Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean
Abstract
1. Molecular Mechanisms for the Photoperiodic Regulation of Flowering in Arabidopsis
1.1. The Photoperiod Pathway in Arabidopsis
1.2. Regulation of FT Expression
1.3. FT Protein Movement
2. Molecular Mechanisms for Photoperiodic Regulation of Flowering Time in Soybean
2.1. Molecular Basis of Soybean E Series Genes for Flowering-Time Regulation
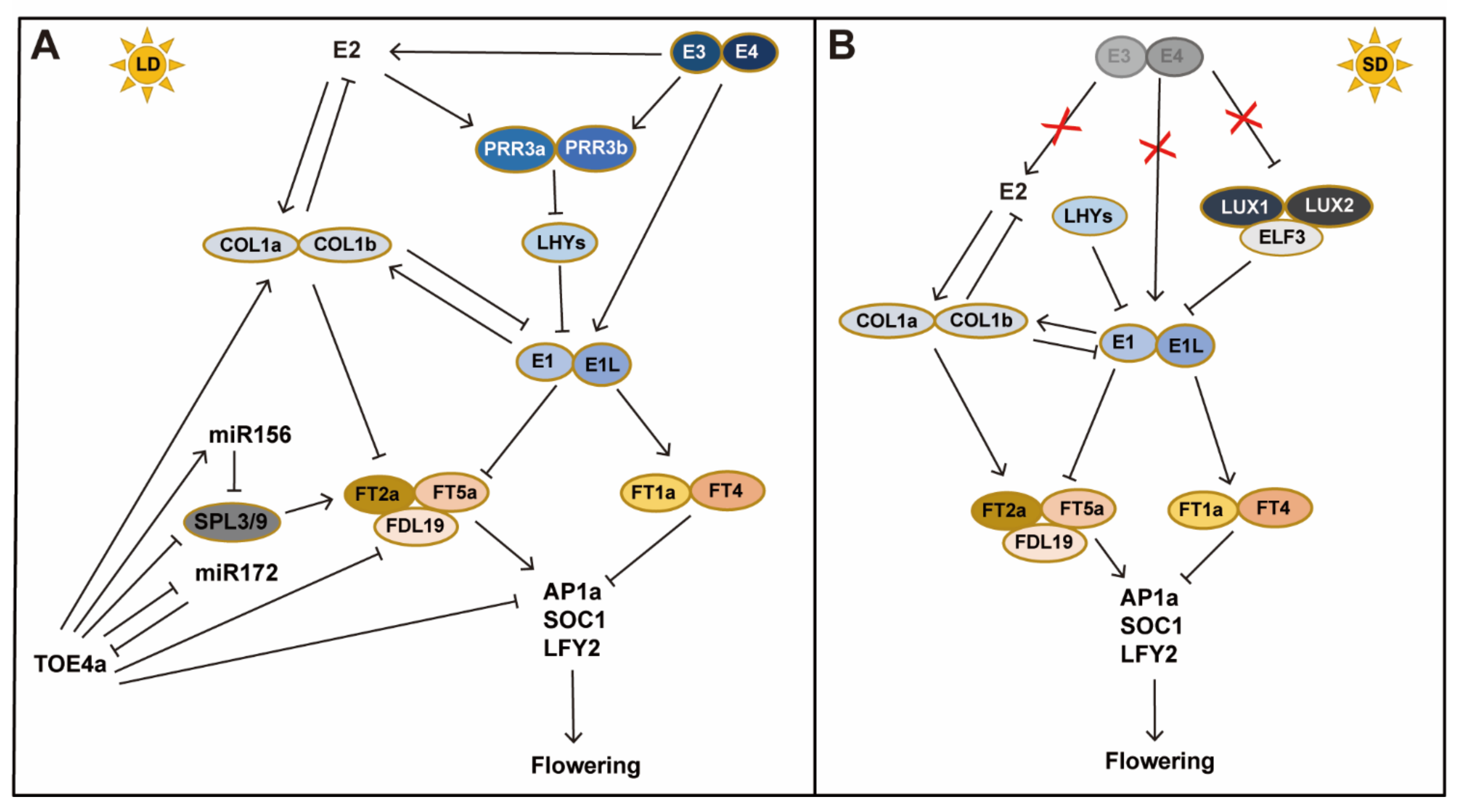
2.2. Core Components in the Photoperiod Pathway in Soybean
2.3. Molecular Mechanisms for Adaptation to Different Latitudes
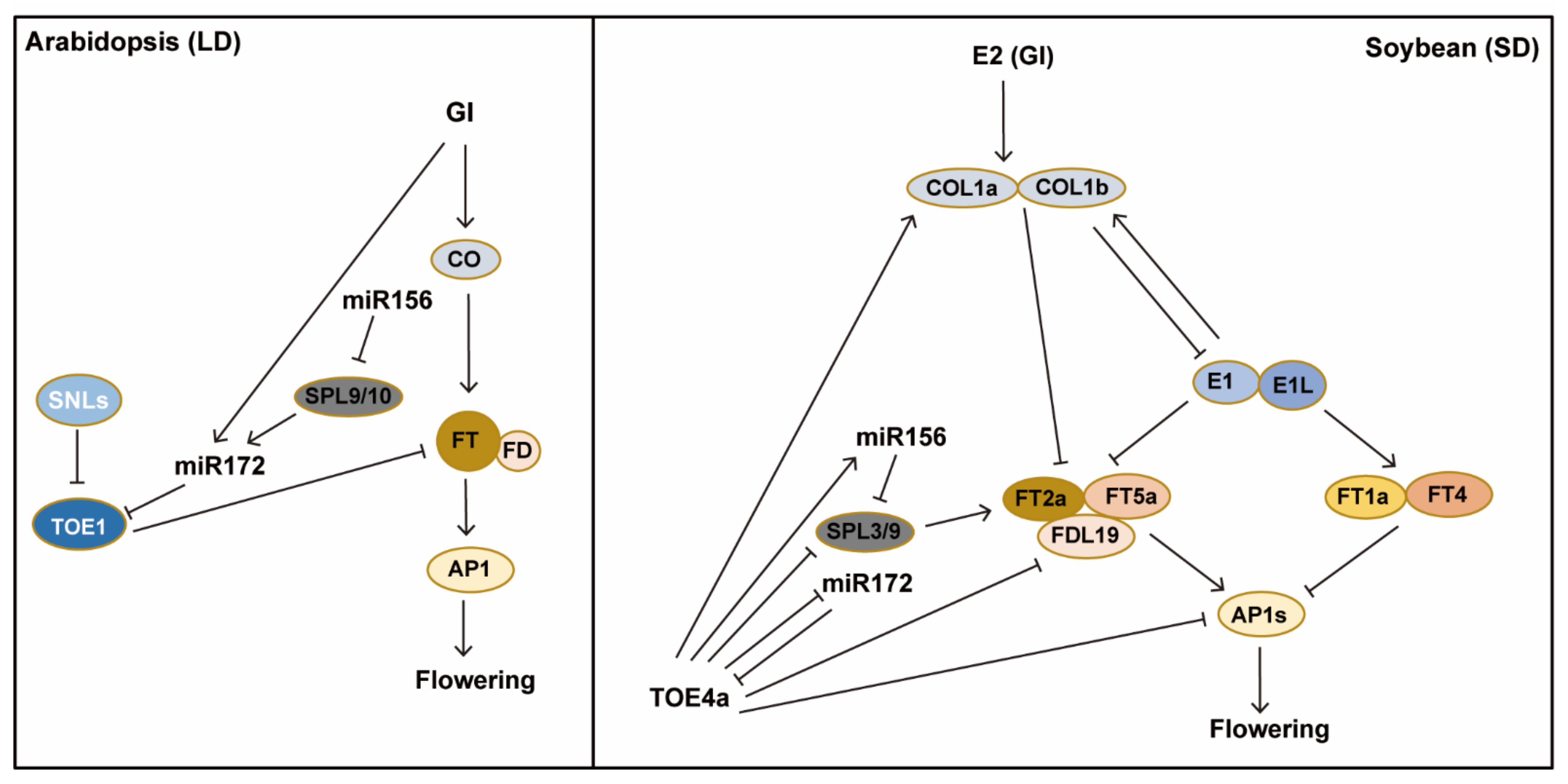
3. Conservation and Divergence of the Photoperiodic Regulation of Flowering in Soybean and Arabidopsis
Author Contributions
Funding
Conflicts of Interest
References
- Cerdan, P.D.; Chory, J. Regulation of flowering time by light quality. Nature 2003, 423, 881–885. [Google Scholar] [CrossRef]
- Lu, J.; Sun, J.; Jiang, A.; Bai, M.; Fan, C.; Liu, J.; Ning, G.; Wang, C. Alternate expression of CONSTANS-LIKE 4 in short days and CONSTANS in long days facilitates day-neutral response in Rosa chinensis. J. Exp. Bot. 2020, 71, 4057–4068. [Google Scholar] [CrossRef]
- Yu, J.W.; Rubio, V.; Lee, N.Y.; Bai, S.; Lee, S.Y.; Kim, S.S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Hoang, Q.T.N.; Han, Y.J.; Kim, J.I. Regulation of Photomorphogenic Development by Plant Phytochromes. Int. J. Mol. Sci. 2019, 20, 6165. [Google Scholar] [CrossRef]
- Sharrock, R.A.; Clack, T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002, 130, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Monte, E.; Alonso, J.M.; Ecker, J.R.; Zhang, Y.; Li, X.; Young, J.; Austin-Phillips, S.; Quail, P.H. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 2003, 15, 1962–1980. [Google Scholar] [CrossRef]
- Du, S.S.; Li, L.; Li, L.; Wei, X.; Xu, F.; Xu, P.; Wang, W.; Xu, P.; Cao, X.; Miao, L.; et al. Photoexcited Cryptochrome2 Interacts Directly with TOE1 and TOE2 in Flowering Regulation. Plant Physiol. 2020, 184, 487–505. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Panda, S.; Liu, X.L.; Strayer, C.A.; Wagner, D.R.; Kay, S.A. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 2001, 13, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- McWatters, H.G.; Bastow, R.M.; Hall, A.; Millar, A.J. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 2000, 408, 716–720. [Google Scholar] [CrossRef]
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 2013, 3, 671–677. [Google Scholar] [CrossRef]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognar, L.; Nagy, F.; Millar, A.J.; Amasino, R.M. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667. [Google Scholar] [CrossRef]
- Feke, A.; Vanderwall, M.; Liu, W.; Gendron, J.M. Functional domain studies uncover novel roles for the ZTL Kelch repeat domain in clock function. PLoS ONE 2021, 16, e0235938. [Google Scholar] [CrossRef]
- Nixdorf, M.; Hoecker, U. SPA1 and DET1 act together to control photomorphogenesis throughout plant development. Planta 2010, 231, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Al Khateeb, W.M.; Schroeder, D.F. DDB2, DDB1A and DET1 exhibit complex interactions during Arabidopsis development. Genetics 2007, 176, 231–242. [Google Scholar] [CrossRef][Green Version]
- Liu, X.L.; Covington, M.F.; Fankhauser, C.; Chory, J.; Wagner, D.R. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 2001, 13, 1293–1304. [Google Scholar] [PubMed]
- Shim, J.S.; Imaizumi, T. Circadian clock and photoperiodic response in Arabidopsis: From seasonal flowering to redox homeostasis. Biochemistry 2015, 54, 157–170. [Google Scholar] [CrossRef]
- Zoltowski, B.D.; Imaizumi, T. Structure and Function of the ZTL/FKF1/LKP2 Group Proteins in Arabidopsis. Enzymes 2014, 35, 213–239. [Google Scholar]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef]
- He, Y.; Chen, T.; Zeng, X. Genetic and Epigenetic Understanding of the Seasonal Timing of Flowering. Plant Commun. 2020, 1, 100008. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Ding, Z.; Doyle, M.R.; Amasino, R.M.; Davis, S.J. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 2007, 176, 1501–1510. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Niwa, Y.; Ito, S.; Nakamichi, N.; Mizoguchi, T.; Niinuma, K.; Yamashino, T.; Mizuno, T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 925–937. [Google Scholar] [CrossRef]
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Roussot, C.; Suarez-Lopez, P.; Corbesier, L.; Vincent, C.; Pineiro, M.; Hepworth, S.; Mouradov, A.; Justin, S.; Turnbull, C.; et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 2004, 131, 3615–3626. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Ruhl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [PubMed]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, A.; Valverde, F.; Pineiro, M.; Jarillo, J.A. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 2012, 24, 982–999. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Zhang, Y.C.; Li, Q.H.; Sang, Y.; Mao, J.; Lian, H.L.; Wang, L.; Yang, H.Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef]
- Laubinger, S.; Marchal, V.; Le Gourrierec, J.; Wenkel, S.; Adrian, J.; Jang, S.; Kulajta, C.; Braun, H.; Coupland, G.; Hoecker, U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 2006, 133, 3213–3222. [Google Scholar] [CrossRef] [PubMed]
- Schepens, I.; Duek, P.; Fankhauser, C. Phytochrome-mediated light signalling in Arabidopsis. Curr. Opin. Plant Biol. 2004, 7, 564–569. [Google Scholar] [CrossRef]
- Zuo, Z.; Liu, H.; Liu, B.; Liu, X.; Lin, C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 2011, 21, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Song, Y.H.; Imaizumi, T. LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Mol. Plant 2012, 5, 573–582. [Google Scholar] [CrossRef]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Tanigawa, Y.; Murakami, T.; Araki, T.; Nagatani, A. PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc. Natl. Acad. Sci. USA 2013, 110, 18017–18022. [Google Scholar] [CrossRef]
- Morris, K.; Thornber, S.; Codrai, L.; Richardson, C.; Craig, A.; Sadanandom, A.; Thomas, B.; Jackson, S. DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell 2010, 22, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Wigge, P.A. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 2007, 17, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Collani, S.; Neumann, M.; Yant, L.; Schmid, M. FT Modulates Genome-Wide DNA-Binding of the bZIP Transcription Factor FD. Plant Physiol. 2019, 180, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, X.; Yuan, W.; Schmitz, R.J.; He, Y. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell 2014, 28, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fu, X.; Wang, Y.; Liu, R.; He, Y. Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nat. Genet. 2018, 50, 1254–1261. [Google Scholar] [CrossRef]
- Yang, Z.; Qian, S.; Scheid, R.N.; Lu, L.; Chen, X.; Liu, R.; Du, X.; Lv, X.; Boersma, M.D.; Scalf, M.; et al. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat. Genet. 2018, 50, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Sgamma, T.; Jackson, A.; Muleo, R.; Thomas, B.; Massiah, A. TEMPRANILLO is a regulator of juvenility in plants. Sci. Rep. 2014, 4, 3704. [Google Scholar] [CrossRef]
- Castillejo, C.; Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Tian, S.; Xie, G.; Liu, R.; Wang, N.; Li, S.; He, Y.; Du, J. TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Mathieu, J.; Yant, L.J.; Murdter, F.; Kuttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009, 7, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Huang, F.; Yuan, W.; Tian, S.; Zheng, Q.; He, Y. SIN3 LIKE genes mediate long-day induction of flowering but inhibit the floral transition in short days through histone deacetylation in Arabidopsis. Plant J. 2019, 100, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Pose, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef]
- Lee, J.H.; Ryu, H.S.; Chung, K.S.; Pose, D.; Kim, S.; Schmid, M.; Ahn, J.H. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 2013, 342, 628–632. [Google Scholar] [CrossRef]
- Gu, X.; Le, C.; Wang, Y.; Li, Z.; Jiang, D.; Wang, Y.; He, Y. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013, 4, 1947. [Google Scholar] [CrossRef]
- Lv, X.; Zeng, X.; Hu, H.; Chen, L.; Zhang, F.; Liu, R.; Liu, Y.; Zhou, X.; Wang, C.; Wu, Z.; et al. Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant Cell 2021, 33, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Guo, Q.; Zha, P.; Lin, R. The chromatin-remodelling factor PICKLE interacts with CONSTANS to promote flowering in Arabidopsis. Plant Cell Environ. 2019, 42, 2495–2507. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Hu, Y.; Zhou, L.; Li, Y.; Hou, X. Temporal-Specific Interaction of NF-YC and CURLY LEAF during the Floral Transition Regulates Flowering. Plant Physiol. 2018, 177, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Gao, Z.; Wang, Y.; Chen, Z.; Zhang, W.; Huang, J.; Yu, H.; He, Y. The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 2018, 95, 17–29. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 2003, 13, 627–637. [Google Scholar] [CrossRef]
- Bu, Z.; Yu, Y.; Li, Z.; Liu, Y.; Jiang, W.; Huang, Y.; Dong, A.W. Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet. 2014, 10, e1004617. [Google Scholar] [CrossRef]
- Xu, Y.; Gan, E.S.; Zhou, J.; Wee, W.Y.; Zhang, X.; Ito, T. Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res. 2014, 42, 10960–10974. [Google Scholar] [CrossRef]
- An, Z.; Yin, L.; Liu, Y.; Peng, M.; Shen, W.H.; Dong, A. The histone methylation readers MRG1/MRG2 and the histone chaperones NRP1/NRP2 associate in fine-tuning Arabidopsis flowering time. Plant J. 2020, 103, 1010–1024. [Google Scholar] [CrossRef]
- Zhou, L.; Lu, Y.; Huang, J.; Sha, Z.; Mo, W.; Xue, J.; Ma, S.; Shi, W.; Yang, Z.; Gao, J.; et al. Arabidopsis CIB3 regulates photoperiodic flowering in an FKF1-dependent way. Biosci. Biotechnol. Biochem. 2021, 85, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Thakur, J.K.; Prasad, M. Histone acetylation dynamics regulating plant development and stress responses. Cell Mol. Life Sci. 2021, 78, 4467–4486. [Google Scholar] [CrossRef]
- Guo, Z.; Li, Z.; Liu, Y.; An, Z.; Peng, M.; Shen, W.H.; Dong, A.; Yu, Y. MRG1/2 histone methylation readers and HD2C histone deacetylase associate in repression of the florigen gene FT to set a proper flowering time in response to day-length changes. New Phytol. 2020, 227, 1453–1466. [Google Scholar] [CrossRef]
- Gu, X.; Wang, Y.; He, Y. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 2013, 11, e1001649. [Google Scholar] [CrossRef]
- Liu, L.; Li, C.; Teo, Z.W.N.; Zhang, B.; Yu, H. The MCTP-SNARE Complex Regulates Florigen Transport in Arabidopsis. Plant Cell 2019, 31, 2475–2490. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, C.; Hou, X.; Xi, W.; Shen, L.; Tao, Z.; Wang, Y.; Yu, H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012, 10, e1001313. [Google Scholar] [CrossRef] [PubMed]
- Karnik, R.; Zhang, B.; Waghmare, S.; Aderhold, C.; Grefen, C.; Blatt, M.R. Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell 2015, 27, 675–694. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Yu, H. Florigen trafficking integrates photoperiod and temperature signals in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1385–1398. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, L.; Shen, L.; Yu, H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat Plants 2016, 2, 16075. [Google Scholar] [CrossRef]
- Susila, H.; Juric, S.; Liu, L.; Gawarecka, K.; Chung, K.S.; Jin, S.; Kim, S.J.; Nasim, Z.; Youn, G.; Suh, M.C.; et al. Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science 2021, 373, 1137–1142. [Google Scholar] [CrossRef]
- Lin, X.; Liu, B.; Weller, J.L.; Abe, J.; Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 2021, 63, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Garner, W.W.; Allard, H.A. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J. Agric. Res. 1920, 18, 553–606. [Google Scholar] [CrossRef]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lu, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef] [PubMed]
- Bernard, R.L. Two Major Genes for Time of Flowering and Maturity in Soybeans. Crop Sci. 1971, 11, 242–244. [Google Scholar] [CrossRef]
- Buzzell, R.I. Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can. J. Genet. Cytol. 1971, 13, 703–707. [Google Scholar] [CrossRef]
- Buzzell, R.I.; Voldeng, H.D. Inheritance of insensitivity to long daylength. Soybean Genet. Newsl. 1980, 7, 26–29. [Google Scholar]
- Mcblain, B.; Bernard, R. A new gene affecting the time of flowering and maturity in soybeans. J. Hered. 1987, 78, 160–162. [Google Scholar] [CrossRef]
- Bonato, E.R.; Vello, N.A. E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 1999, 22, 229–232. [Google Scholar] [CrossRef]
- Cober, E.R.; Voldeng, H.D. Low R:FR Light Quality Delays Flowering of E7E7 Soybean Lines. Crop Sci. 2001, 41, 1823–1826. [Google Scholar] [CrossRef]
- Cober, E.R.; Voldeng, H.D. A New Soybean Maturity and Photoperiod-Sensitivity Locus Linked to and. Crop Sci. 2001, 41, 698–701. [Google Scholar] [CrossRef]
- Kong, F.J.; Nan, H.Y.; Cao, F.F.; Wang, J.L.; Yuan, S.J. A New Dominant Gene E9 Conditions Early Flowering and Maturity in Soybean. Crop Sci. 2014, 54, 2529–2535. [Google Scholar] [CrossRef]
- Samanfar, B.; Molnar, S.J.; Charette, M.; Schoenrock, A.; Dehne, F.; Golshani, A.; Belzile, F.; Cober, E.R. Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 2017, 130, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nan, H.; Chen, L.; Fang, C.; Lu, S. A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed. 2019, 39, 70. [Google Scholar] [CrossRef]
- Ray, J.D.; Hinson, K.; Mankono, J.; Malo, M.F. Genetic Control of a Long-Juvenile Trait in Soybean. Crop Sci. 1995, 35. [Google Scholar] [CrossRef]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef]
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar] [CrossRef]
- Li, X.; Fang, C.; Yang, Y.; Lv, T.; Su, T.; Chen, L.; Nan, H.; Li, S.; Zhao, X.; Lu, S.; et al. Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol. 2021, 31, 3755–3767.e4. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Takeshima, R.; Zhao, C.; Liu, B.; Jun, A.; Kong, F. Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 2017, 68, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Cober, E.R.; Tanner, J.W.; Voldeng, H.D. Genetic Control of Photoperiod Response in Early-Maturing, Near-Isogenic Soybean Lines. Crop Sci. 1996, 36, 601–605. [Google Scholar] [CrossRef]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Ishimoto, M.; Anai, T.; et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Cober, E.R.; Tanner, J.W.; Voldeng, H.D. Soybean Photoperiod-Sensitivity Loci Respond Differentially to Light Quality. Crop Sci. 1996, 36, 606–610. [Google Scholar] [CrossRef]
- Watanabe, S.; Harada, K.; Abe, J. Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed. Sci. 2012, 61, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, Y.; Gao, H.; Qiu, L.; Chang, R.; Chen, S.; He, C. Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 2016, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Takeshima, R.; Zhu, J.; Xu, M.; Sato, M.; Watanabe, S.; Kanazawa, A.; Liu, B.; Kong, F.; Yamada, T.; et al. A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 2016, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Shixiang, L.; Wu, H.; Zhang, Y.; Zhang, X.; Yang, J.; Wang, Y.; Yang, G.; Qiu, H.; Cui, T. Diurnal Expression Pattern, Allelic Variation, and Association Analysis Reveal Functional Features of the E1 Gene in Control of Photoperiodic Flowering in Soybean. PLoS ONE 2015, 10, e0135909. [Google Scholar] [CrossRef]
- Zhai, H.; Lu, S.; Liang, S.; Wu, H.; Zhang, X.; Liu, B.; Kong, F.; Yuan, X.; Li, J.; Xia, Z. GmFT4, a Homolog of FLOWERING LOCUS T, Is Positively Regulated by E1 and Functions as a Flowering Repressor in Soybean. PLoS ONE 2014, 9, e89030. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Liu, B.; Xia, Z.; Sato, S.; Kim, B.M.; Watanabe, S.; Yamada, T.; Tabata, S.; Kanazawa, A.; Harada, K.; et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010, 154, 1220–1231. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Sedivy, E.J.; Price, W.B.; Haider, W.; Hanzawa, Y. Evolutionary trajectories of duplicated FT homologues and their roles in soybean domestication. Plant J. 2017, 90, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Cao, D.; Zhang, D.; Li, Y.; Lu, S.; Tang, L.; Yuan, X.; Liu, B.; Kong, F. GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS ONE 2014, 9, e97669. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jia, Z.; Cao, D.; Jiang, B.; Wu, C.; Hou, W.; Liu, Y.; Fei, Z.; Zhao, D.; Han, T. GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 2011, 6, e29238. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cai, Y.; Qu, M.; Wang, L.; Sun, H.; Jiang, B.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; et al. Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ. 2020, 43, 934–944. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Z.; Liu, Y.; Liu, T.; Li, Q.; Ji, Y.; Li, C.; Fang, C.; Wang, M.; Wu, M.; et al. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef]
- Cao, D.; Li, Y.; Lu, S.; Wang, J.; Nan, H.; Li, X.; Shi, D.; Fang, C.; Zhai, H.; Yuan, X.; et al. GmCOL1a and GmCOL1b Function as Flowering Repressors in Soybean Under Long-Day Conditions. Plant Cell Physiol. 2015, 56, 2409–2422. [Google Scholar] [CrossRef]
- Xu, M.; Yamagishi, N.; Chen, Z.; Takeshima, R.; Abe, J. Soybean-specific E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 2015, 168, 1735–1746. [Google Scholar] [CrossRef] [PubMed]
- Qian-Hao, Z.; Chris, A. Helliwell, Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 2010, 62, 487–495. [Google Scholar]
- Cao, D.; Li, Y.; Wang, J.; Nan, H.; Wang, Y.; Lu, S.; Jiang, Q.; Li, X.; Shi, D.; Fang, C. GmmiR156b overexpression delays flowering time in soybean. Plant Mol. Biol. 2015, 89, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Cao, D.; Huang, Z.; Wang, J.; Lu, S.; Xu, Y.; Liu, B.; Kong, F.; Yuan, X. Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol. Biol. 2015, 88, 343–355. [Google Scholar] [CrossRef]
- Xu, M.; Xu, Z.; Liu, B.; Kong, F.; Tsubokura, Y.; Watanabe, S.; Xia, Z.; Harada, K.; Kanazawa, A.; Yamada, T.; et al. Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 2013, 13, 91. [Google Scholar] [CrossRef]
- Jiang, B.; Nan, H.; Gao, Y.; Tang, L.; Yue, Y.; Lu, S.; Ma, L.; Cao, D.; Sun, S.; Wang, J.; et al. Allelic combinations of soybean maturity Loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS ONE 2014, 9, e106042. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Li, M.W.; Liu, W.; Lam, H.M.; Gendron, J.M. Characterization of Two Growth Period QTLs Reveals Modification of PRR3 Genes During Soybean Domestication. Plant Cell Physiol. 2019, 60, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Yamashino, T.; Mizuno, T. Characterization of circadian-associated APRR3 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L.; et al. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, L.; Cao, X.; Liu, L.; Jiang, B.; Zhang, C.; Jia, H.; Lyu, X.; Su, Y.; Cai, Y.; et al. Cotyledons facilitate the adaptation of early-maturing soybean varieties to high-latitude long-day environments. Plant Cell Environ. 2021, 44, 2551–2564. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J. locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chao, F.; Xu, M.; Zhang, F.; Lu, S.; Nan, H.; Tong, S.; Li, S.; Zhao, X.; Kong, L. Quantitative Trait Locus Mapping of Soybean Maturity Gene E6. Crop Sci. 2017, 57, 2547–2554. [Google Scholar] [CrossRef]
- Fang, C.; Chen, L.; Nan, H.; Kong, L.; Lu, S. Rapid identification of consistent novel QTLs underlying long-juvenile trait in soybean by multiple genetic populations and genotyping-by-sequencing. Mol. Breed. 2019, 39, 1–11. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, N.; Jiang, B.; Li, M.; Wang, H.; Jiang, Z.; Pan, H.; Xia, Q.; Ma, Q.; Han, T.; et al. A Single Nucleotide Deletion in J. Encoding GmELF3 Confers Long Juvenility and Is Associated with Adaption of Tropic Soybean. Mol. Plant 2017, 10, 656–658. [Google Scholar] [CrossRef]
- Fang, C.; Liu, J.; Zhang, T.; Su, T.; Li, S.; Cheng, Q.; Kong, L.; Li, X.; Bu, T.; Li, H.; et al. A recent retrotransposon insertion of J. caused E6 locus facilitating soybean adaptation into low latitude. J. Integr. Plant Biol. 2021, 63, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Bu, T.; Lu, S.; Wang, K.; Dong, L.; Li, S.; Xie, Q.; Xu, X.; Cheng, Q.; Chen, L.; Fang, C.; et al. A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef]
- Hicks, K.A.; Albertson, T.M.; Wagner, D.R. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 2001, 13, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Dong, L.; Su, T.; Li, T.; Gan, Z.; Nan, H.; Lu, S.; Fang, C.; Kong, L.; Li, H.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 2019, 19, 562. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, L.; Zeng, L.; Zhang, C.; Ma, H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015, 29, 975–987. [Google Scholar] [CrossRef]
- Chen, L.; Nan, H.; Kong, L.; Yue, L.; Yang, H.; Zhao, Q.; Fang, C.; Li, H.; Cheng, Q.; Lu, S.; et al. Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 2020, 62, 1868–1879. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Komiya, R.; Ikegami, A.; Tamaki, S.; Yokoi, S.; Shimamoto, K. Hd3a and RFT1 are essential for flowering in rice. Development 2008, 135, 767–774. [Google Scholar] [CrossRef]
- Chardon, F.; Damerval, C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Zhai, H.; Wu, H.; Xu, K.; Watanabe, S.; Harada, K. The Synchronized Efforts to Decipher the Molecular Basis for Soybean Maturity Loci E1, E2, and E3 That Regulate Flowering and Maturity. Front. Plant Sci. 2021, 12, 632754. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Y.; Zhang, M.; Zhan, X.; Shen, X.; Yu, P.; Chen, D.; Liu, Q.; Sinumporn, S.; Hussain, K.; et al. The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Biophys. Res. Commun. 2018, 495, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; You, C.; Li, C.; Long, T.; Chen, G.; Byrne, M.E.; Zhang, Q. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12915–12920. [Google Scholar] [CrossRef] [PubMed]

| Gene in Soybean | Function | Arabidopsis Ortholog | Function | ||
|---|---|---|---|---|---|
| E1 | Inhibition | [85] | None | ||
| E1La | Inhibition | [119] | None | ||
| E1Lb | Inhibition | [106,119] | None | ||
| GmGI (E2) | Inhibition | [106] | GI | Promotion | [25,28] |
| GmphyA3 (E3) | Inhibition | [102] | PHYA | Promotion | [5] |
| GmphyA2 (E4) | Inhibition | [103] | PHYA | Promotion | [5] |
| GmFT2a (E9) | Promotion | [93,108,111] | FT | Promotion | [43,44] |
| GmFT5a | Promotion | [111] | FT | Promotion | [43,44] |
| GmFT4 (E10) | Inhibition | [94,110] | FT | Promotion | [43,44] |
| GmFT1a | Inhibition | [116] | FT | Promotion | [43,44] |
| GmCOL1a/1b | Inhibition | [118] | CO | Promotion | [26] |
| miR156 | Inhibition | [121] | miR156 | Inhibition | [54,55] |
| miR172 | Promotion | [122] | miRNA172 | Promotion | [53,54] |
| GmTOE4a | Inhibition | [122] | TOE1 | Inhibition | [7,141] |
| GmPRR3a (Tof11) | Inhibition | [97,125] | APRR3 | Inhibition | [127] |
| GmPRR3b (Tof12) | Inhibition | [97,125] | APRR3 | Inhibition | [127] |
| GmELF3 (J) | Promotion | [130,133] | ELF3 | Inhibition | [137] |
| GmLUX1 | Promotion | [135] | LUX | Inhibition | [136] |
| GmLUX2 | Promotion | [135] | LUX | Inhibition | [136] |
| LHY1a (Tof16) | Promotion | [98] | LHY | Inhibition | [139] |
| LHY1b/c/d | Promotion | [98] | LHY | Inhibition | [139] |
| GmFDL19 | Promotion | [113] | FD | Promotion | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Yin, M.; He, Y. Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean. Int. J. Mol. Sci. 2022, 23, 466. https://doi.org/10.3390/ijms23010466
Luo X, Yin M, He Y. Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean. International Journal of Molecular Sciences. 2022; 23(1):466. https://doi.org/10.3390/ijms23010466
Chicago/Turabian StyleLuo, Xiao, Mengnan Yin, and Yuehui He. 2022. "Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean" International Journal of Molecular Sciences 23, no. 1: 466. https://doi.org/10.3390/ijms23010466
APA StyleLuo, X., Yin, M., & He, Y. (2022). Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean. International Journal of Molecular Sciences, 23(1), 466. https://doi.org/10.3390/ijms23010466






