Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice
Abstract
:1. Introduction
2. Results
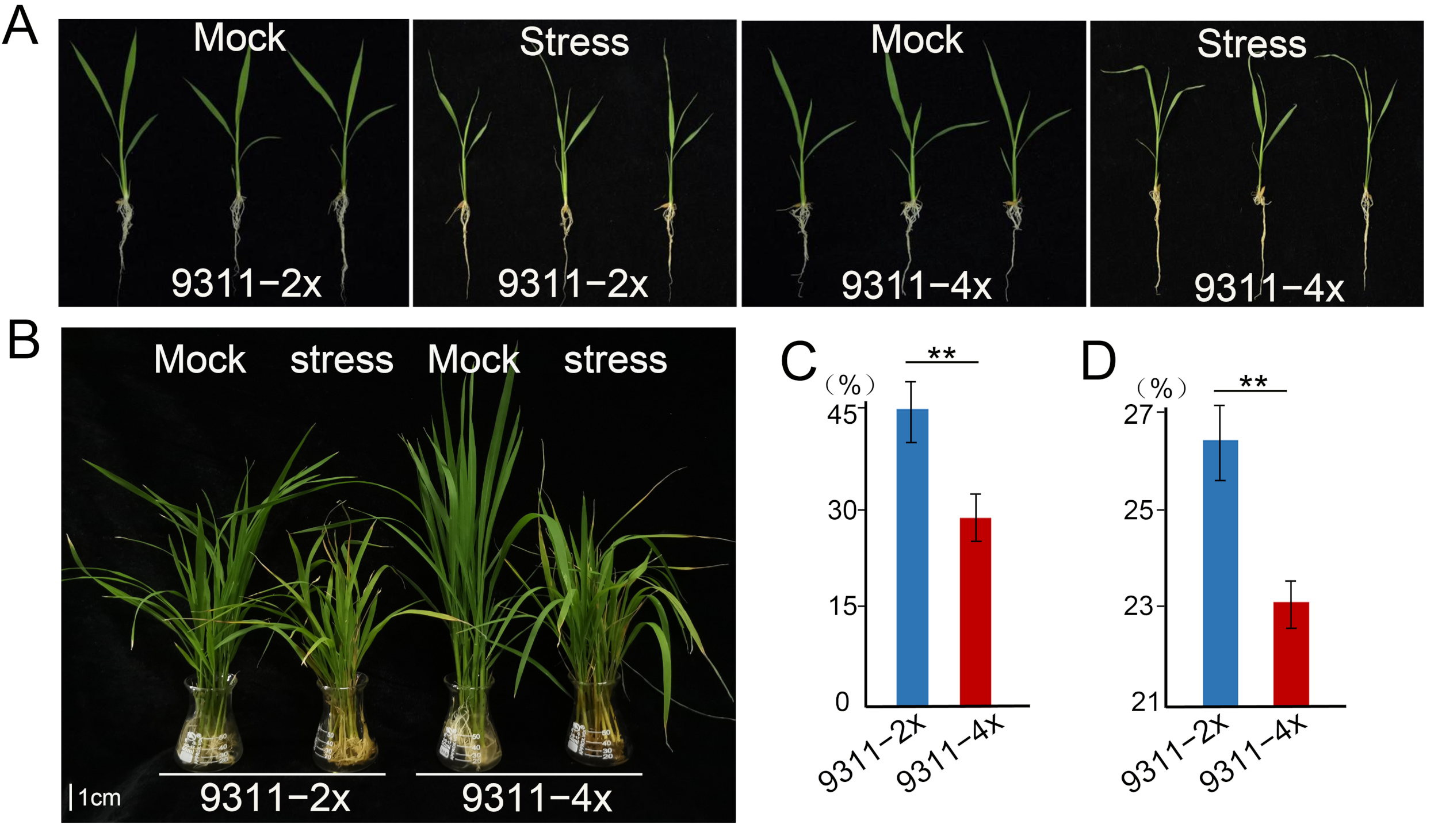
2.1. Effect of Polyploidization on Phenotype
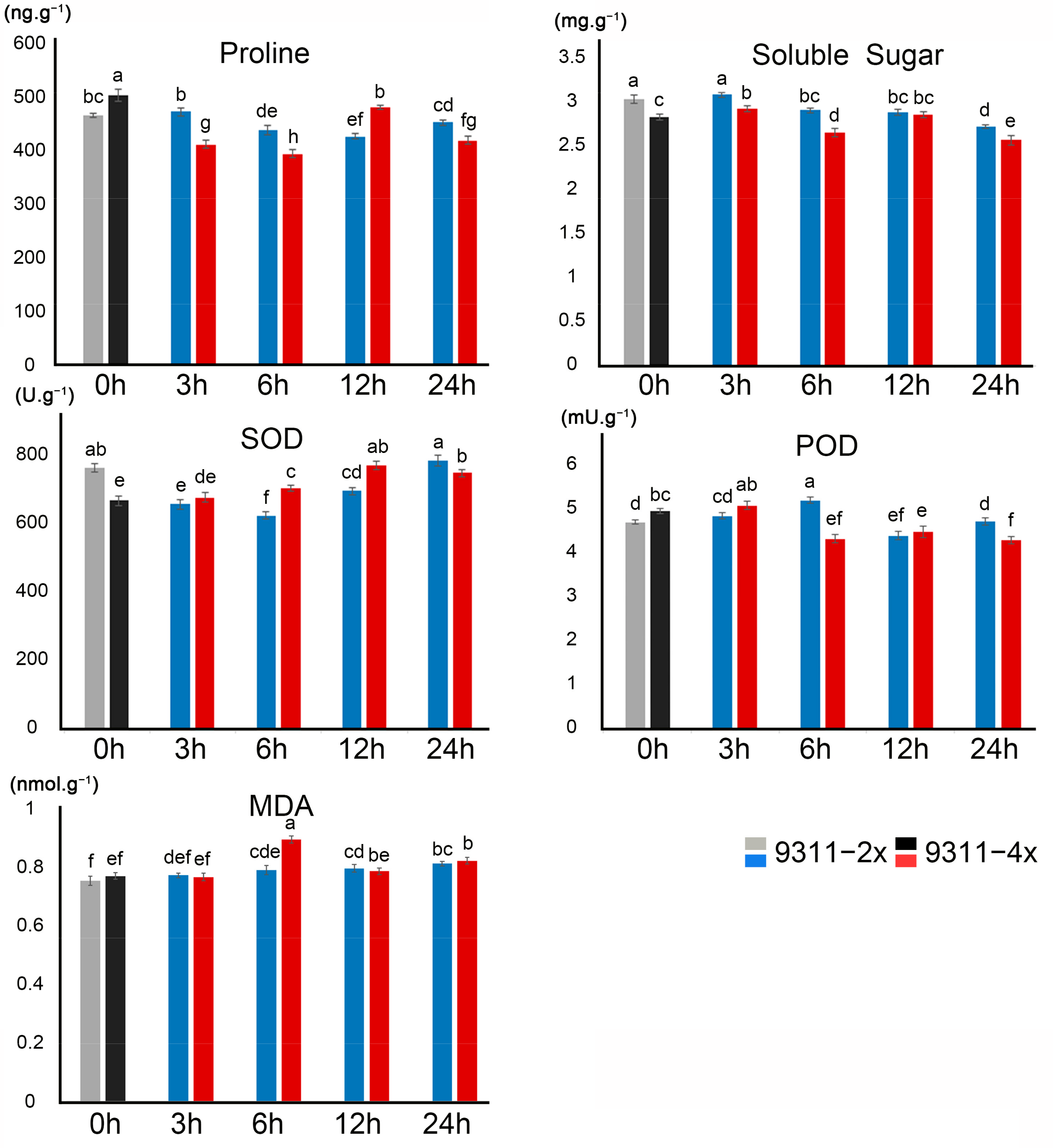
2.2. Effect of Polyploidization on Physiological Parameters
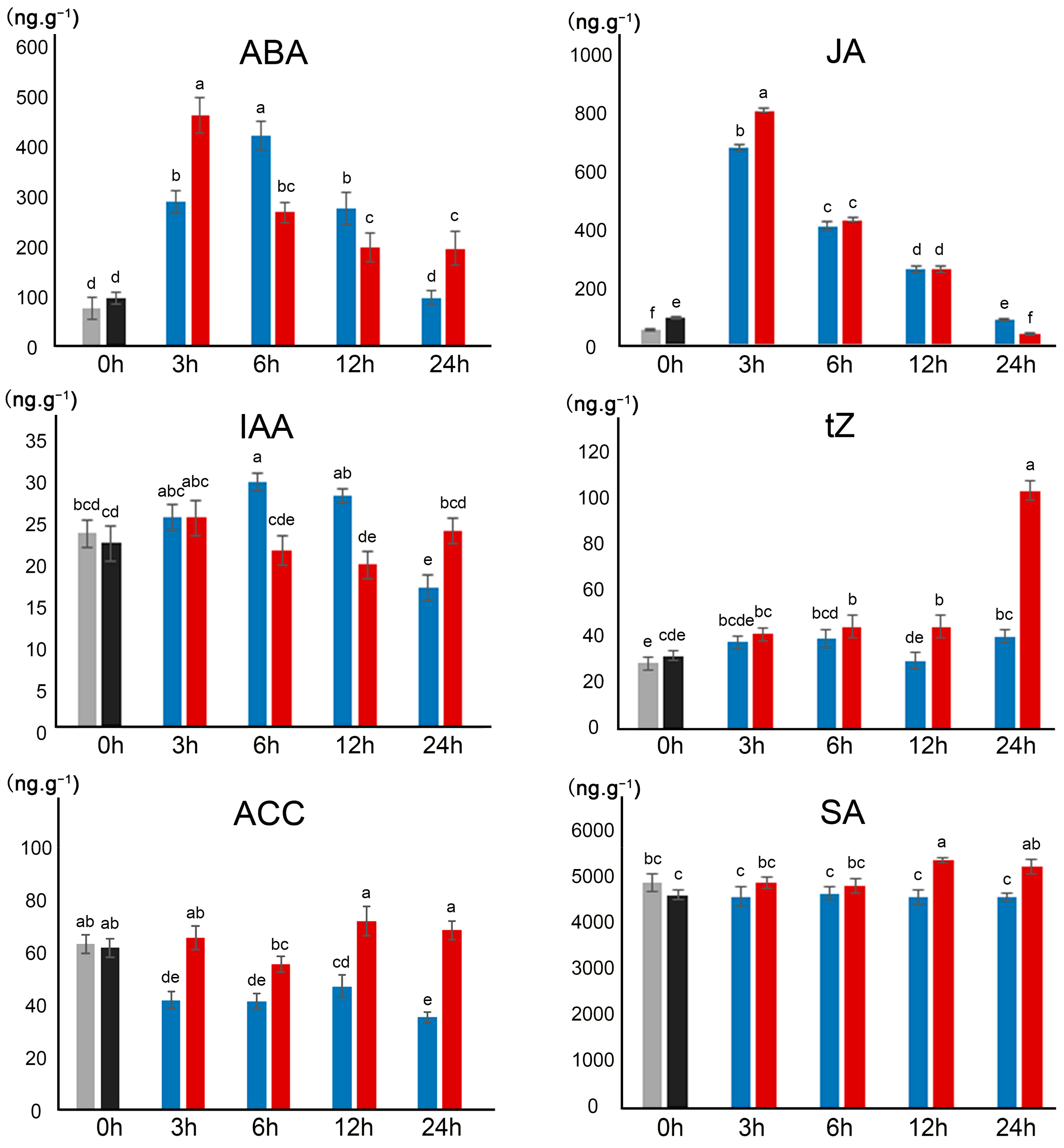
2.3. Effect of Polyploidization on Hormone Levels
2.4. Effect of Polyploidization on Gene Expression
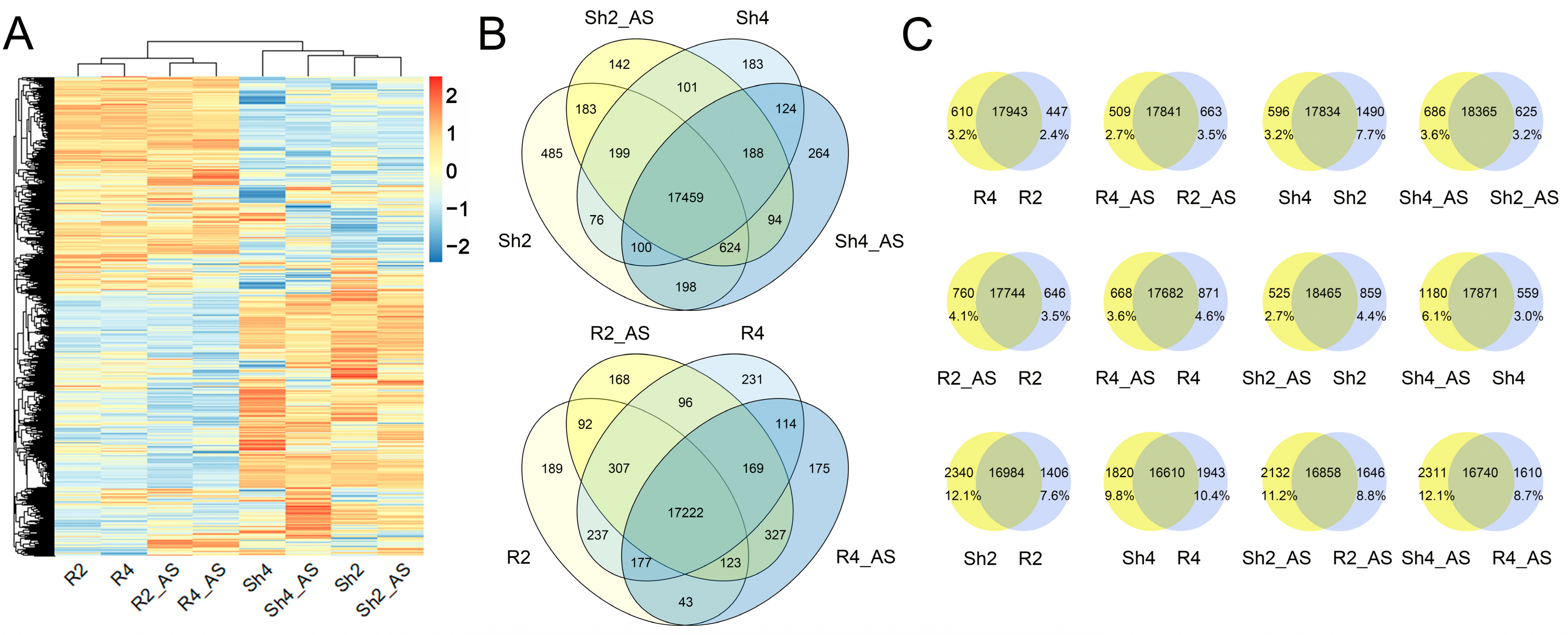
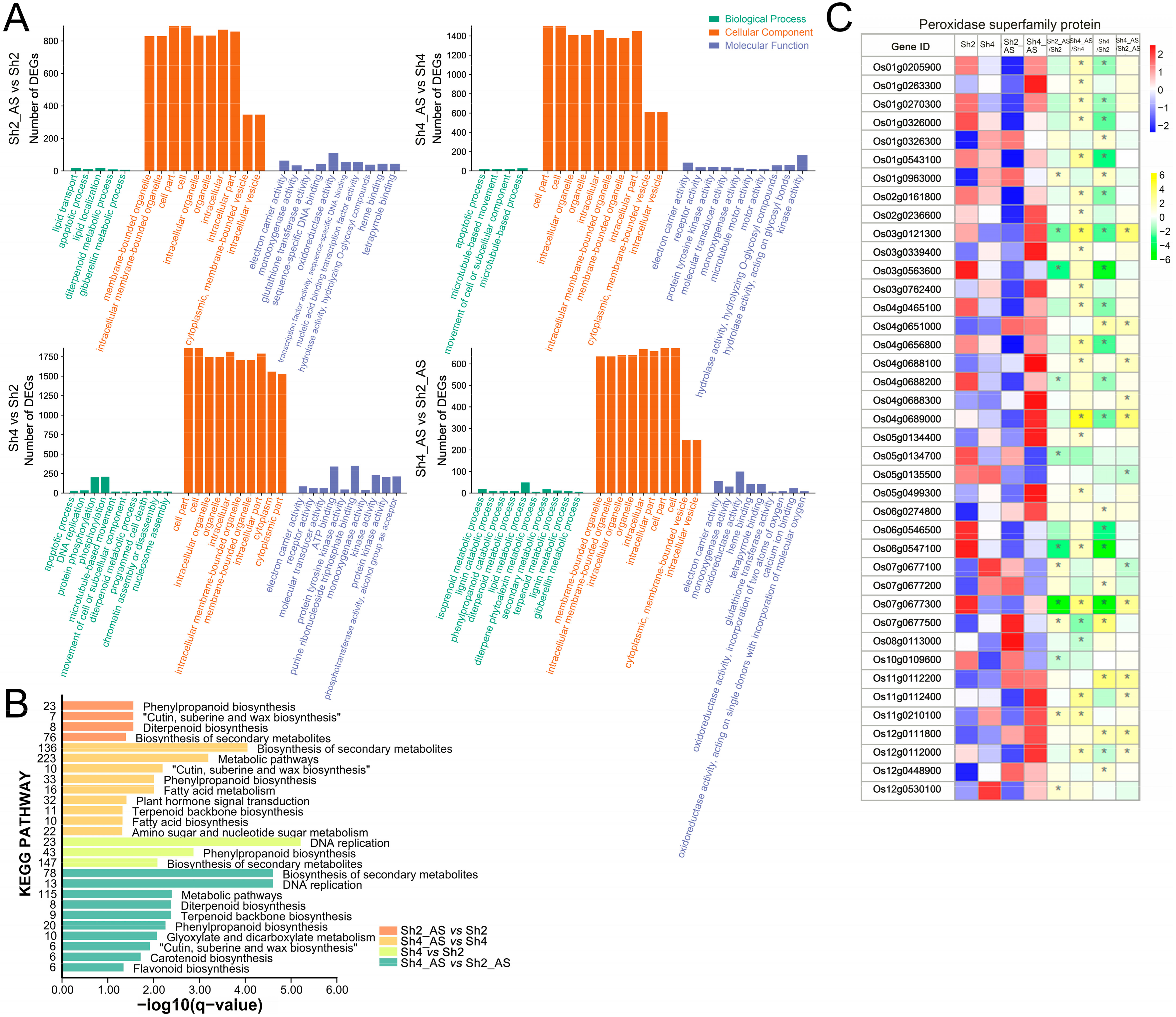
2.5. Differentially Expressed Genes (DEGs) between Diploid and Autotetraploid Rice under Alkaline Treatment
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses of DEGs between Diploid and Autotetraploid Rice
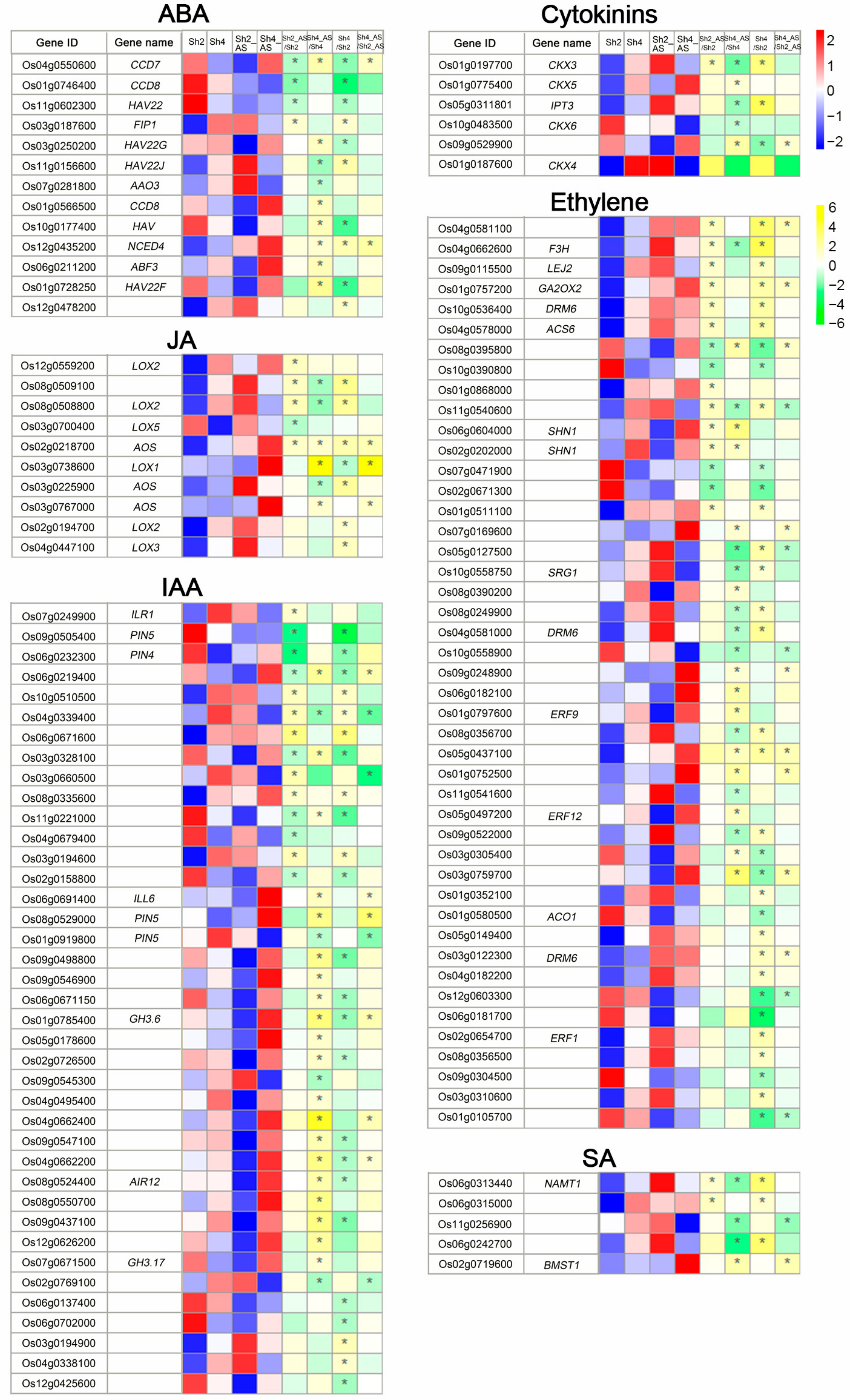
2.7. Phytohormone Related DEGs Vary with Ploidy Level
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Measurements of Small-Molecule Organic Compounds, Enzyme Activity, and Hormone Levels
4.3. RNA Isolation and Sequencing Analysis
4.4. Quantitative Real-Time Polymerase Chain Reaction
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Ashman, T.L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef]
- Masterson, J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H. Yesterday’s polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001, 2, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [Green Version]
- Valentine, D.H.; Stebbins, G.L. Variation and Evolution in Plants. J. Ecol. 1951, 39, 434. [Google Scholar] [CrossRef]
- Song, Q.; Chen, Z.J. Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 2015, 24, 101–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Lin, F.; Zhou, Y.; Wang, J.; Sun, S.; Wang, B.; Zhang, Z.; Li, G.; Lin, X.; Wang, X.; et al. Genomic mosaicism due to homoeologous exchange generates extensive phenotypic diversity in nascent allopolyploids. Natl. Sci. Rev. 2021, 8, nwaa277. [Google Scholar] [CrossRef]
- Comai, L.; Tyagi, A.P.; Winter, K.; Holmes-Davis, R.; Reynolds, S.H.; Stevens, Y.; Byers, B. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 2000, 12, 1551–1568. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tian, L.; Madlung, A.; Lee, H.S.; Chen, M.; Lee, J.J.; Watson, B.; Kagochi, T.; Comai, L.; Chen, Z.J. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 2004, 167, 1961–1973. [Google Scholar] [CrossRef] [Green Version]
- Mcclintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Schoenfelder, K.P.; Fox, D.T. The expanding implications of polyploidy. J. Cell Biol. 2015, 209, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ali, A.; Hou, F.; Wu, T.; Guo, D.; Zeng, X.; Wang, F.; Zhao, H.; Chen, X.; Xu, P.; et al. Effects of ploidy variation on promoter DNA methylation and gene expression in rice (Oryza sativa L.). BMC Plant Biol. 2018, 18, 314. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Sun, J.; Wei, C.; Zhang, P.; Liu, X. Studies on the abnormality of embryosac and pollen fertility in autotetraploid rice during diferent growing seasons. Pak. J. Bot. 2010, 42, 7–19. [Google Scholar] [CrossRef]
- Wu, J.; Shahid, M.Q.; Chen, L.; Chen, Z.; Wang, L.; Liu, X.; Lu, Y. Polyploidy Enhances F₁ Pollen Sterility Loci Interactions That Increase Meiosis Abnormalities and Pollen Sterility in Autotetraploid Rice. Plant Physiol. 2015, 169, 2700–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Kaur, H.; Sirhindi, G.; Bhardwaj, R.; Alyemeni, M.N.; Siddique, K.H.M.; Ahmad, P. 28-homobrassinolide regulates antioxidant enzyme activities and gene expression in response to salt- and temperature-induced oxidative stress in Brassica juncea. Sci. Rep.-UK 2018, 8, 8735. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Aziz, U.; Alsahli, A.A.; Alyemeni, M.N.; Ahmad, P. Influence of Exogenous Salicylic Acid and Nitric Oxide on Growth, Photosynthesis, and Ascorbate-Glutathione Cycle in Salt Stressed Vigna angularis. Biomolecules 2020, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Dai, Y.; Zheng, C.; Yang, Y.; Chen, W.; Wang, Q.; Chandrasekaran, U.; Du, J.; Liu, W.; Shu, K. The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 2021, 229, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Sun, J.; He, L.; Li, T. Response of seedling growth and physiology of Sorghum bicolor (L.) Moench to saline-alkali stress. PLoS ONE 2019, 14, e220340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Liu, M.; Liu, X.; Cheng, X.; Liang, Z. Silicon priming created an enhanced tolerance in Alfalfa (Medicago sativa L.) seedlings in response to high alkaline stress. Front. Plant Sci. 2018, 9, 716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, M.; Wang, X.; Chang, D.; Wang, S.; Hong, D.; Fan, H.; Wang, K. Application of compound material alleviates saline and alkaline stress in cotton leaves through regulation of the transcriptome. BMC Plant Biol. 2020, 20, 462. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Oster, J.; Shainberg, I.; Abrol, I. Reclamation of Salt-Affected Soils. Agric. Drain. 1999, 38, 659–691. [Google Scholar] [CrossRef]
- Guo, M.; Li, S.; Tian, S.; Wang, B.; Zhao, X. Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba). PLoS ONE 2017, 12, e185732. [Google Scholar] [CrossRef] [Green Version]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Yang, G.; Yu, Z.; Gao, L.; Zheng, C. SnRK2s at the Crossroads of Growth and Stress Responses. Trends Plant Sci. 2019, 24, 672–676. [Google Scholar] [CrossRef]
- Yu, J.; Niu, L.; Yu, J.; Liao, W.; Xie, J.; Lv, J.; Feng, Z.; Hu, L.; Dawuda, M. The Involvement of Ethylene in Calcium-Induced Adventitious Root Formation in Cucumber under Salt Stress. Int. J. Mol. Sci. 2019, 20, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef]
- Urano, K.; Maruyama, K.; Ogata, Y.; Morishita, Y.; Takeda, M.; Sakurai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Kobayashi, M.; et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009, 57, 1065–1078. [Google Scholar] [CrossRef]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef]
- Khan, A.; Kamran, M.; Imran, M.; Al-Harrasi, A.; Al-Rawahi, A.; Al-Amri, I.; Lee, I.J.; Khan, A.L. Silicon and salicylic acid confer high-pH stress tolerance in tomato seedlings. Sci. Rep. 2019, 9, 19788. [Google Scholar] [CrossRef] [Green Version]
- Hyoung, S.; Cho, S.H.; Chung, J.H.; So, W.M.; Cui, M.H.; Shin, J.S. Cytokinin oxidase PpCKX1 plays regulatory roles in development and enhances dehydration and salt tolerance in Physcomitrella patens. Plant Cell Rep. 2020, 39, 419–430. [Google Scholar] [CrossRef]
- Urano, K.; Maruyama, K.; Jikumaru, Y.; Kamiya, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Analysis of plant hormone profiles in response to moderate dehydration stress. Plant J. 2017, 90, 17–36. [Google Scholar] [CrossRef]
- Ma, S.; Lv, L.; Meng, C.; Zhou, C.; Fu, J.; Shen, X.; Zhang, C.; Li, Y. Genome-Wide Analysis of Abscisic Acid Biosynthesis, Catabolism, and Signaling in Sorghum Bicolor under Saline-Alkali Stress. Biomolecules 2019, 9, 823. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Liang, W.; Li, Y.; Li, M.; Ma, B.; Liu, C.; Ma, F.; Li, C. Transcriptome analysis reveals the effects of alkali stress on root system architecture and endogenous hormones in apple rootstocks. J. Integr. Agr. 2019, 18, 2264–2271. [Google Scholar] [CrossRef]
- Wang, L.; Cao, S.; Wang, P.; Lu, K.; Song, Q.; Zhao, F.; Chen, Z.J. DNA hypomethylation in tetraploid rice potentiates stress-responsive gene expression for salt tolerance. Proc. Natl. Acad. Sci. USA 2021, 118, 1. [Google Scholar] [CrossRef]
- Nakamori, E. On the Occurrence of the Tetraploid Plant of Rice, Oryza sativa L. Proc. Imp. Acad. 1933, 9, 340–341. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Feng, Z.; Zhang, X.; Song, Z.; Cai, D. A New Way of Rice Breeding: Polyploid Rice Breeding. Plants 2021, 10, 422. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, A.F.; Avery, A.G. Methods of inducing doubling of chromosomes in plants. J. Hered. 1937, 28, 393–411. [Google Scholar] [CrossRef]
- Park, C.; Lim, C.W.; Lee, S.C. The Pepper CaOSR1 Protein Regulates the Osmotic Stress Response via Abscisic Acid Signaling. Front. Plant Sci. 2016, 7, 890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Peat, T.S.; Böttcher, C.; Newman, J.; Lucent, D.; Cowieson, N.; Davies, C. Crystal Structure of an Indole-3-Acetic Acid Amido Synthetase from Grapevine Involved in Auxin Homeostasis. Plant Cell 2012, 24, 4525–4538. [Google Scholar] [CrossRef] [Green Version]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, L.; Yu, Y.; Quan, R.; Zhang, Z.; Zhang, H.; Huang, R. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 2011, 68, 88–99. [Google Scholar] [CrossRef]
- Cao, Z.Z.; Lin, X.Y.; Yang, Y.J.; Guan, M.Y.; Xu, P.; Chen, M.X. Gene identification and transcriptome analysis of low cadmium accumulation rice mutant (lcd1) in response to cadmium stress using MutMap and RNA-seq. BMC Plant Biol. 2019, 19, 250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Gan, L.; Chen, D.; Zhang, Y.; Zhang, Y.; Liu, X.; Chen, S.; Wei, Z.; Tong, L.; Song, Z.; et al. Integration of small RNA, degradome and proteome sequencing in Oryza sativa reveals a delayed senescence network in tetraploid rice seed. PLoS ONE 2020, 15, e242260. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, X.; Li, X.; Zhou, H.; Wang, S.; Yuan, Z.; Zhang, Y.; Li, S.; You, A.; Zhou, L.; et al. A Genome Doubling Event Reshapes Rice Morphology and Products by Modulating Chromatin Signatures and Gene Expression Profiling. Rice 2021, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, L.; Zhang, H.; Yang, Z.; Wang, H.; Wen, S.; Zhang, C.; Rustgi, S.; von Wettstein, D.; Liu, B. Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl. Acad. Sci. USA 2014, 111, 11882–11887. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Shi, J.; Wang, Z.; Zhang, W.; Yang, H. H2S pretreatment mitigates the alkaline salt stress on Malus hupehensis roots by regulating Na+/K+ homeostasis and oxidative stress. Plant Physiol. Bioch. 2020, 156, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Wu, C.; Zhang, L.; Chen, X. Contrasting photosynthesis and photoinhibition in tetraploid and its autodiploid honeysuckle (Lonicera japonica Thunb.) under salt stress. Front. Plant Sci. 2015, 6, 227. [Google Scholar] [CrossRef] [Green Version]
- Ponce, K.S.; Guo, L.; Leng, Y.; Meng, L.; Ye, G. Advances in Sensing, Response and Regulation Mechanism of Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 2254. [Google Scholar] [CrossRef]
- Marco, F.; Alcázar, R.; Tiburcio, A.F.; Carrasco, P. Interactions between polyamines and abiotic stress pathway responses unraveled by transcriptome analysis of polyamine overproducers. Omics J. Integr. Biol. 2011, 15, 775–781. [Google Scholar] [CrossRef] [Green Version]
- Fuhrer, J.; Kaur-Sawhney, R.; Shih, L.M.; Galston, A.W. Effects of Exogenous 1,3-Diaminopropane and Spermidine on Senescence of Oat Leaves: II. Inhibition of Ethylene Biosynthesis and Possible Mode of Action. Plant Physiol. 1982, 70, 1597–1600. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Li, N.; Xu, C.; Zhong, S.; Lin, X.; Yang, J.; Zhou, T.; Yuliang, A.; Wu, Y.; Chen, Y.R.; et al. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 2014, 111, 10642–10647. [Google Scholar] [CrossRef] [Green Version]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Chen, Y.; Chen, J.; Dai, X.; Zhang, W. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Shi, H.; Hu, C.; Luo, M.; Xu, C.; Wang, S.; Li, N.; Tang, W.; Zhou, Y.; Wang, C.; et al. Transcriptome differences in response mechanisms to low-nitrogen stress in two wheat varieties. Int. J. Mol. Sci. 2021, 22, 12278. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, D.; Wang, Z.; Xun, H.; Ma, J.; Wang, H.; Huang, W.; Liu, Y.; Lin, X.; Li, N.; et al. Mutation of the RDR1 gene caused genome-wide changes in gene expression, regional variation in small RNA clusters and localized alteration in DNA methylation in rice. BMC Plant Biol. 2014, 14, 177. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.; Zhang, C. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Fan, X.; Lin, Y.; Li, Z.; Wang, Y.; Zhou, Y.; Meng, W.; Peng, Z.; Zhang, C.; Ma, J. Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice. Int. J. Mol. Sci. 2022, 23, 5561. https://doi.org/10.3390/ijms23105561
Wang N, Fan X, Lin Y, Li Z, Wang Y, Zhou Y, Meng W, Peng Z, Zhang C, Ma J. Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice. International Journal of Molecular Sciences. 2022; 23(10):5561. https://doi.org/10.3390/ijms23105561
Chicago/Turabian StyleWang, Ningning, Xuhong Fan, Yujie Lin, Zhe Li, Yingkai Wang, Yiming Zhou, Weilong Meng, Zhanwu Peng, Chunying Zhang, and Jian Ma. 2022. "Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice" International Journal of Molecular Sciences 23, no. 10: 5561. https://doi.org/10.3390/ijms23105561
APA StyleWang, N., Fan, X., Lin, Y., Li, Z., Wang, Y., Zhou, Y., Meng, W., Peng, Z., Zhang, C., & Ma, J. (2022). Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice. International Journal of Molecular Sciences, 23(10), 5561. https://doi.org/10.3390/ijms23105561





