OsBIC1 Directly Interacts with OsCRYs to Regulate Leaf Sheath Length through Mediating GA-Responsive Pathway
Abstract
:1. Introduction
2. Results
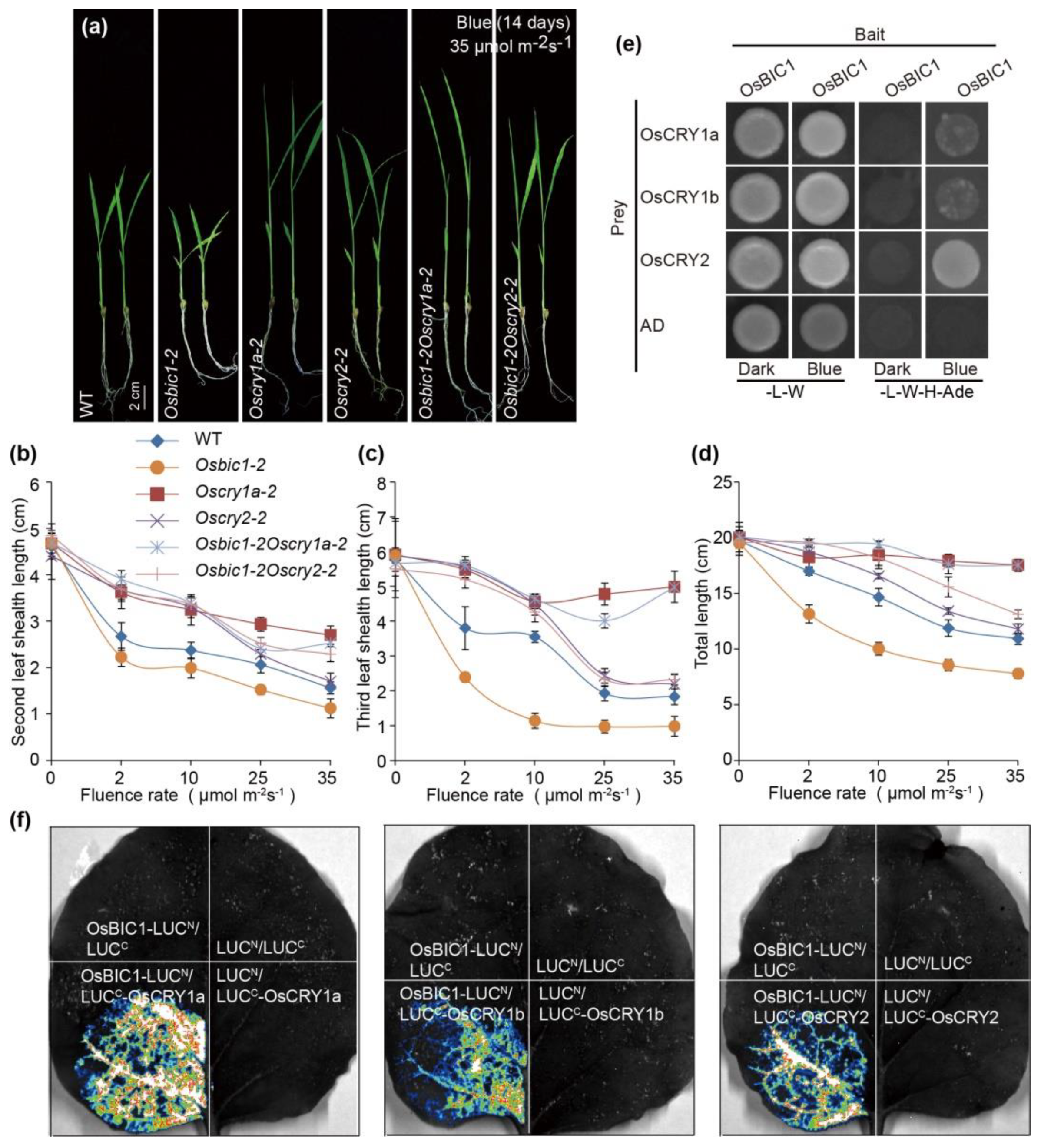
2.1. OsBICs Promoted Leaf Sheath Elongation in Blue Light Specific Manner
2.2. OsBIC1 Directly Interacted with OsCRYs to Regulate Blue Light-Induced Leaf Sheath Growth
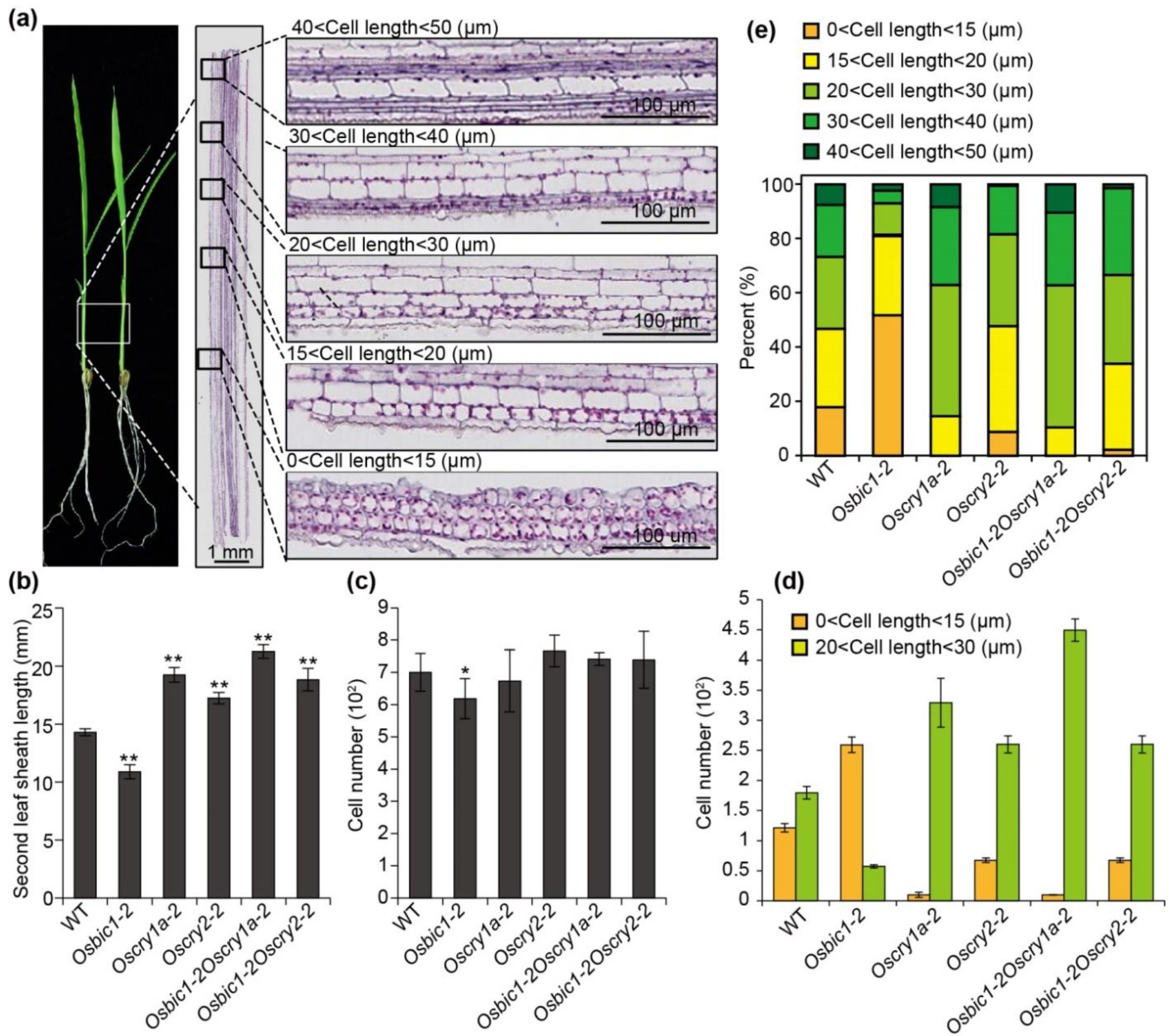
2.3. OsBIC1 and OsCRYs Regulated Leaf Sheath Length by Regulating Epidermal Cell Length
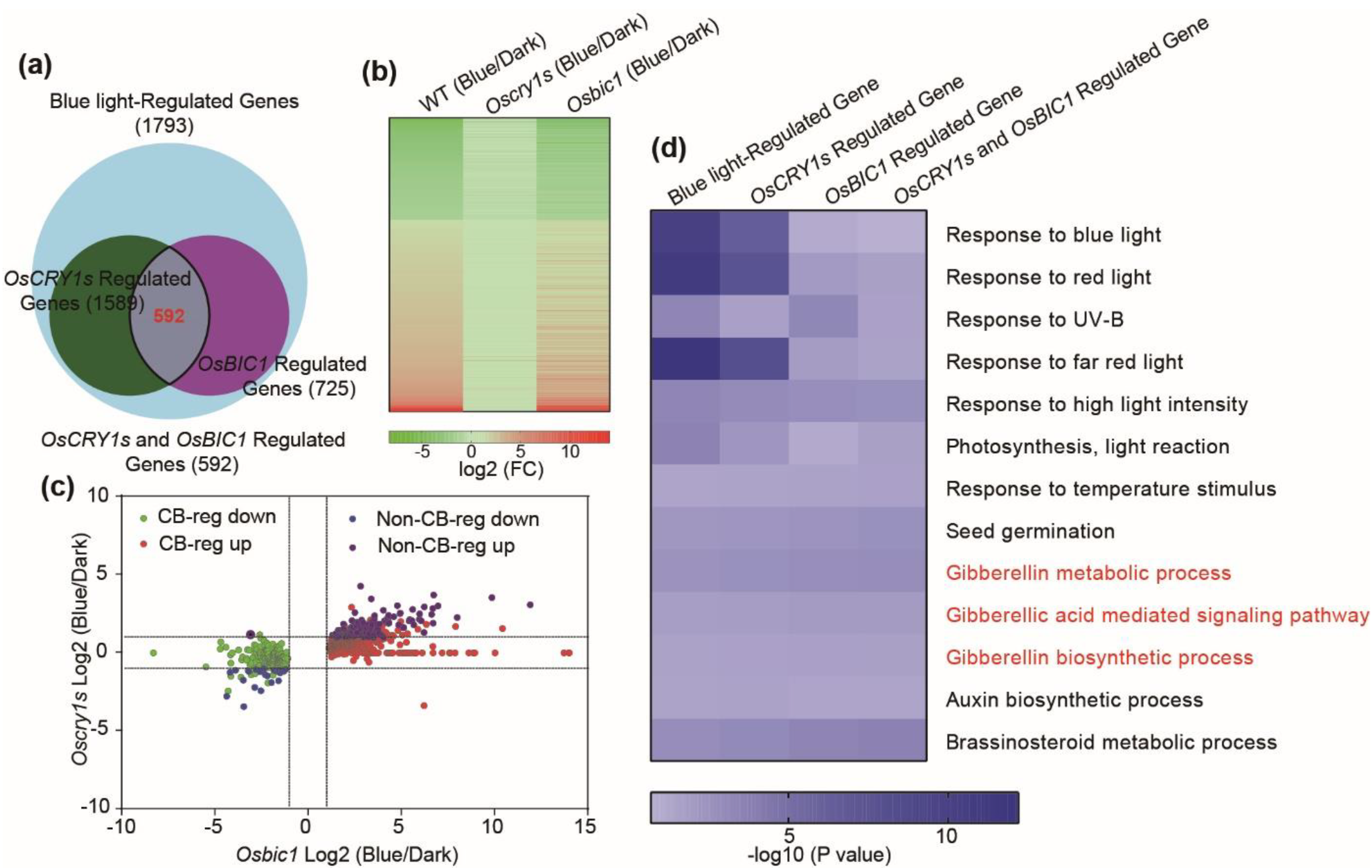
2.4. OsBIC1 and OsCRY1s Regulated Similar Transcriptome Changes under Blue Light Conditions
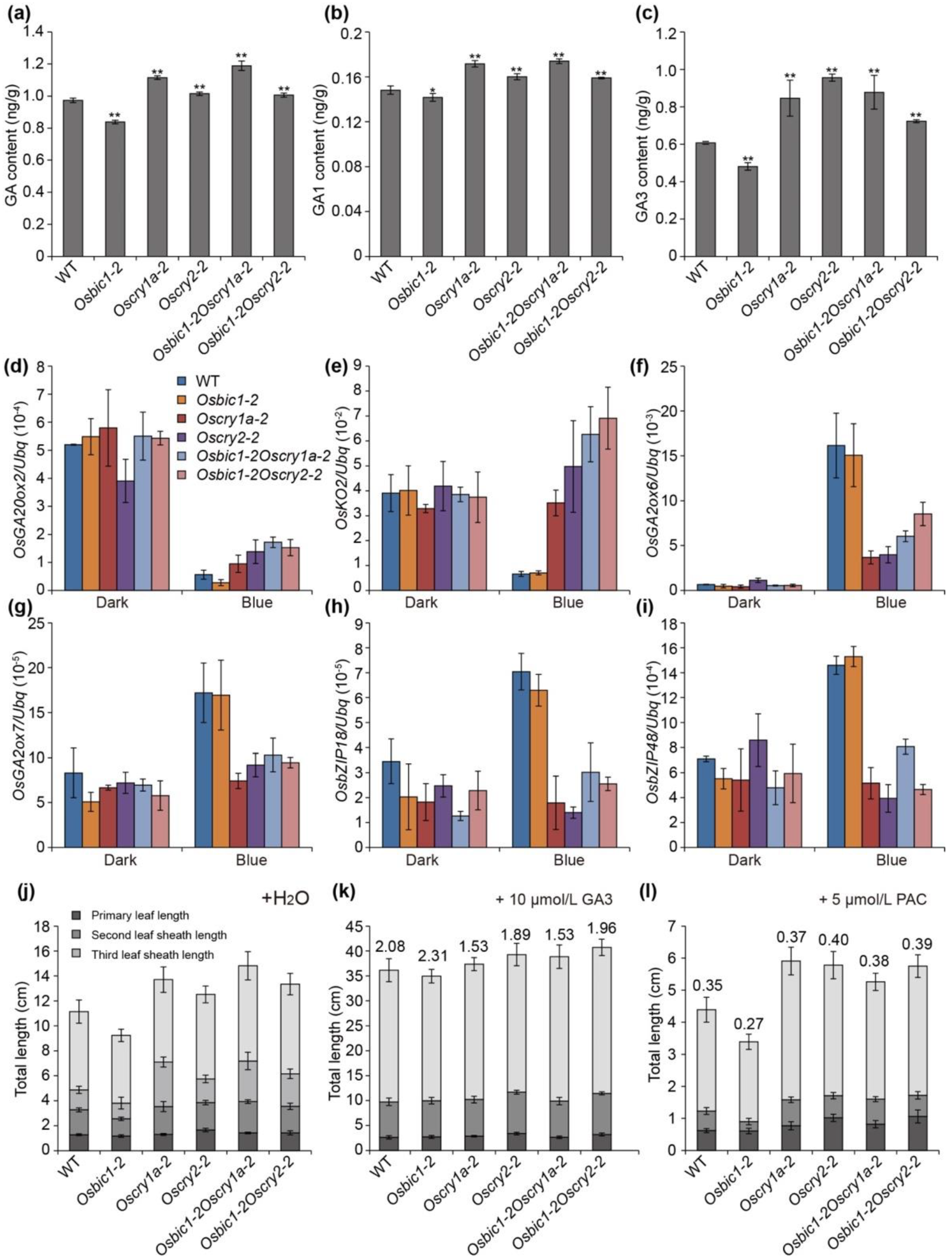
2.5. OsBIC1 and OsCRYs Regulated Leaf Sheath Length through the GA-Responsive Pathway
3. Discussion
4. Materials and Methods
4.1. Primers and Accession Numbers
4.2. Plasmid Construction, Plant Materials and Growth Conditions
4.3. Yeast Two-Hybrid Assays
4.4. Firefly Luciferase Complementation Imaging Assays in N. benthamiana
4.5. Longitudinal Sections Histological Analysis
4.6. Sample Collection and qRT-PCR Analysis
4.7. RNA-seq and Data Analysis
4.8. GA Content Analysis Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, C.; Shalitin, D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 2003, 54, 469–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, M.; Cashmore, A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, P.D.; Batschauer, A.; Hays, J.B. PHH1, a novel gene from Arabidopsis thaliana that encodes a protein similar to plant blue-light photoreceptors and microbial photolyases. Mol. Gen. Genet. 1996, 253, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, H.; Guo, H.; Mockler, T.; Chen, J.; Cashmore, A.R. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 1998, 95, 2686–2690. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zuo, Z.; Wang, X.; Gu, L.; Yoshizumi, T.; Yang, Z.; Yang, L.; Liu, Q.; Liu, W.; Han, Y.J.; et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 2016, 354, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, Q.; Han, Y.J.; Liu, Q.; Gu, L.; Yang, Z.; Su, J.; Liu, B.; Zuo, Z.; He, W.; et al. A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 2017, 92, 426–436. [Google Scholar] [CrossRef]
- Wang, Q.; Zuo, Z.; Wang, X.; Liu, Q.; Gu, L.; Oka, Y.; Lin, C. Beyond the photocycle-how cryptochromes regulate photoresponses in plants? Curr. Opin. Plant Biol. 2018, 45, 120–126. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Xiang, Y.; Zhu, H.; Xu, H.; Zhang, Z.; Zhang, C.; Zhang, L.; Ma, Z. Wheat cryptochromes: Subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol. 2008, 149, 760–774. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Zhu, H.; Xu, H.; Zhang, Z.; Zhang, C.; Zhang, L.; Ma, Z. Composition and phylogenetic analysis of wheat cryptochrome gene family. Mol. Biol. Rep. 2009, 37, 825–832. [Google Scholar] [CrossRef]
- Ishibashi, N.; Setoguchi, H. Polymorphism of DNA sequences of cryptochrome genes is not associated with the photoperiodic flowering of wild soybean along a latitudinal cline. J. Plant Res. 2012, 125, 483–488. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Li, H.; Wang, Q.; Liu, B.; Lin, C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 2013, 25, 4405–4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrero, J.M.; Downie, B.A.; Xu, Q.; Gubler, F. A Role for Barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 2014, 26, 1094–1104. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, N.; Hirano, T.; Iwasaki, T.; Yamamoto, N. Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol. 2003, 133, 1494–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, F.; Shinomura, T.; Tanabata, T.; Shimada, H.; Takano, M. Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 2006, 47, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Gong, S.F.; Li, Q.H.; Sang, Y.; Yang, H.Q. Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J. 2006, 46, 971–983. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Lian, H.L.; Zhang, L.D.; Mao, Z.L.; Li, X.M.; Xu, F.; Li, L.; Yang, H.Q. Transcriptome analyses reveal the involvement of both C and N termini of cryptochrome 1 in its regulation of phytohormone-responsive gene expression in Arabidopsis. Front. Plant Sci. 2016, 7, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szekeres, M.; Nemeth, K.; Koncz-Kalman, Z.; Mathur, J.; Kauschmann, A.; Altmann, T.; Redei, G.P.; Nagy, F.; Schell, J.; Koncz, C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 1996, 85, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Richards, D.E.; King, K.E.; Ait-Ali, T.; Harberd, N.P. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 67–88. [Google Scholar] [CrossRef] [Green Version]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Hsing, Y.I.; Kitano, H.; Yamaguchi, I.; et al. Gibberellin insensitive DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef]
- Thomas, S.G.; Rieu, I.; Steber, C.M. Gibberellin metabolism and signaling. Vitam. Horm. 2005, 72, 289–338. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, X.; Foo, E.; Symons, G.M.; Lopez, J.; Bendehakkalu, K.T.; Xiang, J.; Weller, J.L.; Liu, X.; Reid, J.B.; et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007, 145, 106–118. [Google Scholar] [CrossRef] [Green Version]
- He, S.B.; Wang, W.X.; Zhang, J.Y.; Xu, F.; Lian, H.L.; Li, L.; Yang, H.Q. The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol. Plant 2015, 8, 822–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, F.; Inagaki, N.; Hanada, A.; Yamaguchi, S.; Kamiya, Y.; Miyao, A.; Hirochika, H.; Takano, M. Cryptochrome and phytochrome cooperatively but independently reduce active gibberellin content in rice seedlings under light irradiation. Plant Cell Physiol. 2012, 53, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, H.; Li, R.; Hu, R.; Fan, C.; Chen, F.; Wang, Z.; Liu, X.; Fu, Y.; Lin, C. Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl. Acad. Sci. USA 2008, 105, 21028–21033. [Google Scholar] [CrossRef] [Green Version]
- Lyu, X.; Cheng, Q.; Qin, C.; Li, Y.; Xu, X.; Ji, R.; Mu, R.; Li, H.; Zhao, T.; Liu, J.; et al. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 2021, 14, 298–314. [Google Scholar] [CrossRef]
- Itoh, H.; Tatsumi, T.; Sakamoto, T.; Otomo, K.; Toyomasu, T.; Kitano, H.; Ashikari, M.; Ichihara, S.; Matsuoka, M. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 2004, 54, 533–547. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Asano, K.; Yamasaki, M.; Takuno, S.; Miura, K.; Katagiri, S.; Ito, T.; Doi, K.; Wu, J.; Ebana, K.; Matsumoto, T.; et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11034–11039. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, K.T.; Liu, S.H.; Wang, I.W.; Chen, L.J. Phalaenopsis orchid miniaturization by overexpression of OsGA2ox6, a rice GA2-oxidase gene. Bot. Stud. 2020, 61, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tang, D.; Shen, Y.; Qin, B.; Hong, L.; You, A.; Li, M.; Wang, X.; Yu, H.; Gu, M.; et al. Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J. Genet. Genom. 2010, 37, 23–36. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Nakajima, M.; Katoh, E.; Ohmiya, H.; Asano, K.; Saji, S.; Hongyu, X.; Ashikari, M.; Kitano, H.; Yamaguchi, I.; et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007, 19, 2140–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueguchi-Tanaka, M.; Nakajima, M.; Motoyuki, A.; Matsuoka, M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 2007, 58, 183–198. [Google Scholar] [CrossRef] [Green Version]
- Burman, N.; Bhatnagar, A.; Khurana, J.P. OsbZIP48, a HY5 Transcription factor ortholog, exerts pleiotropic effects in light-regulated development. Plant Physiol. 2018, 176, 1262–1285. [Google Scholar] [CrossRef] [PubMed]
- Mockler, T.C.; Guo, H.; Yang, H.; Duong, H.; Lin, C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 1999, 126, 2073–2082. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, S.; Wu, W.; Chen, D.; Zhan, X.; Zhu, A.; Zhang, Y.; Cheng, S.; Cao, L.; Lou, X.; et al. Dissection of genetic architecture of rice plant height and heading date by multiple-strategy-based association studies. Sci. Rep. 2016, 6, 29718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, C.T.; Paek, N.C. Gibberellic Acid: A key phytohormone for spikelet fertility in rice grain production. Int. J. Mol. Sci. 2016, 17, 794. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, M.; Itoh, H.; Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 2003, 35, 104–115. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mi, X.F.; Shan, J.X.; Li, X.M.; Xu, J.L.; Lin, H.X. The QTL GNP1 Encodes GA20ox1, Which Increases Grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016, 12, e1006386. [Google Scholar] [CrossRef] [PubMed]
- Deveshwar, P.; Prusty, A.; Sharma, S.; Tyagi, A.K. Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front Genet. 2020, 11, 586462. [Google Scholar] [CrossRef] [PubMed]
- Weller, J.L.; Hecht, V.; Vander Schoor, J.K.; Davidson, S.E.; Ross, J.J. Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 2009, 21, 800–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [Green Version]
- Hirano, K.; Kouketu, E.; Katoh, H.; Aya, K.; Ueguchi-Tanaka, M.; Matsuoka, M. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012, 71, 443–453. [Google Scholar] [CrossRef]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Tourinan, N.; Legris, M.; Minguet, E.G.; Costigliolo-Rojas, C.; Nohales, M.A.; Iniesto, E.; Garciotaa-Leomicronn, M.; Paciotan, M.; Heucken, N.; Blomeier, T.; et al. COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 13792–13799. [Google Scholar] [CrossRef]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.L.; He, S.B.; Zhang, Y.C.; Zhu, D.M.; Zhang, J.Y.; Jia, K.P.; Sun, S.X.; Li, L.; Yang, H.Q. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011, 25, 1023–1028. [Google Scholar] [CrossRef] [Green Version]
- Saijo, Y.; Sullivan, J.A.; Wang, H.; Yang, J.; Shen, Y.; Rubio, V.; Ma, L.; Hoecker, U.; Deng, X.W. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003, 17, 2642–2647. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Maier, A.; Lee, J.H.; Laubinger, S.; Saijo, Y.; Wang, H.; Qu, L.J.; Hoecker, U.; Deng, X.W. Biochemical characterization of Arabidopsis complexes containing constitutively PHOTOMORPHOGENIC1 and suppressor of PHYA proteins in light control of plant development. Plant Cell 2008, 20, 2307–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Z.; Liu, H.; Liu, B.; Liu, X.; Lin, C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 2011, 21, 841–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNellis, T.W.; von Arnim, A.G.; Deng, X.W. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 1994, 6, 1391–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, H.S.; Yang, J.Y.; Ishikawa, M.; Bolle, C.; Ballesteros, M.L.; Chua, N.H. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 2003, 423, 995–999. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Cornejo, M.J.; Luth, D.; Blankenship, K.M.; Anderson, O.D.; Blechl, A.E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 1993, 23, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Hua, D.; Wang, C.; He, J.; Liao, H.; Duan, Y.; Zhu, Z.; Guo, Y.; Chen, Z.; Gong, Z. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 2012, 24, 2546–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Wang, X.; Zhang, L.; Zhang, C.; Yu, C.; Zhao, T.; Liu, B.; Li, H.; Liu, J. OsBIC1 Directly Interacts with OsCRYs to Regulate Leaf Sheath Length through Mediating GA-Responsive Pathway. Int. J. Mol. Sci. 2022, 23, 287. https://doi.org/10.3390/ijms23010287
Li C, Wang X, Zhang L, Zhang C, Yu C, Zhao T, Liu B, Li H, Liu J. OsBIC1 Directly Interacts with OsCRYs to Regulate Leaf Sheath Length through Mediating GA-Responsive Pathway. International Journal of Molecular Sciences. 2022; 23(1):287. https://doi.org/10.3390/ijms23010287
Chicago/Turabian StyleLi, Cong, Xin Wang, Liya Zhang, Chunyu Zhang, Chunsheng Yu, Tao Zhao, Bin Liu, Hongyu Li, and Jun Liu. 2022. "OsBIC1 Directly Interacts with OsCRYs to Regulate Leaf Sheath Length through Mediating GA-Responsive Pathway" International Journal of Molecular Sciences 23, no. 1: 287. https://doi.org/10.3390/ijms23010287
APA StyleLi, C., Wang, X., Zhang, L., Zhang, C., Yu, C., Zhao, T., Liu, B., Li, H., & Liu, J. (2022). OsBIC1 Directly Interacts with OsCRYs to Regulate Leaf Sheath Length through Mediating GA-Responsive Pathway. International Journal of Molecular Sciences, 23(1), 287. https://doi.org/10.3390/ijms23010287





