Abstract
Recent investigations have shown the possibility of artificial induction of RNA interference (RNAi) via plant foliar treatments with naked double-stranded RNA (dsRNA) to silence essential genes in plant fungal pathogens or to target viral RNAs. Furthermore, several studies have documented the downregulation of plant endogenous genes via external application of naked gene-specific dsRNAs and siRNAs to the plant surfaces. However, there are limited studies on the dsRNA processing and gene silencing mechanisms after external dsRNA application. Such studies would assist in the development of innovative tools for crop improvement and plant functional studies. In this study, we used exogenous gene-specific dsRNA to downregulate the gene of chalcone synthase (CHS), the key enzyme in the flavonoid/anthocyanin biosynthesis pathway, in Arabidopsis. The nonspecific NPTII-dsRNA encoding the nonrelated neomycin phosphotransferase II bacterial gene was used to treat plants in order to verify that any observed effects and processing of AtCHS mRNA were sequence specific. Using high-throughput small RNA (sRNA) sequencing, we obtained six sRNA-seq libraries for plants treated with water, AtCHS-dsRNA, or NPTII-dsRNA. After plant foliar treatments, we detected the emergence of a large number of AtCHS- and NPTII-encoding sRNAs, while there were no such sRNAs after control water treatment. Thus, the exogenous AtCHS-dsRNAs were processed into siRNAs and induced RNAi-mediated AtCHS gene silencing. The analysis showed that gene-specific sRNAs mapped to the AtCHS and NPTII genes unevenly with peak read counts at particular positions, involving primarily the sense strand, and documented a gradual decrease in read counts from 17-nt to 30-nt sRNAs. Results of the present study highlight a significant potential of exogenous dsRNAs as a promising strategy to induce RNAi-based downregulation of plant gene targets for plant management and gene functional studies.
1. Introduction
RNA interference (RNAi) is a natural regulatory mechanism that functions in plants and other eukaryotes to defend them against pathogens and transposable elements and to downregulate the expression of endogenous protein-coding genes [,,]. RNAi involves sequence-specific degradation of target mRNAs or translation inhibition induced by small RNAs (sRNAs), which are generally divided into two main categories, microRNA (miRNAs) and small interfering RNA (siRNAs) pathways [,]. The siRNA-mediated RNAi pathway is generally based on the recognition and processing of dsRNA precursors that are converted into siRNAs, i.e., 20–24-nucleotide (nt)-long RNA duplexes, by a ribonuclease DICER. These siRNAs are then incorporated into the RNA-induced silencing complex (RISC) to sequester the single-stranded siRNA guide strand and induce gene silencing in a sequence-specific manner via cleavage, destabilization, or translation repression of homologous mRNAs.
It is known that different sRNAs that are generated in plant cells can spread throughout the plant via the plant vascular system and can be transferred to plant pathogens via extracellular vesicles [,,]. Recent studies have indicated that plants can uptake and process externally applied synthetic dsRNAs [,,,,,] or siRNAs [,,,,] that have been artificially designed and supplied to activate RNAi in plants and plant pathogens and silence target genes in a sequence-specific manner. Both plants and infecting pathogens were capable of dsRNA uptake, and this eventually triggered RNAi-mediated silencing of the pathogen virulence-related genes or targeted virus RNA for degradation [,,,,,]. These dsRNAs were capable of systemic spreading within the plant and were transferred to the pathogens/pest. Currently, external dsRNA or siRNA application is being developed as a promising tool for plant pathogen protection and is currently termed spray-induced gene silencing (SIGS) [,,]. It appears that the application of dsRNA or siRNAs to external plant surfaces is a promising instrument of plant RNA-based “vaccination” that can become an alternative to transgenic plants and virus-induced gene silencing (VIGS) [,,,].
Currently, there is a high number of studies reporting on successful external dsRNA application to the plant surfaces for RNAi induction and plant pathogen protection (reviewed in [,,]). However, there is a limited number of investigations reporting on the silencing of plant endogenous genes after exogenous dsRNA or siRNA application. According to our literature analysis, there are five investigations and a patent that showed that external plant treatments with naked dsRNAs led to downregulation of plant endogenous genes, including silencing of the 3-phosphate synthase (EPSPS) gene in tobacco and amaranth leaves [], MYB1 gene in the orchid flower buds [], Mob1A, WRKY23, and Actin genes in Arabidopsis and rice [], two sugar transporter genes STP1 and STP2 in tomato seedlings [], a downy mildew susceptibility gene LBDIf7, and a glutathione S-transferase GST40 gene in grapevine [,]. According to the data, external plant dsRNA treatments led to the dsRNA uptake, reduced mRNA levels of the gene targets, and some phenotypic or biochemical changes.
We also found that there were quite limited studies where RNA-seq or other approaches would be applied to analyze the processing of exogenous dsRNAs applied to plant surfaces, while such studies are required to uncover the molecular mechanisms of exogenously induced RNAi. Several studies performed RNA-seq to detect virus-specific sRNAs occurring in plant tissues with and without dsRNA treatments [,] or to analyze the sRNA population in plant leaves treated with dsRNA targeting fungal genes [,,]. Using RNA-seq, Uslu et al. [] found that naked GFP-dsRNAs and GFP-hpRNAs did not induce GFP transgene silencing in N. benthamiana and were not processed into siRNAs. However, high-pressure spraying of GFP-encoding synthetic siRNAs was efficient in inducing GFP silencing and GFP-siRNA amplification through the process of transitivity in N. benthamiana according to RNA-seq analysis [,]. To the best of our knowledge, there were no studies analyzing dsRNA processing and post-treatment siRNA fraction after exogenously induced silencing of a plant endogenous gene. Such studies would allow the development of promising tools to manage plant gene expression for crop improvement and plant gene functional studies.
In this study, we aimed to analyze sRNAs occurring after plant treatments with dsRNAs targeting the gene of chalcone synthase (CHS), the key enzyme in the flavonoid/anthocyanin biosynthesis pathway, in Arabidopsis. Recently, we demonstrated that exogenous dsRNA and siRNA can efficiently downregulate mRNA levels of several genes involved in anthocyanin biosynthesis in A. thaliana, including the AtCHS gene, AtMYBL2 gene encoding an R3-type single-MYB protein, and AtANAC032 gene encoding a NAC-type transcription factor []. The plants treated with the AtCHS-specific dsRNAs exhibited lowered anthocyanin content and lighter leaf color in comparison with A. thaliana treated with water or NPTII-dsRNA, while the plants treated with dsRNAs targeting the MYBL2 and ANAC032 transcriptional repressors of anthocyanin biosynthesis accumulated higher levels of anthocyanins and showed darker leaf color []. In the present study, we showed that plant foliar treatments with both AtCHS- and NPTII-dsRNAs led to the emergence of a large number of AtCHS- and NPTII-specific sRNAs that have not been detected after control water treatment. The data revealed that the exogenous AtCHS-encoding dsRNA downregulated mRNA levels of the AtCHS gene and was processed into specific siRNAs, while NPTII-dsRNA did not induce AtCHS silencing and was presumably degraded. Results of the present study highlight a significant potential of exogenous dsRNAs as a promising strategy to induce RNAi-based downregulation of plant gene targets for plant management and gene functional studies.
2. Results
To analyze the effect of exogenous dsRNAs on the composition sRNA population, PCR and the in vitro transcription protocol were used to produce dsRNA of the CHS gene of A. thaliana and the bacterial NPTII gene. We synthesized the nonrelated NPTII-dsRNAs to verify whether any observed influence of exogenous dsRNAs on AtCHS mRNA levels and sRNA profiles were sequence specific. Large fragments of AtCHS cDNA and NPTII gene were amplified by PCR (Figure 1a). The obtained PCR products, containing T7 promoters at both ends, were used as templates for in vitro transcription (Figure 1b). For external application, the synthesized AtCHS- and NPTII-dsRNAs were diluted in water to a final concentration of 0.35 µg/µL. The AtCHS- and NPTII-dsRNAs (100 µL of each dsRNA per individual plant, i.e., 35 µg) or water (100 µL of sterile filtered water per individual plant) were applied on the leaf surface (Figure 1c) on both the adaxial and abaxial sides of four-week-old rosettes of A. thaliana by spreading with sterile individual soft brushes [,]. Two whole four-week-old rosettes of A. thaliana were treated per each type of exposure: water (rosettes WC-1a, WC-1b), AtCHS-dsRNA (rosettes dsCHS-3a, dsCHS-3b), and NPTII-dsRNA treatment (rosettes NPTII-4a, NPTII-4b) (Figure 1c). Then, we incubated the treated A. thaliana rosettes under anthocyanin-inducing conditions (+7 °C, and 23 h light) for two days, in order to induce AtCHS expression and anthocyanin biosynthesis, since under standard cultivation conditions, AtCHS mRNA and anthocyanin levels were low [].
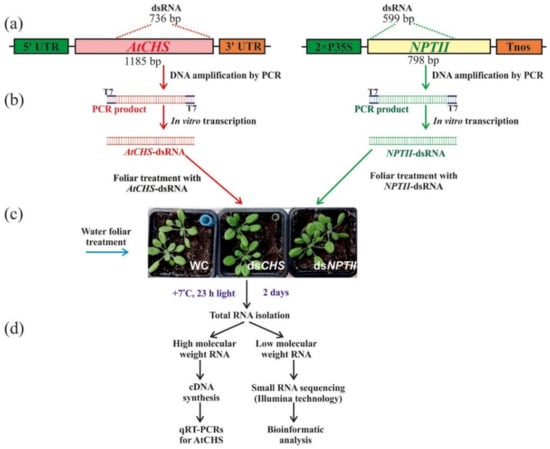
Figure 1.
Schematic representation of the experiment conducted in the study to verify the effects of external dsRNA treatments in Arabidopsis thaliana on small RNAs profiles and AtCHS gene expression. (a) Representation of AtCHS- and NPTII-coding regions with positions of the AtCHS- and NPTII-specific dsRNA; (b) PCRs with the T7 promoter appended to both PCR primers for AtCHS and NPTII partial amplification, followed by in vitro dsRNA production; (c) plant foliar treatments with AtCHS- and NPTII-specific dsRNA; (d) low- and high-molecular-weight RNA isolation and processing. 2xP35S—the double 35S promoter of the cauliflower mosaic virus (CaMV); AtCHS—the chalcone synthase gene from A. thaliana; NPTII—the neomycin phosphotransferase II (NPTII) gene; Tnos—nopaline synthase terminator. T7–T7 promoter.
Importantly, we treated the four-week-old rosettes of A. thaliana at a late day time (21:00–21:30) under low soil moisture conditions, since appropriate plant age, late day time, and low soil moisture (at the time of dsRNA application) were important parameters for successful gene suppression in transgenic A. thaliana according to our analysis [,]. An analysis of the dsRNA concentration effect on transgene silencing in A. thaliana was performed previously [], indicating that 35 µg of a transgene-encoding dsRNA resulted in the highest transgene silencing efficiency as compared to other dsRNA concentrations. Then, high-molecular-weight (HMW) and low-molecular-weight (LMW) RNA fractions were isolated two days after the dsRNA treatment to study whether exogenous application of the naked AtCHS and NPTII-dsRNAs on the foliar surface of wild-type A. thaliana could lead to any changes in the mRNA transcript levels of AtCHS gene and sRNAs in comparison with the control water treatment (Figure 1d). HMW RNA was used for cDNA synthesis and qRT-PCR analysis of the AtCHS gene expression, while LMW RNA was used for small RNA sequencing using Illumina technology.
First, we analyzed whether exogenous AtCHS and NPTII-dsRNAs affect mRNA levels of the AtCHS gene (Figure 2). Using qRT-PCR, we showed that the exogenous plant treatment with AtCHS-dsRNA significantly decreased AtCHS mRNA levels by four times, while the nonrelated NPTII-dsRNA did not significantly affect AtCHS expression (Figure 2).
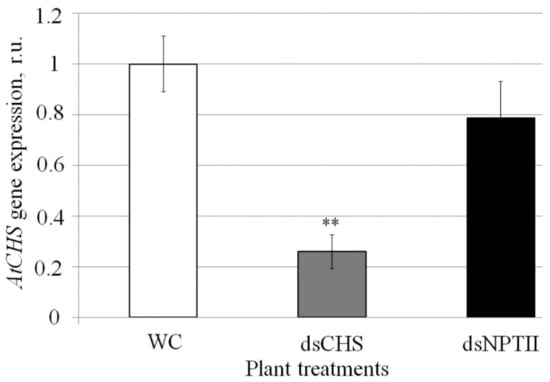
Figure 2.
The effect of external AtCHS- and NPTII-encoding dsRNAs on AtCHS mRNA level in Arabidopsis thaliana analyzed by quantitative real-time PCR. WC—A. thaliana treated with sterile water; dsCHS—A. thaliana treated with AtCHS-dsRNA; dsNPTII—A. thaliana treated with NPTII-dsRNA; dpt—days post-treatment. A. thaliana plants were grown under anthocyanin-inducing (+7 °C, 23 h light) conditions for two days after treatment with sterile water or synthetic dsRNA. qRT-PCR data are presented as the mean ± SE. **—significantly different from WC at p ≤ 0.01 according to Student’s t-test.
Next, using the Illumina NovaSeq 6000 instrument, we obtained six RNA-seq libraries: two for water-treated plants, two for AtCHS-dsRNA-treated plants, and two for NPTII-dsRNA-treated plants (Table 1). In total, we obtained 526,174,120 reads, including 168,404,990 reads for the water-treated plants, 198,443,550 reads for AtCHS-dsRNA-treated plants, and 159,325,580 reads for NPTII-dsRNA-treated plants. After removing low-quality reads and unqualified reads (adapters, reads shorter than 17 and longer than 30 nucleotides, nongenomic reads (except for NPTII), ribosomal and chloroplast DNA sequences), 5.9–14.1 million clean reads were obtained for each sample (Table 1). Subsequently, the length of the sRNAs in each sample was determined. The distribution of the total sRNA population in length is shown in Figure 3. In all samples, 21-nt, 23-nt, and 24-nt sRNAs exhibited the highest abundance, while the frequency of other sRNA size classes was considerably lower. We also noticed that plant treatment with AtCHS-dsRNA resulted in a considerably increased level of 21-nt sRNAs, while the content of 23-nt and 24-nt sRNAs was in turn decreased in comparison to other treatments (Figure 3). After preprocessing, we obtained 56,944,787 reads, which were about 11% of the initial number. As a result, for further analysis, we used 25,247,201 reads for water treatment, 12,357,084 reads for dsCHS treatment, and 19,340,502 reads for dsNPTII treatment.

Table 1.
Samples analyzed by sRNA-seq and read numbers obtained after high-throughput sequencing (Illumina NovaSeq 6000 instrument). WC—A. thaliana treated with sterile water; dsCHS—A. thaliana treated with AtCHS-dsRNAs; dsNPTII—A. thaliana treated with NPTII-dsRNA.
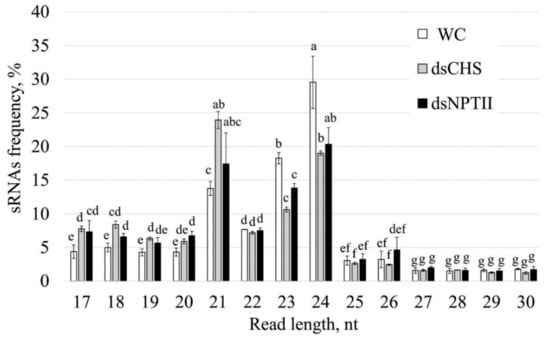
Figure 3.
Length size distribution of total small RNAs in Arabidopsis thaliana after treatment with water or dsRNA. WC—A. thaliana plants treated with sterile water; dsCHS—A. thaliana treated with AtCHS-dsRNA; dsNPTII—A. thaliana treated with NPTII-dsRNA. The data are presented as the mean ± SE. Means followed by the same letter were not different using Student’s t-test. p < 0.05 was considered statistically significant.
Then, we analyzed the abundance and composition of 17–30-nt sRNAs aligned to the AtCHS and NPTII sequences (Figure 4). The analysis revealed that there were 1.75% of AtCHS- specific sRNAs and 0.85% of NPTII-specific sRNAs in A. thaliana treated with the corresponding dsRNAs among all obtained reads after preprocessing, while there were no such sequences in the water-treated control plants (Figure 4a). The data revealed that the length size distribution of the sRNAs mapping to AtCHS or NPTII (Figure 4) was different from the length size distribution of the total sRNA fraction (Figure 3). A large fraction of the AtCHS- or NPTII-specific sRNAs was presented by small-sized sRNAs ranging from 17 to 20 nt and reached more than half of all target-specific sRNAs (Figure 4). The data revealed that the shorter the length of both the AtCHS- or NPTII-specific sRNAs, the more read counts were detected, and the proportion of the AtCHS- or NPTII-specific sRNAs gradually decreased from the 17-nt sRNAs to 30-nt sRNAs.
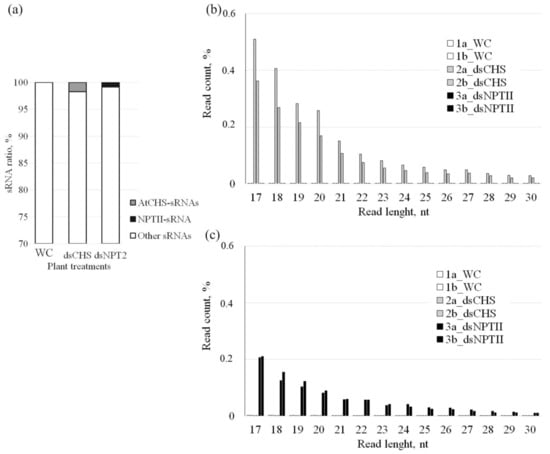
Figure 4.
The effect of AtCHS-dsRNA and NPTII-dsRNA on the proportion and length size distribution of AtCHS-encoding 17–30-nt sRNAs among all detected small RNAs. (a) The proportion of AtCHS- and NPTII-encoding 17–30-nt sRNAs among all detected small RNAs. WC—A. thaliana plants treated with sterile water; dsCHS—A. thaliana treated with AtCHS-dsRNA; dsNPTII—A. thaliana treated with NPTII-dsRNA; (b) length size distribution of AtCHS-encoding 17–30-nt sRNAs; (c) length size distribution of NPTII-encoding 17–30-nt sRNAs. 1a_WC, 1b_WC—two A. thaliana plants treated with sterile water; 2a_dsCHS, 2b_dsCHS—two A. thaliana plants treated with AtCHS-dsRNA; 3a_dsNPTII, 3b_dsNPTII—two A. thaliana plants treated with NPTII-dsRNA.
We also analyzed the sRNA distribution patterns across both the CHS and NPTII genes (Figure 5a,b). The majority of sRNAs were not uniformly mapped along both the CHS and NPTII genes and covered the gene sequences unevenly with high frequencies of reads at particular positions, including 6–8 peak read count positions for the sense strand and 5–6 peak read count positions for the antisense strand. The AtCHS gene sense sequence was covered by 8 peaks of read counts (528–559 nt, 714–745 nt, 745–776 nt, 776–807 nt, 869–900 nt, 962–993 nt, 993–1024 nt, 1055–1086 nt), while the antisense sequence was covered by 6 peaks (776–807 nt, 838–869 nt, 931–962 nt, 1086–1117 nt, 1148–1179 nt) (Figure 5a). Similarly, the NPTII sense sequence was covered by 7 peaks (1–31 nt, 31–61 nt, 91–121 nt, 151–181 nt, 241–271 nt, 331–361 nt, 451–481 nt). In the mentioned intervals, the number of reads prevailed over read counts at other positions. For example, there are almost no sRNAs mapping to the sense NPTII strand at 391–451 nt position (Figure 5a). The antisense sequence of the NPTII gene was covered by a fewer number of peaks with 5 peaks of read counts (1–31 nt, 61–91 nt, 121–151 nt, 391–421 nt, 481–511 nt). Notably, the peak positions on the sense strand did not coincide with the peak positions on the antisense strand (Figure 5). Most reads mapped to the sense strands of both the AtCHS and NPTII genes. For the AtCHS gene, there were 1.4–1.5 times more sequences identical to the sense than for the antisense gene strand (Figure 5a). Similarly, for the NPTII gene, reads mapped to the sense strand 2.5–2.7 times higher than to the antisense NPTII strand (Figure 5b).
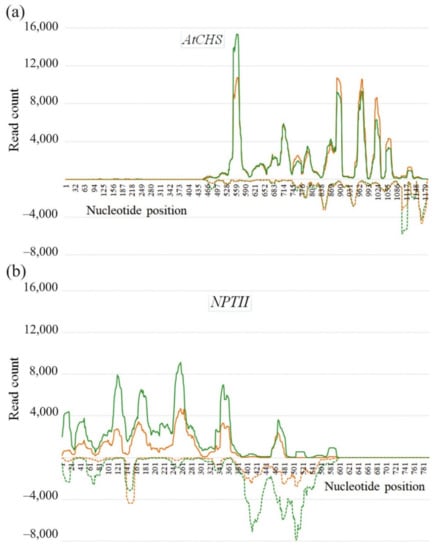
Figure 5.
Strand-specific distribution of the 17–30-nt AtCHS- and NPTII-encoding sRNAs detected in Arabidopsis thaliana after dsRNA treatments along the AtCHS (a) and NPTII (b) gene coding regions. Read depth was counted by the number of reads on each position. Sense strand is plotted with a solid line above the x-axis, and antisense strand is plotted with a dotted line below the x-axis. Green and orange colors indicate two biological replicates.
3. Discussion
The increasing human population and discussions about the safety of transgenic plants promote the development of new strategies to regulate plant properties without genomic manipulations. Plant foliar treatment with dsRNA has considerable potential as an innovative approach for gene regulation in plants and for plant pathogen control to ensure low-risk and environmentally friendly plant management. Recently, we demonstrated that foliar treatments of A. thaliana with gene-specific dsRNAs and siRNAs significantly reduced expression of the target genes, including AtCHS and two genes AtMybL2 and AtANAC032 encoding transcriptional repressors of anthocyanin biosynthesis []. In this study, before sRNA-seq, we analyzed AtCHS mRNA levels and confirmed that AtCHS mRNA levels sharply decreased after external plant treatments with the AtCHS-specific dsRNA, while AtCHS expression was not considerably affected after application of the nonspecific NPTII-dsRNA.
In the current literature, there is a lack of data on the plant uptake and processing of exogenously applied dsRNAs. Using confocal microscopy and some other approaches, several investigations have shown that exogenous dsRNA is taken up by the plant and is transferred via the plant vascular system [,,,,]. According to the studies, these dsRNAs have been systemically translocated within the plant and were also detectable inside infecting fungal pathogens or insect pests [,]. However, the mechanism of uptake of exogenously applied dsRNAs and siRNAs still remains unclear. Song et al. [] found that exogenous dsRNA was absorbed more efficiently via the wounded than the healthy surface of wheat coleoptiles. Based on their microscopic studies, the authors proposed that the used dsRNA entered the damaged cells of the wounded coleoptile surface and was transferred via the tracheary elements. Confocal microscopy revealed that fluorescently labeled dsRNAs applied onto the plant leaves can be detected in leaf veins, parenchyma cells, and companion cells, as well as in trichomes and stomata [,]. Therefore, there is a possibility of dsRNA stomatal uptake in plants. It is likely that externally applied synthetic dsRNAs and siRNAs could enter plant tissues and cells, exploiting the same natural mechanisms as extracellular nucleic acids originating from plant microbial pathogens, insects, or viruses. Pathogenesis-related extracellular RNA and DNA derived from plant microbial and virus pathogens are known to induce plant innate immunity and regulate self- and non-self-recognition in plants [,,,,]. It is proposed that extracellular pathogen- and virus-related DNA and RNA molecules are perceived as genuine microbe- and pathogen-associated molecular patterns (MAMPs and PAMPs) in plants and act via pattern recognition receptors (PRRs) to induce pattern-triggered plant immune (PTI) signaling. However, there are again scarce data on the plant perception of pathogenesis-related DNA and RNA. To date, no specific receptors responsible for the recognition and uptake of extracellular DNA and RNA have been identified in plants. The findings by Niehl et al. [] shed light on this issue, revealing that applied virus-related dsRNA induced PTI responses in Arabidopsis via a somatic embryogenesis receptor-like kinase 1 (SERK1), and not through antiviral DICER-like proteins (DCL). This study suggested that dsRNA-mediated PTI involved membrane-associated processes and operated independently of RNA silencing.
According to Uslu et al. [], naked GFP-dsRNAs and GFP-hpRNAs did not induce GFP silencing in N. benthamiana and were not processed into siRNAs, indicating insufficient dsRNA uptake by plant cells. However, high-pressure spraying of GFP-encoding synthetic siRNAs was efficient in inducing GFP silencing in N. benthamiana [,]. Uslu et al. [] performed RNA-seq to investigate the profile and amounts of sRNAs mapping to GFP after exogenous N. benthamiana treatments with three synthetic 22-nt siRNAs. According to the study, the 22-nt siRNA targeting the 3′ of the GFP transgene was less efficient in inducing silencing when compared with the siRNAs targeting the 5′ and middle region of the GFP. Application of the siRNAs induced GFP silencing and GFP-siRNA amplification through the process of transitivity. It has been shown that dsRNA treatments considerably lowered viral-derived sRNAs and improved plant virus resistance [,]. However, the profile of sRNAs derived from externally added virus-specific dsRNAs (not from infecting virus) still requires investigation. The sRNA-seq analysis of plant tissues treated with fungal-derived dsRNA revealed a passage of the dsRNA via the plant vascular system and processing into siRNAs by the fungal DICER-LIKE 1 after uptake by the pathogen [,].
Although a number of studies have demonstrated that exogenous gene-specific dsRNAs can induce downregulation of target plant endogenous genes [,,,,,], there are no studies (to the best of our knowledge) where RNA-seq would be applied to analyze post-application processing of exogenous dsRNAs targeting plant genes. In the present study, we firstly showed that foliar application of both AtCHS- and NPTII-dsRNAs led to the emergence of a large number of sRNAs mapping to the AtCHS and NPTII, which has not been detected after the control water treatments. The data indicate that exogenous AtCHS-dsRNAs are processed into siRNAs and eventually target the plant AtCHS gene leading to AtCHS downregulation, while NPTII-dsRNAs were presumably degraded. The prevailing frequencies of the 21-nt, 23-nt, and 24-nt sRNAs in the total sRNA fraction confirmed the previously obtained data of other scientists that the main part of the sRNA pool is represented by molecules ranging in size from 21 to 24 nt [,]. The length size distribution of the total sRNA pool after exogenous dsRNA application was also similar to the data by Power et al. [], showing that 21-nt and 24-nt sRNAs prevailed. Notably, while there were three peaks in the sRNA amounts of 21-nt, 23-nt, and 24-nt sizes in the total sRNA fraction, we did not detect a considerable increase in the frequency of certain sizes of the AtCHS- and NPTII sRNAs. The length size distribution of the AtCHS- and NPTII-derived sRNAs was similar to that reported by Koch et al. [] for CYP3-dsRNA-derived sRNAs in terms of the distribution of sRNA levels gradually decreasing from small-sized sRNAs to longer ones. This distribution of sRNA lengths suggested that the dsRNA-derived sRNAs may gradually degrade, leading to this gradually decreasing sRNA length. Thus, the exogenously applied dsRNAs might be processed according to different laws as compared with the naturally formed dsRNAs, which are processed by DICER to 21–24-nt siRNAs. For externally applied dsRNA, it is possible that a significant portion of the dsRNA is destroyed nonsystematically, because there is no specific sRNA size fraction, and the smallest sRNA size prevails. The present study revealed the CHS- and NPTII-specific sRNAs unevenly distributed across both the CHS and NPTII genes, with peaks of read counts at particular positions on both sense and antisense strands. Koch et al. [] also detected uneven distribution of CYP3-dsRNA-derived sRNAs mapping to CYP3-dsRNA sequence. Further studies are needed to investigate whether this irregular sRNA mapping pattern is a result of rapid degradation of certain sRNAs or whether the read count peaks represent hotspots of secondary siRNAs generated by a transitivity-related process.
Taken together, the data show that exogenous dsRNAs are processed into siRNAs whose functional fraction induces RNAi and downregulates mRNA levels of the target gene. The results reported in this work highlight the potential of exogenous dsRNA application for silencing of specific plant genes and modulating desired plant traits.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
The seeds of wild-type A. thaliana (cv. Columbia) were vapor-phase sterilized and plated on solid ½ Murashige and Skoog (MS) medium as described []. The plates were placed at 22 °C for 1 week in a growth chamber (Sanyo MLR-352, Panasonic, Osaka, Japan) at a light intensity of ~120 μmol m−2 s−1 over a 16 h daily light period. Then, 1-week-old seedlings of A. thaliana were planted in pots 7 cm × 7 cm containing 100 g of commercially available rich soil “Universal Soil” (Fasko, Moscow, Russia). The soil consisted of riding peat, lowland peat, sand, limestone (dolomite) flour, and complex mineral fertilizer with microelements. The content of nutrients available to plants (mg/kg) was not less than: nitrogen—350; phosphorous—400; potassium—500; pH—6–7. The soil was well irrigated by filtered water applied at the bottom of the pots. Then, the plants were grown in the chamber at 22 °C under plastic wrap for additional three weeks without additional irrigation. Plant dsRNA treatments were performed using four-week-old rosettes of A. thaliana. After the RNA treatments, the four-week-old plants were incubated for additional seven days under anthocyanin-inducing (+7 °C, 23 h daily light period) conditions in a growth chamber (KS-200, Smolenskoe SKTB SPU, Smolensk, Russia) without further irrigation to induce AtCHS expression and anthocyanin accumulation.
4.2. Isolation and Sequencing of AtCHS mRNA Transcripts
Full-length coding cDNA sequences of AtCHS (AT5G13930.1, 1188 bp) were amplified by RT-PCR using total RNA samples extracted from the adult leaves of A. thaliana as described []. The RT-PCR products were subcloned into pJET1.2/blunt and sequenced as described previously [].
4.3. dsRNA Synthesis and Application
The AtCHS- and NPTII-dsRNAs were synthesized using the T7 RiboMAX™ Express RNAi System (Promega, Madison, WI, USA). For this purpose, a large cDNA fragment of AtCHS (736 bp out of 1188 bp) was amplified by PCR for in vitro transcription and dsRNA production. We amplified a large fragment of NPTII (GenBank AJ414108, 599 bp out of 798 bp) using pZP-RCS2-nptII plasmid []. The T7 promoter sequence was introduced into both the 5′ and 3′ ends of the amplified AtCHS or NPTII in a single PCR for each gene using primers listed in Table S1 (Figure 1). The PCRs were performed in the Bis-M1105 Thermal Cycler programmed according to T7 RiboMAX™ Express RNAi System instructions. PCR was carried out in a final volume of 30 µL, containing 1X Taq reaction buffer with 3 mM MgCl2, 0.5 µL plasmid DNA (50 ng), 200 µM dNTPs, 0.2 µM of each primer, and 2.0 units of Taq DNA polymerase (Evrogen, Moscow, Russia). After an initial denaturation at 95 °C for 5 min, the first 5 cycles were performed as follows: 95 °C for 10 s, 63 °C (AtCHS) or 65 °C (NPTII) for 10 s, 72 °C for 38 s (AtCHS) or 38 s (NPTII), followed by 40 cycles each of 95 °C for 10 s and 72 °C for 48 s (AtCHS) or 48 s (NPTII). After a final extension at 72 °C for 5 min, PCR fragments were loaded on 1% agarose gel and purified by the Cleanup Standard kit (Evrogen). Then, the obtained PCR products were used as templates for in vitro transcription and dsRNA synthesis following the manufacturer’s protocol. The resultant dsRNAs were analyzed by gel electrophoresis and spectrophotometry to estimate dsRNA purity, integrity, and amount.
The AtCHS- and NPTII-dsRNAs were applied to the foliar surface of four-week-old rosettes of wild-type A. thaliana by spreading with individual soft brushes (natural pony hair) sterilized by autoclaving (Figure 1) as described [,]. For each dsRNA treatment, 35 µg of the dsRNA were diluted in 100 µL of nuclease-free water and applied to all leaves of one rosette on both the adaxial and abaxial sides. Two rosettes were treated in total per each type of condition (35 µg of dsRNA per each rosette). The water and dsRNA treatments were performed at a late day time (21:00–21:30) under low soil moisture conditions using four-week-old rosettes of A. thaliana, since the appropriate late day time, low soil moisture, and plant age at the time of dsRNA application were important parameters for successful gene suppression in A. thaliana according to our recent analysis [,]. Soil water content before dsRNA treatment was 50–60%.
4.4. RNA Isolation
For RNA isolations, two whole four-week-old rosettes of A. thaliana were collected two days after treatments per each type of treatments: water treatment (rosettes WC-1a, WC-1b), AtCHS-dsRNA treatment (rosettes dsCHS-3a, dsCHS-3b), and NPTII-dsRNA treatment (rosettes NPTII-4a, NPTII-4b). High-molecular-weight (HMW) and low-molecular-weight (LMW) RNA fractions were isolated using a modified cetyltrimethylammonium bromide (CTAB)–lithium chloride (LiCl)-based protocol []. First, we began isolation of total RNA using the CTAB-polyvinylpyrrolidone K30 (PVP) extraction buffer prepared as described []. Tissue samples of A. thaliana (a whole 4-week-old rosette of 70–80 mg) were homogenized in the extraction buffer and incubated for 5 min at 65 °C in a water bath. After the above incubation, 700 µL of chloroform was added. The samples were then centrifuged at full speed (13,200 rpm, +4 °C) for 15 min (5415R, Eppendorf, Germany). Approximately, 1 mL of the water phase was mixed with 250 µL of 10 M LiCl (Panreac, Spain) and incubated at +4 °C overnight. After the above incubation, the samples were centrifuged at full speed (13,200 rpm, +4 °C) for 15 min.
To separate HMW RNAs, the pellets were air-dried and dissolved in 100 µL of sterile distilled water for the following precipitation with a 2.5 volume of precooled absolute ethanol at −20 °C overnight. Then, the HMW RNA samples were centrifuged at full speed (13,200 rpm, +4 °C) for 15 min. The air-dried pellets were resuspended in an appropriate volume of sterile distilled water and used for complementary DNA (cDNA) preparation.
After LiCl precipitation, 400 µL of the resulting upper aqueous phase was transferred into another clean 1.5 mL collection tube to separate LMW RNAs. The LMW RNAs were precipitated with 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volume of precooled absolute ethanol at −20 °C overnight.
4.5. Reverse Transcription and qRT-PCRs
cDNAs were synthesized using 1.5 µg of total RNA as described []. The 1 µL samples of reverse transcription products were then amplified by PCR and verified in the absence of DNA contamination using primers for the AtCHS gene (AT5G13930.1) listed in Table S1. The qRT-PCRs were performed with SYBR Green I Real-Time PCR dye and a real-time PCR kit (Evrogen, Moscow, Russia) as described [] using two internal controls (GAPDH and UBQ) selected in previous studies as relevant reference genes for qRT-PCRs on Arabidopsis []. Amplification was carried out in a DTprime real-time thermocycler (DNA Technology, Moscow, Russia). PCR conditions consisted of 2 min at 95 °C, followed by 50 cycles of 10 s at 95 °C and 25 s at 62 °C. The expression was calculated by the 2−ΔΔCT method []. The data were processed with the RealTime_PCR v.7.3 (DNA Technology, Russia). All gene identification numbers and used primers are listed in Table S1.
4.6. Small RNA Sequencing
The ethanol-precipitated LMW RNA fractions were sent for high-throughput sequencing using Illumina technology to Evrogen (Moscow, Russia). Incoming sample quality control was performed by horizontal electrophoresis in agarose gel. RNA was prepared (100 ng per biological replicate) for sequencing using the Qiagen Small RNA Sample Prep kit (Qiagen, Dusseldorf, Germany). The quality of the obtained libraries was checked using Fragment Analyzer (Agilent, CA, USA). Quantitative analysis was performed by qPCR. After quality control and DNA quantity estimation, the library pool was sequenced on an Illumina NovaSeq 6000 instrument (1 × 100 bp single-read sequencing). When sequencing cDNA libraries, the loading concentration was 300 pM, and the volume was 27 µL in accordance with the standard Illumina recommendations. The FASTQ files were obtained using bcl2fastq Conversion Software v2.20 (Illumina, San Diego, CA, USA). The recording format of the quality data string is Phred 33. As a result, 526,174,120 reads were obtained. General information about the read numbers is presented in Table 1. sRNA library sequences were deposited to the National Center for Biotechnology Information (NCBI) under Accession Number PRJNA827691 and in the database of the Laboratory of Biotechnology, Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences, Russia (https://biosoil.ru/downloads/biotech/RNAseq/Arabidopsis/2021-03-20012551-data1(Our-RNAseq-1(2))/) (accessed on 8 May 2022).
4.7. Bioinformatic Analysis
Adaptor sequences (5′AACTGTAGGCACCATCAAT, 5′AGATCGGAAGAGCACACGT) were trimmed, and low-quality reads and reads shorter than 17 and longer than 30 nucleotides were removed from obtained high-throughput sequencing data with the BBDuk program []. Using the Bowtie program (0 mismatch available) [], reads aligned to ribosomal and chloroplast DNA sequences and reads unaligned to the A. thaliana TAIR10 genome [] were excluded from analysis (except for the NPTII gene). Then, using the same program, we analyzed the sRNAs aligned to AtCHS and NPTII sequences. Small RNA coverage data on AtCHS and NPTII gene sequences were obtained using BEDTools software []. The following data processing and line charts/bar graphs were plotted using Microsoft Excel based on only 17 to 30 nt long reads.
4.8. Statistical Analysis
The data are presented as mean ± standard error (SE) and were tested by paired Student’s t-test. The p < 0.05 level was selected as the point of minimal statistical significance in all analyses. Two whole four-week-old rosettes of A. thaliana were collected two days after treatments per each type of treatment (two biological replicates per each type of analysis).
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/ijms23105325/s1.
Author Contributions
A.S.D. was responsible for research design, data analysis, and paper preparation. K.V.K. performed dsRNA treatments, RNA isolation, data analysis, and data interpretation. N.N.N. performed bioinformatic analysis. A.R.S. was involved in plant management and qRT-PCRs. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Russian Science Foundation (19-74-10023).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
sRNA library sequences were deposited to the National Center for Biotechnology Information (NCBI) under Accession Number PRJNA827691 and in the database of the Laboratory of Biotechnology, Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences, Russia (https://biosoil.ru/downloads/biotech/RNAseq/Arabidopsis/2021-03-20012551-data1(Our-RNAseq-1(2))/) (accessed on 8 May 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, A.; Gautam, V.; Singh, S.; Sarkar Das, S.; Verma, S.; Mishra, V.; Mukherjee, S.; Sarkar, A.K. Plant small RNAs: Advancement in the understanding of biogenesis and role in plant development. Planta 2018, 248, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA interference: A natural immune system of plants to counteract biotic stressors. Cells 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.H.; Guo, H.S. RNA silencing: From discovery and elucidation to application and perspectives. J. Integr. Plant Biol. 2022, 64, 476–498. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.J.; Yan, X.N.; Gu, C.X.; Yuan, X.F. Biogenesis, trafficking, and function of small RNAs in plants. Front. Plant Sci. 2022, 13, 825477. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Dean, R.A. Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 2020, 21, 589–601. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Aleynova, O.A.; Kalachev, A.V.; Suprun, A.R.; Ogneva, Z.V.; Kiselev, K.V. Induction of transgene suppression in plants via external application of synthetic dsRNA. Int. J. Mol. Sci. 2019, 20, 1585. [Google Scholar] [CrossRef] [Green Version]
- Pampolini, F.; Rodrigues, T.B.; Leelesh, R.S.; Kawashima, T.; Rieske, L.K. Confocal microscopy provides visual evidence and confirms the feasibility of dsRNA delivery to emerald ash borer through plant tissues. J. Pest Sci. 2020, 93, 1143–1153. [Google Scholar] [CrossRef]
- Holeva, M.C.; Sklavounos, A.; Rajeswaran, R.; Pooggin, M.M.; Voloudakis, A.E. Topical application of double-stranded RNA targeting 2b and CP genes of Cucumber mosaic virus protects plants against local and systemic viral infection. Plants 2021, 10, 963. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kalachev, A.V.; Dubrovina, A.S. External dsRNA downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 6749. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kostetsky, E.Y.; Dubrovina, A.S. The specificity of transgene suppression in plants by exogenous dsRNA. Plants 2022, 11, 715. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; McMillan, J.N.; Cardoza, V.; Maegele, I.; Dadami, E.; Runne, M.; Krczal, G.; Wassenegger, M. Induction of silencing in plants by high-pressure spraying of in vitro-synthesized small RNAs. Front. Plant Sci. 2016, 7, 1327. [Google Scholar] [CrossRef] [Green Version]
- Dalakouras, A.; Jarausch, W.; Buchholz, G.; Bassler, A.; Braun, M.; Manthey, T.; Krczal, G.; Wassenegger, M. Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petiole absorption. Front. Plant Sci. 2018, 9, 1253. [Google Scholar] [CrossRef] [Green Version]
- Dubrovina, A.S.; Aleynova, O.A.; Suprun, A.R.; Ogneva, Z.V.; Kiselev, K.V. Transgene suppression in plants by foliar application of in vitro-synthesized small interfering RNAs. Appl. Microbiol. Biotechnol. 2020, 104, 2125–2135. [Google Scholar] [CrossRef]
- Uslu, V.V.; Dalakouras, A.; Steffens, V.A.; Krczal, G.; Wassenegger, M. High-pressure sprayed siRNAs influence the efficiency but not the profile of transitive silencing. Plant J. 2022, 109, 1199–1212. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- McLoughlin, A.G.; Wytinck, N.; Walker, P.L.; Girard, I.J.; Rashid, K.Y.; de Kievit, T.; Fernando, W.G.D.; Whyard, S.; Belmonte, M.F. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 2018, 9, 7320. [Google Scholar] [CrossRef]
- Power, I.L.; Faustinelli, P.C.; Orner, V.A.; Sobolev, V.S.; Arias, R.S. Analysis of small RNA populations generated in peanut leaves after exogenous application of dsRNA and dsDNA targeting aflatoxin synthesis genes. Sci. Rep. 2020, 10, 13820. [Google Scholar] [CrossRef]
- Wang, M.; Jin, H. Spray-induced gene silencing: A powerful innovative strategy for crop protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Morozov, S.Y.; Solovyev, A.G.; Kalinina, N.O.; Taliansky, M.E. Double-stranded RNAs in plant protection against pathogenic organisms and viruses in agriculture. Acta Nat. 2019, 11, 13–21. [Google Scholar] [CrossRef]
- Niu, D.; Hamby, R.; Sanchez, J.N.; Cai, Q.; Yan, Q.; Jin, H. RNAs—A new frontier in crop protection. Curr. Opin. Biotechnol. 2021, 70, 204–212. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically modified organism-free RNA interference: Exogenous application of RNA molecules in plants. Plant. Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [Green Version]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef] [Green Version]
- Das, P.R.; Sherif, S.M. Application of exogenous dsRNAs-induced RNAi in agriculture: Challenges and triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef]
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA Interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650. [Google Scholar] [CrossRef]
- Sammons, R.; Ivashuta, S.; Liu, H.; Wang, D.; Feng, P.; Kouranov, A.; Andersen, S. Polynucleotide Molecules for Gene Regulation in Plants. U.S. Patent 20110296556 A1, 1 December 2011. [Google Scholar]
- Lau, S.E.; Schwarzacher, T.; Othman, R.Y.; Harikrishna, J.A. dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 2015, 15, 194. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef]
- Warnock, N.D.; Wilson, L.; Canet-Perez, J.V.; Fleming, T.; Fleming, C.C.; Maule, A.G.; Dalzell, J.J. Exogenous RNA interference exposes contrasting roles for sugar exudation in host-finding by plant pathogens. Int. J. Parasitol. 2016, 46, 473–477. [Google Scholar] [CrossRef] [Green Version]
- Marcianò, D.; Ricciardi, V.; Fassolo, E.M.; Passera, A.; BIANCO, P.A.; Failla, O.; Casati, P.; Maddalena, G.; De Lorenzis, G.; Toffolatti, S.L. RNAi of a putative grapevine susceptibility gene as a possible downy mildew control strategy. Front. Plant Sci. 2021, 12, 667319. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Guaschino, M.; Pagliarani, C.; De Rosso, M.; Lovisolo, C.; Chitarra, W. Spray-induced gene silencing targeting a glutathione S-transferase gene improves resilience to drought in grapevine. Plant Cell Environ. 2022, 45, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Song, X.S.; Gu, K.X.; Duan, X.X.; Xiao, X.M.; Hou, Y.P.; Duan, Y.B.; Wang, J.X.; Yu, N.; Zhou, M.G. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol. Plant Pathol. 2018, 19, 2543–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uslu, V.V.; Bassler, A.; Krczal, G.; Wassenegger, M. High-pressure-sprayed double stranded RNA does not induce RNA interference of a reporter gene. Front. Plant Sci. 2020, 11, 534391. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Dubrovina, A.S. Physiological conditions and dsRNA application approaches for exogenously induced RNA interference in Arabidopsis thaliana. Plants 2021, 10, 264. [Google Scholar] [CrossRef] [PubMed]
- Biedenkopf, D.; Will, T.; Knauer, T.; Jelonek, L.; Furch, A.C.U.; Busche, T.; Koch, A. Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare. ExRNA 2020, 2, 12. [Google Scholar] [CrossRef]
- Yakushiji, S.; Ishiga, Y.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Bacterial DNA activates immunity in Arabidopsis thaliana. J. Gen. Plant Pathol. 2009, 75, 227–234. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Carteni, F.; Bonanomi, G.; Senatore, M.; Termolino, P.; Giannino, F.; Incerti, G.; Rietkerk, M.; Lanzotti, V.; Chiusano, M.L. Inhibitory effects of extracellular self-DNA: A general biological process? New Phytol. 2015, 206, 127–132. [Google Scholar] [CrossRef]
- Bhat, A.; Ryu, C.M. Plant perceptions of extracellular DNA and RNA. Mol. Plant 2016, 9, 956–958. [Google Scholar] [CrossRef]
- Lee, B.; Park, Y.S.; Lee, S.; Song, G.C.; Ryu, C.M. Bacterial RNAs activate innate immunity in Arabidopsis. New Phytol. 2016, 209, 785–797. [Google Scholar] [CrossRef]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [Green Version]
- Tzfira, T.; Tian, G.W.; Lacroix, B.; Vyas, S.; Li, J.; Leitner-Dagan, Y.; Krichevsky, A.; Taylor, T.; Vainstein, A.; Citovsky, V. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 2005, 57, 503–516. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Dubrovina, A.S.; Shumakova, O.A.; Karetin, Y.A.; Manyakhin, A.Y. Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep. 2013, 32, 431–442. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Kiselev, K.V.; Ogneva, Z.V.; Dubrovina, A.S. The grapevine calmodulin-like protein gene CML21 is regulated by alternative splicing and involved in abiotic stress response. Int. J. Mol. Sci. 2020, 21, 7939. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. The role of calcium-dependent protein kinase genes VaCPK1 and VaCPK26 in the response of Vitis amurensis (in vitro) and Arabidopsis thaliana (in vivo) to abiotic stresses. Russ. J. Genet. 2019, 55, 319–329. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bushnell, B. BBTools, a Suite of Fast, Multithreaded Bioinformatics Tools Designed for Analysis of DNA and RNA Sequence Data. Version 38.86. 2015. Available online: https://github.com/BioInfoTools/BBMap (accessed on 8 May 2022).
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, 1202–1210. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).