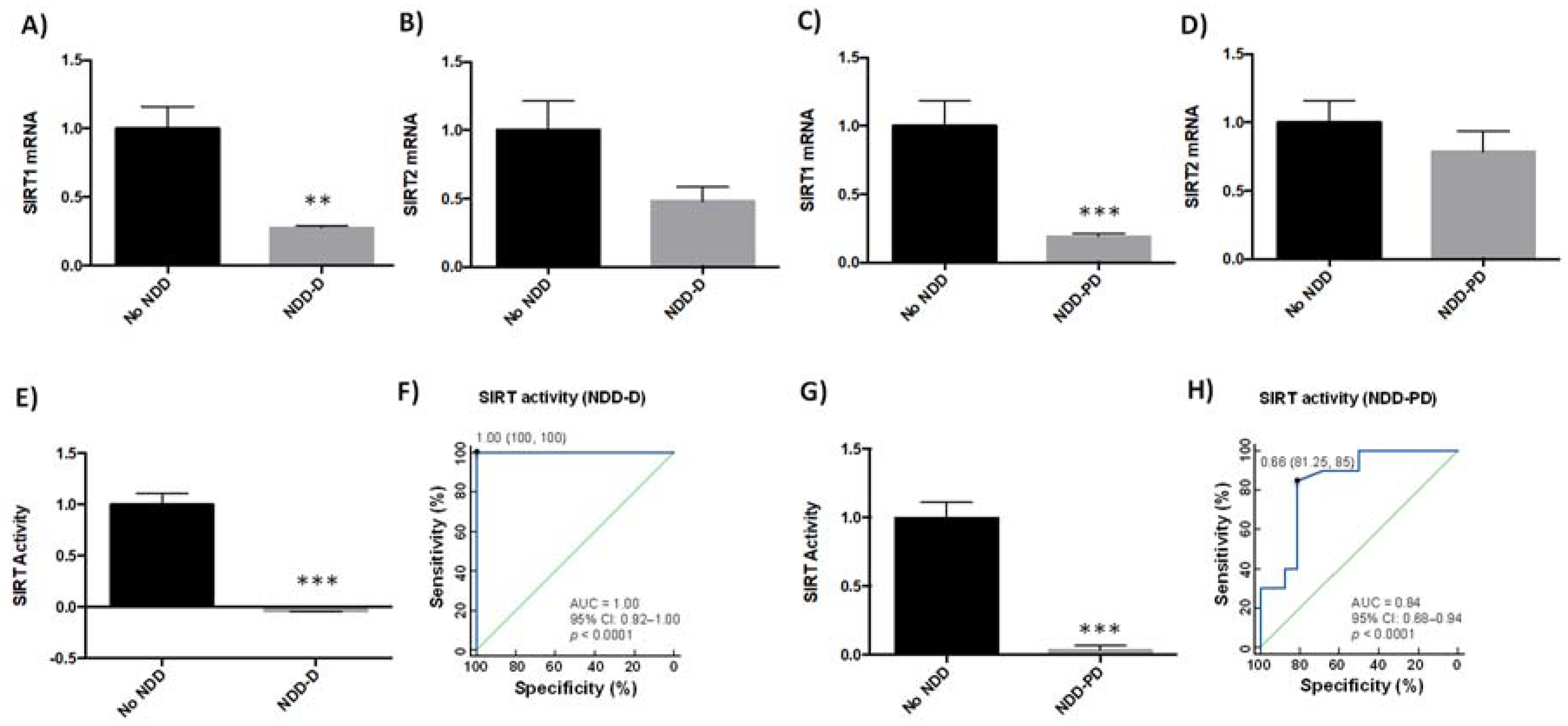
2.2. SIRT Expression and Activity Are Reduced in Patients with Neurodegenerative Disorders
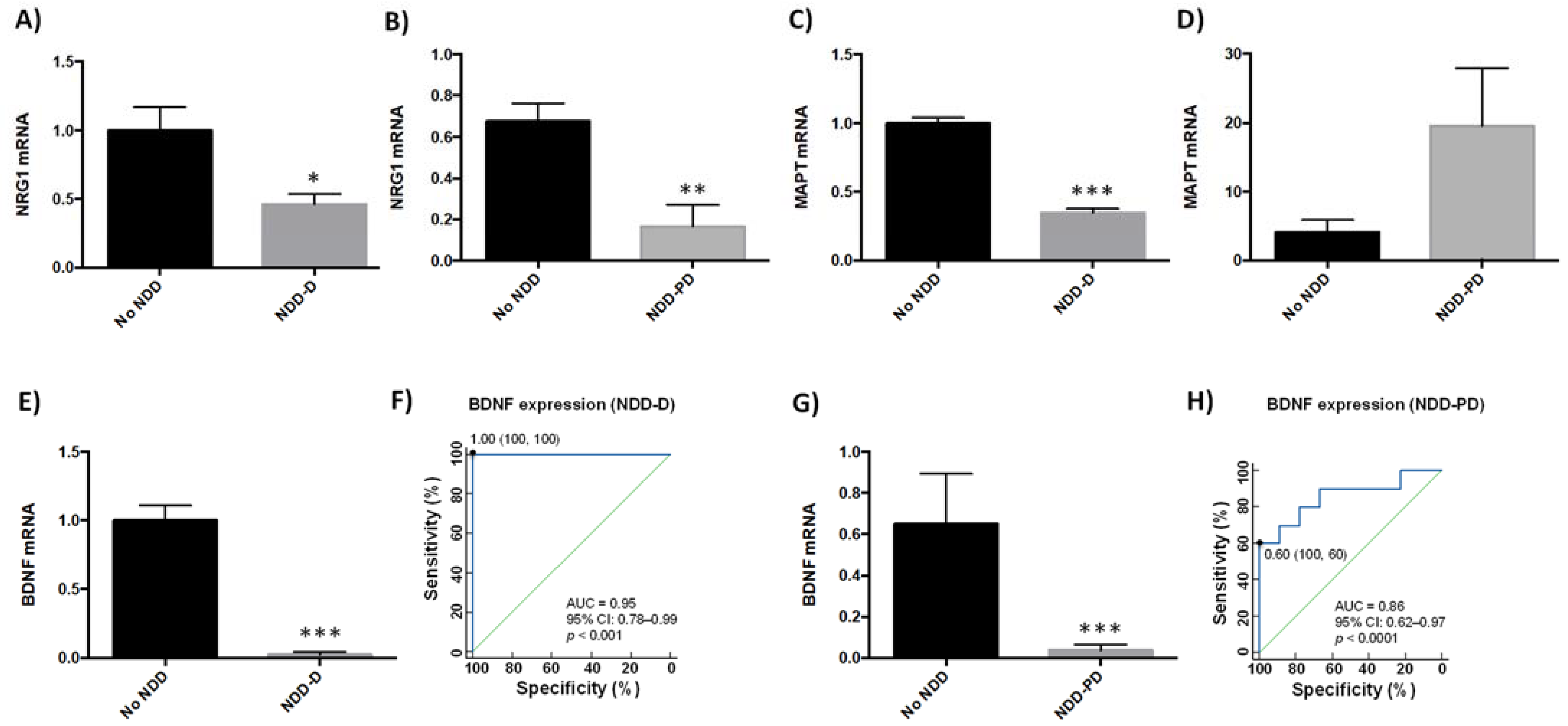
2.3. Neurodegeneration-Related Gene Expression Is Altered in Neurodegenerative Disorders
3. Discussion
4. Materials and Methods
4.1. Patients and Selection Criteria
4.2. DNA Extraction
4.3. RNA Extraction
4.4. Nuclear Protein Extraction
4.5. Quantification of Global DNA Methylation (5mC)
4.6. Quantification of SIRT Activity
4.7. Quantitative Real-Time RT-PCR
4.8. Genotyping
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cacabelos, R.; Teijido, O.; Carril, J.C. Can cloud-based tools accelerate Alzheimer’s disease drug discovery? Expert Opin. Drug Discov. 2016, 11, 215–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacabelos, R. Pharmacogenomics of Alzheimer’s and Parkinson’s Diseases. Neurosci. Lett. 2018, 726, 133807. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Fernandez-Novoa, L.; Lombardi, V.; Kubota, Y.; Takeda, M. Molecular genetics of Alzheimer’s disease and aging. Methods Find. Exp. Clin. Pharm. 2005, 27 (Suppl. A), 1–573. [Google Scholar]
- Teijido, O.; Cacabelos, R. Pharmacoepigenomic Interventions as Novel Potential Treatments for Alzheimer’s and Parkinson’s Diseases. Int. J. Mol. Sci. 2018, 19, E3199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacabelos, R. Pharmacogenomics in Alzheimer’s disease. Methods Mol. Biol. 2008, 448, 213–357. [Google Scholar] [PubMed]
- Ciceri, F.; Rotllant, D.; Maes, T. Understanding Epigenetic Alterations in Alzheimer’s and Parkinson’s Disease: Towards Targeted Biomarkers and Therapies. Curr. Pharm. Des. 2017, 23, 839–857. [Google Scholar] [CrossRef]
- Dauer, W.; Przedborski, S. Parkinson’s disease: Mechanisms and models. Neuron 2003, 39, 889–909. [Google Scholar] [CrossRef] [Green Version]
- Spillantini, M.G.; Scmidt, M.L.; Lee, V.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewi bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Urdinguio, R.; Sanchez-Mut, J.; Esteller, M. Epigenetic mechanisms in neurological diseases: Genes, syndromes, and therapies. Lancet Neurol. 2009, 8, 1056–1072. [Google Scholar] [CrossRef]
- Sweatt, J. The emerging field of neuroepigenetics. Neuron 2013, 80, 624–632. [Google Scholar] [CrossRef] [Green Version]
- Cacabelos, R.; Tellado, I.; Cacacbelos, P. The Epigenetic Machinery in the Life Cycle and Pharmacoepigenetics; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Delgado-Morales, R.; Esteller, M. Opening up the DNA methylome of dementia. Mol. Psychiatry 2017, 22, 485–496. [Google Scholar] [CrossRef]
- Lahiri, D.; Maloney, B. The “LEARn” (latent early-life associated regulation) model: An epigenetic pathway linking metabolic and cognitive disorders. J. Alzheimers Dis. 2012, 30, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Maloney, B.; Lahiri, D. Epigenetics of dementia: Understanding the disease as a transformation rather than a state. Lancet Neurol. 2016, 15, 760–774. [Google Scholar] [CrossRef]
- Cacabelos, R.; Carril, J.C.; Cacabelos, N.; Kazantsev, A.G.; Vostrov, A.V.; Corzo, L.; Cacabelos, P.; Goldgaber, D. Sirtuins in Alzheimer´s disease: SIRT2-related genophenotypes and implications for pharmacoepigenetics. Int. J. Mol. Sci. 2019, 20, 1249. [Google Scholar] [CrossRef] [Green Version]
- Alberti, K.G.; Zimmet, P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetes Med. 1998, 15, 539–553. [Google Scholar] [CrossRef]
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018, 11, 292. [Google Scholar] [CrossRef] [Green Version]
- Bird, A. The essentials of DNA methylation. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef]
- Gräff, J.; Mansury, I. Epigenetic codes in cognition and behaviour. Behav. Brain Res. 2008, 1, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Cross, S.; Bird, A. Gene silencing by methyl-CpG-binding proteins. Novartis Found. Symp. 1998, 214, 6–16. [Google Scholar] [PubMed]
- Nan, X.; Campoy, F.; Bird, A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997, 88, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Hermann, A.; Gowher, J.; Jeltsch, A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol. Life Sci. 2004, 61, 2571–2587. [Google Scholar] [CrossRef]
- Guo, J.; Su, Y.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; Zhu, H.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Gowher, H.; Liebert, K.; Hermann, A.; Xu, G.; Jeltsch, A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005, 80, 13341–13348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Su, Y.; Zhong, C.; Ming, G.; Song, J. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2014, 16, 258–264. [Google Scholar] [CrossRef]
- Li, X.; Feng, Y.; Wang, X.-X.; Truong, D.; Wu, Y.-C. The critical role of SIRT1 in Parkinson´s Disease: Mechanism and therapeutic consideration. Aging Dis. 2020, 11, 1608–1622. [Google Scholar] [CrossRef]
- Grabowska, W.; Sikora, E.; Bielak-Zmijewska, A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology 2017, 18, 447–476. [Google Scholar] [CrossRef] [Green Version]
- Amjad, S.; Nisar, S.; Bhat, A.A.; Shah, A.R.; Frenneaux, M.P.; Fakhro, K.; Haris, M.; Reddy, R.; Patay, Z.; Baur, J.; et al. Role of NAD+ in regulating cellular and metabolic signaling pathways. Mol. Metab. 2021, 49, 101195. [Google Scholar] [CrossRef]
- Donmez, G.; Outeiro, T.F. SIRT1 and SIRT2: Emerging targets in neurodegeneration. EMBO Mol. Med. 2013, 5, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Michasn, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadmiri, N.E.; Said, N.; Slassi, I.; Moutawakil, B.E.; Nadifi, S. Biomarkers for Alzheimer Disease: Classical and Novel Candidates’ Review. Neuroscience 2018, 370, 181–190. [Google Scholar] [CrossRef]
- Martinez-Iglesias, O.; Carrera, I.; Carril, J.C.; Fernández-Novoa, L.; Cacabelos, N.; Cacabelos, R. DNA Methylation in Neurodegenerative and Cerebrovascular disorders. Int. J. Mol. Sci. 2020, 21, 2220. [Google Scholar] [CrossRef] [Green Version]
- Stocker, H.; Möllers, T.; Perna, L.; Brenner, H. The genetic risk of Alzheimer’s disease beyond APOE e4: Systematic review of Alzheimer´s genetic risk scores. Transl. Psychiatry 2018, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H. Mechanisms and disease implications of sirtuin-mediated autophagic regulation. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Tiwari, P.; Kaur, A.; Singh, T.G. Sirtuin Acetylation and Deacetylation: A Complex Paradigm in Neurodegenerative Disease. Mol. Neurobiol. 2021, 58, 3903–3917. [Google Scholar] [CrossRef] [PubMed]
- Cooper-Knock, J.; Kirby, J.; Ferraiuolo, L.; Heath, P.R.; Rattray, M.; Sjaw, P.J. Gene expression profiling in human neurodegenerative disease. Nat. Rev. Neurol. 2012, 8, 518–530. [Google Scholar] [CrossRef]
- Xu, J.; de Winter, F.; Farrokhi, C.; Rockenstein, E.; Mante, M.; Adame, A.; Cook, J.; Jin, X.; Masliah, E.; Lee, K.-L. Neuregulin 1 improves cognitive deficits and neuropathology in an Alzheimer’s disease model. Sci. Rep. 2016, 6, 31692. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.; Hong, B.-H.; Kim, Y.-J.; Yang, E.-J.; Choi, I.; Kim, H.; Ahn, S.; Baik, T.-K.; Woo, R.-S.; Kim, H.-S. Neuregulin-1 attenuates cognitive function impairments in a transgenic mouse model of Alzheimer’s disease. Cell Death Dis. 2016, 7, e2117. [Google Scholar] [CrossRef] [Green Version]
- Hama, Y.; Yabe, I.; Wakabayashi, K.; Kano, T.; Hirotani, M.; Iwakura, Y.; Utsumi, J.; Sasaki, H. Level of plasma neuregulin-1 SMDF is reduced in patients with idiopatic Parkinson’s disease. Neurosci. Lett. 2015, 5, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Caillet-Boudin, M.-L.; Buée, L.; Sergeant, N.; Lefebvre, B. Regulation of human MAPT gene expression. Mol. Neurodegener. 2015, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, J.T.; de Labio, R.W.; Rasmussen, L.T.; de Oliveira, L.C.; Chen, E.; Villares, J.; Tureck, G.; Smith, M.; Payao, S. CDK5 and MAPT Gene Expression in Alzheimer’s Disease Brain Samples. Curr. Alzheimer Res. 2018, 15, 182–186. [Google Scholar] [CrossRef]
- Tobin, J.E.; Latourelle, J.C.; Lew, M.-F.; Klein, C.; Suchowersky, O.; Shill, H.A.; Golbe, L.I.; Mark, M.H.; Growdon, J.H.; Wooten, G.F. Haplotypes and gene expression imlicate the MAPT region for Parkinson disease: The GenePD Study. Neurology 2008, 71, 28–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacobbo, B.L.; Doorduin, J.; Klein, H.C.; Dierckx, R.; Bromberg, E.; de Vries, E. Brain-Derived Neurotrophic Factor in Brain Disorders: Focus on Neuroinflammation. Mol. Neurobiol. 2019, 56, 3295–3312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colucci-D´Amato, L.; Speranza, L.; Volpicelli, F. Neurotrophic Factor BDNF, Physiological Functions and Therapeutic Potential in Depression, Neurodegeneration and Brain Cancer. Int. J. Mol. Sci. 2020, 21, 7777. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Esteller, M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012, 13, 679–692. [Google Scholar] [CrossRef]
- Lu, H.; Liu, X.; Deng, Y.; Qing, H. DNA methylation, a hand behind neurodegenerative diseases. Front. Aging Neurosci. 2013, 5, 85. [Google Scholar] [CrossRef] [Green Version]
- Kalaria, R. Similarities between Alzheimer’s disease and vascular dementia. J. Neurol. Sci. 2002, 203, 29–34. [Google Scholar] [CrossRef]
- Pook, M. DNA methylation and trinucleotide repeat expansion diseases. DNA Methylation- Genom. Technol. 2012, 10, 194–209. [Google Scholar]
- Francesco, A.D.; Arosio, B.; Falconi, A.; Bonaventura, M.M.D.; Karimi, M.; Mari, D.; Casati, M.; Maccarrobe, M.; D´Affario, C. Global changes in DNA methylation in Alzheimer’s disease peripheral blood mononuclear cells. Brain Behav. Immun. 2015, 45, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Masliah, E.; Dumaop, W.; Galasko, D.; Desplats, P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA methylation biomarkers in aging and age-related diseases. Front Genet. 2020, 11, 171. [Google Scholar] [CrossRef]
- Unniktishnan, A.; Freeman, W.; Jackson, J.; Wren, J.; Porter, H.; Richardson, A. The role of DNA methylation in epigenetics and aging. Pharmacol. Ther. 2019, 195, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Landgrave-Gómez, J.; Mercado-Gómez, O.; Guevara-Guzmán, R. Epigenetic mechanisms in neurological and neurodegenerative diseases. Front. Cell Neurosci. 2015, 9, 58. [Google Scholar] [PubMed] [Green Version]
- Mastroeni, D.; Grover, A.; Delvaux, E.; Whiteside, C.; Coleman, P.; Rogers, J. Epigenetic changes in Alzheimer’s disease: Decrements in DNA methylation. Neurobiol. Aging 2010, 31, 2025–2037. [Google Scholar] [CrossRef] [Green Version]
- Chouliaras, L.; Mastroeni, D.; Delvaux, E.; Grover, A.; Kenis, G.; Hof, P.; Steinbusch, H.; Coleman, P.; Rutten, B.; Hove, D. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging 2013, 34, 2091–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condliffe, D.; Wong, A.; Troakes, C.; Proitsi, P.; Patel, Y.; Chouliaras, L.; Fernandes, C.; Cooper, J.; Lovestone, S.; Schalkwyk, L.; et al. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer’s disease brain. Neurobiol. Aging 2014, 35, 1850–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, K.X.; Milic, J.; El-Khodor, B.; Dhana, K.; Nano, J.; Pulido, T.; Kraja, B.; Zaciragic, A.; Bramer, W.M.; Troup, J.; et al. The Role of DNA Methylation and Histone Modifications in Neurodegenerative Diseases: A Systematic Review. PLoS ONE 2016, 11, e0167201. [Google Scholar] [CrossRef]
- Ciccarone, F.; Tagliatesta, S.; Caiafa, P.; Zampieri, M. DNA methylation dynamics in aging: How far are we from understanding the mechanisms? Mech. Ageing Dev. 2018, 174, 3–17. [Google Scholar] [CrossRef]
- Xu, X.; Cui, D. DNA methyltransferases, DNA methylation and age-associated cognitive function. Int. J. Mol. Sci. 2018, 19, 1315. [Google Scholar]
- Oliveira, A.M.M.; Hemstedt, T.J.; Bading, H. Rescue of aging-associated decline in DnmtDNMT3a2 expression restores cognitive abilities. Nat. Neurosci. 2012, 15, 1111. [Google Scholar] [CrossRef]
- Miller, C.A.; Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levenson, J.M.; Roth, T.L.; Lubin, F.D.; Miller, C.A.; Juang, I.-C.; Desai, P.; Malone, L.M.; Swearr, J.D. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006, 281, 15763–15773. [Google Scholar] [CrossRef] [Green Version]
- Wülner, U.; Kaut, O.; de Boni, L.; Piston, D. DNA methylation in Parkinson’s disease. J. Neurosci. 2016, 139, 108–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lashley, T.; Gami, P.; Valizadeh, N.; Lo, A.; Revesz, T.; Balazs, R. Alterations in global DNA methylation and hydroxymethylation are not detected in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2016, 41, 497–506. [Google Scholar] [CrossRef]
- Phipps, A.J.; Vickers, J.C.; Taberlay, P.C.; Woodhouse, A. Neurofilament-labeled pyramidal neurons and astrocytes are deficient in DNA methylation marks in Alzheimer’s disease. Neurobiol. Aging 2016, 45, 30–42. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- JAffe, A.E.; Irizarry, R.A. Accounting for cellular heterogeinity is critical in epigenome-wide association studies. Genome Biol. 2014, 15, R31. [Google Scholar] [CrossRef] [PubMed]
- Julien, C.; Tremblay, C.; Emond, V.; Lebbadi, M.; Salem, N., Jr.; Bennet, D.A.; Calon, F. Sirtuin1 reduction parallels the accumulation of tau in Alzheimer’s disease. J. Neuropath Exp. Neutol. 2009, 48, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joao, F.; Gong, Z. The beneficial roles of SIRT1 in Neuroinflammation-related Diseases. Oxidative Med. Cell Long. 2020, 2020, 6782872. [Google Scholar] [CrossRef]
- Donmez, G.; Guarante, L. Aging and disease: Connection to sirtuins. Aging Cell. 2010, 9, 285–290. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Zhu, K.; Chi, S.; Ewang, C.; Xie, A. Emerging role of sirtuin 2 in Parkinson’s disease. Front. Aging Neurosci. 2020, 11, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strarheam, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.C.; McLean, P.J. Sirtuin2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007, 317, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.; Rahman, M.S.; Saha, S.K.; Saikot, F.K.; Deep, A.; Kim, K.H. Function of the SIRT3 mitochondrial deacetylase in cellular physiology, cancer, and neurodegenerative disease. Aging Cell. 2017, 16, 4–16. [Google Scholar] [CrossRef]
- Yang, W.; Zou, Y.; Zhang, M.; Zhao, N.; Tian, Q.; Gu, M.; Liu, W.; Shi, R.; Lü, Y.; Yu, W. Mitochondrial SIRT3 expression is decreased in APP/PS1 double transgenic mouse model of Alzheimer´s Disease. Neurochem. Res. 2015, 40, 1576–1582. [Google Scholar] [CrossRef]
- Mohamed Nasir, N.-F.; Zainuddin, A.; Shamsuddin, S. Emerging roles of Sirtuin 6 in Alzheimer’s disease. J. Mol. Neurosci. 2018, 64, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Kaluski, S.; Portillo, M.; Besnard, A.; Stein, D.; Einav, M.; Zhong, L.; Ueberham, U.; Arendt, T.; Mostolavsky, R.; Sahay, A. Neuroprotective Functions for the Histone Deacetylase SIRT6. Cell Rep. 2017, 18, 3052–3062. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Hu, C.; Chen, S.; Shen, H.; Jiang, Q.; Huang, P.; Zhao, W. Neuregulin-1 protects mouse cerebellum against oxidative stress and neuroinflammation. Brain Res. 2017, 1670, 32–43. [Google Scholar] [CrossRef]
- Carlsoon, T.; Schindler, F.R.; Höllerhage, M.; Depboylu, C.; Arias-Carrion, O.; Schnurrbusch, S.; Rösler, T.W.; Wozny, W.; Schwall, G.P.; Groebe, K.; et al. Systemic administration of neuregulin-1b1 protects dopaminergic neurons in a mouse model of Parkinson’s disease. J. Neurochem. 2011, 117, 1066–1074. [Google Scholar] [CrossRef]
- Zuccato, C.; Cathaneo, E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat. Rev. Neurol. 2009, 5, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.-S.; Shen, L.-L.; Zhu, C.; Bu, X.-L.; Liu, C.-H.; Yao, Z.-Q.; Zhang, L.-L.; Zhou, H.-D.; Walker, D.-G.; Tan, J.; et al. Brain-derived neurotrophic factor in neurodegenerative diseases. Transl. Psychiatry 2016, 5, 311–329. [Google Scholar]
- Gezen-Ak, D.; Dursun, E.; Hanagasi, H.; Bilgiç, B.; Lohman, E.; Araz, Ö.S.; Atasoy, I.L.; Alaylıoğlu, M.; Önal, B.; Gürvit, H.; et al. BDNF, TNFa, HSP90, CFH, and IL-10 serum levels in patients with early or late onset Alzheimer’s disease or mild cognitive impairment. J. Alzheimer’s Dis. 2013, 37, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.; Ho, C.; Tam, W.; Kua, E.H.; Ho, R.C.M. Decreased serum brain-derived neurotrophic factor (BDNF) levels in patients with Alzheimer´s disease (AD): A systematic review and meta-analysis. Int. J. Mol. Sci. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eyileten, C.; Sharif, L.; Wicik, Z.; Jacubik, D.; Jarosz-Popek, J.; Soplinska, A.; Postula, M.; Czlonkowska, A.; Kaplon-Cieslicka, A.; Mirowska-Guzel, D. The relation of the brain-derived neurotrophic factor with microRNAs in neurodegenerative diseases and ischemic stroke. Mol. Neurobiol. 2021, 58, 329–347. [Google Scholar] [CrossRef] [PubMed]
- Ventiglia, M.; Zanardini, R.; Bonomino, C.; Zanetti, O.; Volpe, D.; Pasqualetti, P.; Gennarelli, M.; Bocchio-Chiavetto, L. Serum brain-derived neurotrophic factor levels in different neurological diseases. BioMed Res. Int. 2013, 2013, 901082. [Google Scholar]
- Miranda, M.; Morici, J.F.; Zanoni, M.B.; Bekinschtein, P. Brain-Derived Neurotrophic Factor: A key molecule for memory in the healthy and the pathological brain. Front. Cell Neurosci. 2019, 13, 363. [Google Scholar] [CrossRef] [PubMed]
- Porritt, M.-J.; Batchelor, P.E.; Howells, D.W. Inhibiting BDNF expression by antisense oligonucleotide infusion causes loss of nigral dopaminergic neurons. Exp. Neurol. 2005, 192, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Scalzao, P.; Kümmer, A.; Bretas, T.L.; Cardoso, F.; Teixeira, A.L. Serum levels of brain-derived neurotrophic factor correlate with motor impairment in Parkinson’s disease. J. Neurol. 2010, 257, 540–545. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, H.; Wang, C.; Ming, F.; Shi, X.; Yang, M. Serum level of brain.derived neurotrophic factor in Parkinson’s disease: A meta-analysis. Progr. Neuro-Psychopharm Biol. Psych. 2019, 88, 168–174. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Zhu, C.-H.; Liu, D.-S.; Wang, Y.; Zhang, J.-B.; Wang, S.-P.; Song, Y.-N. Rehabilitation training improves cognitive disorder after cerebrovascular accident by improving BDNF Bcl-2 and Bax expressions in regulating the JMK pathway. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 3807–3821. [Google Scholar] [PubMed]
- Turner, R.S.; Stubbs, T.; Davies, D.A.; Albensi, B.C. Potential New Approaches for Diagnosis of Alzheimer’s Disease and Related Dementias. Front. Neurol. 2020, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sun, X.; Jiang, H.; Yu, S.; Robins, C.; Armstrong, M.; Li, R.; Mei, Z.; Shi, X.; Gerasimov, E.; et al. A machine learning approach to brain epigenetic analysis reveals kinases associated with Alzheimer’s disease. Nat. Commun. 2021, 12, 4472. [Google Scholar] [CrossRef]
- Fangyu, L.I.; Hua, H.E. Assesing the accuracy of diagnostic test. Shanghai Arch. Psych. 2018, 30, 207–2012. [Google Scholar]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
| Total | Healthy | Dementia | PD | Others | |||
|---|---|---|---|---|---|---|---|
| 5mC study (Figure 1) | Gender | N | 60 | 25 | 21 | 7 | 7 |
| Age | 47 ± 2.95 | 64 ± 3.34 | 70 ± 3.38 | 34 ± 8.13 | |||
| Male | 11 | 6 | 4 | 5 | |||
| Female | 14 | 15 | 3 | 2 | |||
| APOE Genotype | 3.3 | 17 | 13 | 5 | 6 | ||
| 3.4 | 5 | 4 | 1 | ||||
| 4.4 | 2 | 3 | |||||
| 2.3 | 1 | 1 | 1 | 1 | |||
| 2.4 | |||||||
| Sirtuin activity study (Figure 2) | Gender | ||||||
| N | 50 | 15 | 16 | 19 | |||
| Age | 65.73 ± 2.23 | 65.44 ± 1.97 | 65.21 ± 2.07 | ||||
| male | 6 | 6 | 9 | ||||
| Female | 9 | 10 | 10 | ||||
| APOE Genotype | 3.3 | 12 | 6 | 10 | |||
| 3.4 | 2 | 7 | 8 | ||||
| 4.4 | 2 | ||||||
| 2.3 | 1 | 1 | 1 | ||||
| 2.4 | |||||||
| Gene expression study (Figure 1, Figure 2, Figure 3) | Gender | ||||||
| N | 24 | 9 | 6 | 9 | |||
| Age | 67.71 ± 8.28 | 67.9 ± 6.1 | 79.89 ± 5.06 | ||||
| male | 4 | 4 | 4 | ||||
| Female | 5 | 2 | 5 | ||||
| APOE Genotype | 3.3 | 5 | 3 | 2 | |||
| 3.4 | 2 | 1 | 5 | ||||
| 4.4 | 1 | 1 | |||||
| 2.3 | 1 | 1 | 1 | ||||
| 2.4 | 1 |
| Gene | Reference |
|---|---|
| DNMT3A | Hs1027162_m1 |
| SIRT1 | Hs109006_m1 |
| SIRT2 | Hs1560239_m1 |
| NRG1 | Hs110158_m1 |
| MAPT | Hs00902194_m1 |
| BDNF | Hs329549_m1 |
| GAPDH | Hs2786634_g1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Iglesias, O.; Naidoo, V.; Cacabelos, N.; Cacabelos, R. Epigenetic Biomarkers as Diagnostic Tools for Neurodegenerative Disorders. Int. J. Mol. Sci. 2022, 23, 13. https://doi.org/10.3390/ijms23010013
Martínez-Iglesias O, Naidoo V, Cacabelos N, Cacabelos R. Epigenetic Biomarkers as Diagnostic Tools for Neurodegenerative Disorders. International Journal of Molecular Sciences. 2022; 23(1):13. https://doi.org/10.3390/ijms23010013
Chicago/Turabian StyleMartínez-Iglesias, Olaia, Vinogran Naidoo, Natalia Cacabelos, and Ramón Cacabelos. 2022. "Epigenetic Biomarkers as Diagnostic Tools for Neurodegenerative Disorders" International Journal of Molecular Sciences 23, no. 1: 13. https://doi.org/10.3390/ijms23010013
APA StyleMartínez-Iglesias, O., Naidoo, V., Cacabelos, N., & Cacabelos, R. (2022). Epigenetic Biomarkers as Diagnostic Tools for Neurodegenerative Disorders. International Journal of Molecular Sciences, 23(1), 13. https://doi.org/10.3390/ijms23010013








