Genetic Elements Orchestrating Lactobacillus crispatus Glycogen Metabolism in the Vagina
Abstract
:1. Introduction
2. Results
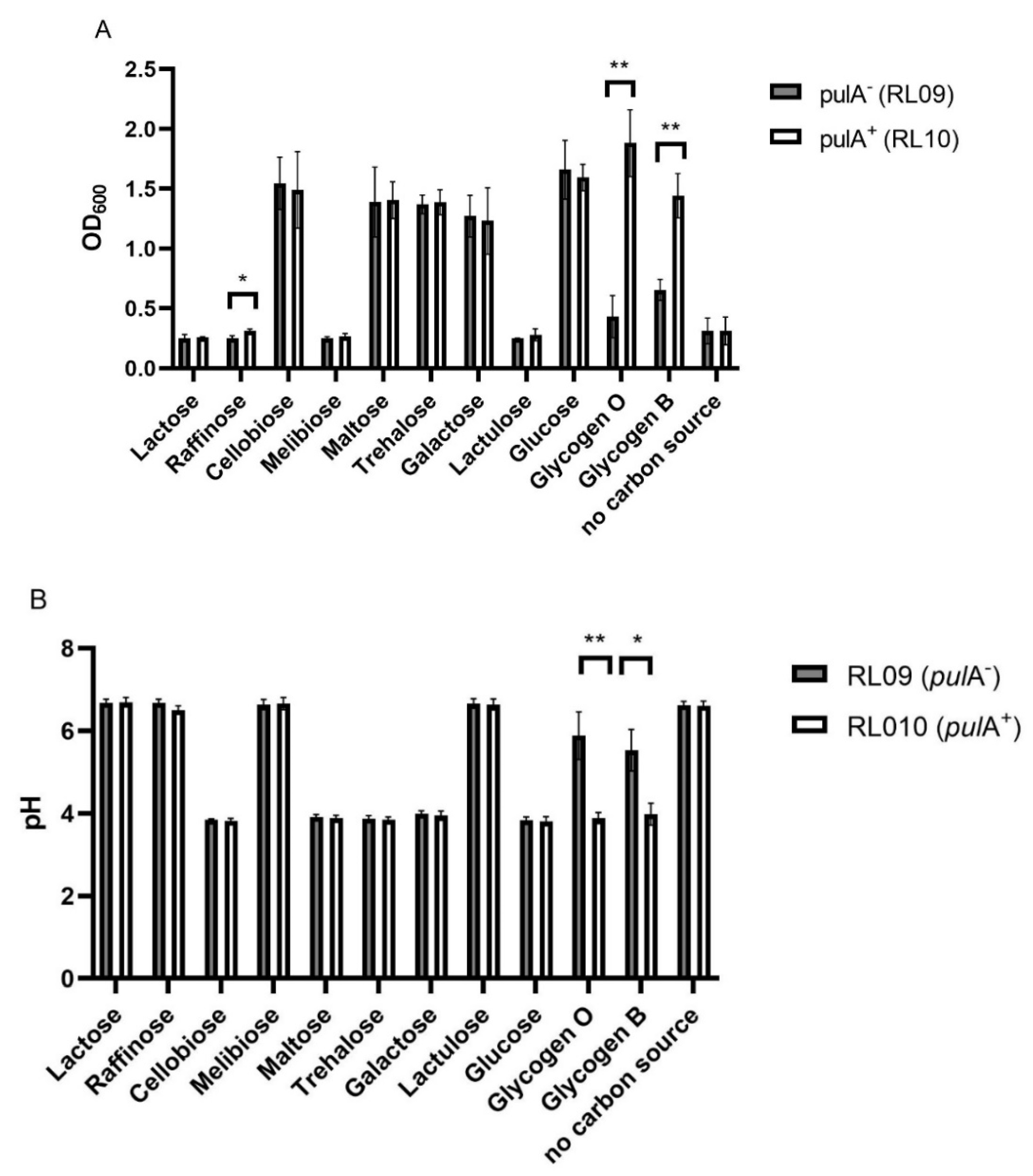
2.1. Substrate Utilization of PulA-Sufficient and Deficient Lactobacillus crispatus Strains
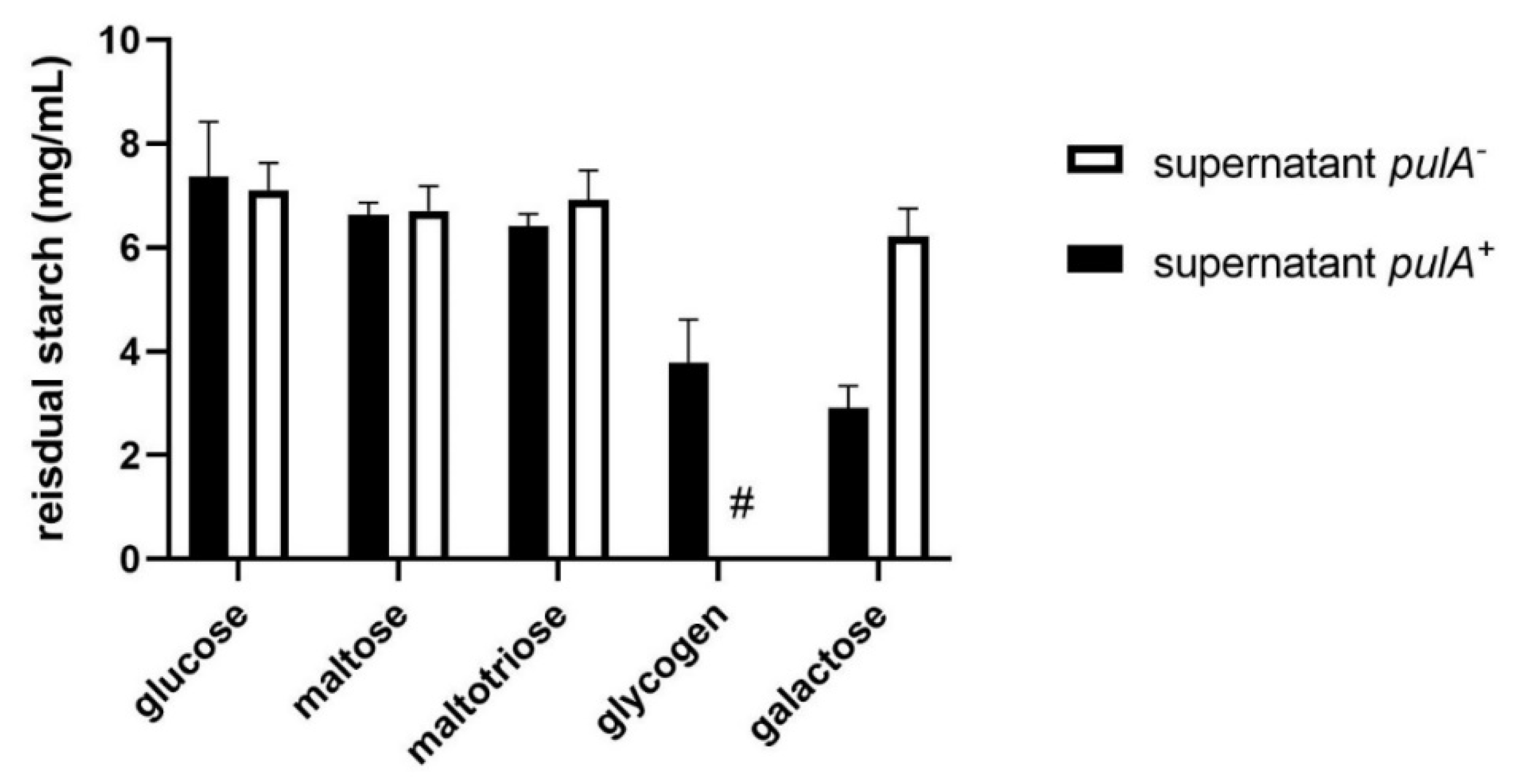
2.2. Starch Degradation Activity of PulA Sufficient and Deficient L. crispatus Strains on Various Carbon Sources
2.3. S-Layer Enrichment and Proteome Analysis
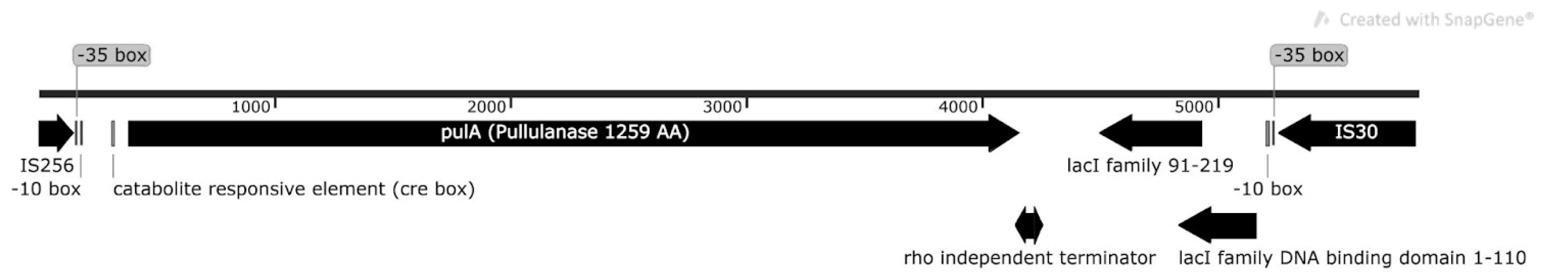
2.4. Metagenome Analysis
3. Discussion
4. Materials and Methods
4.1. Standard Cultivation Conditions
4.2. PCR and Sequencing of Signal Peptide Region L. crispatus Amylopullulanase
4.3. Serial Propagation on Various Carbohydrates
4.4. The Starch Degradation Assay
4.5. Enrichment for Surface Layer (Associated) Proteins
4.6. Sample Preparation for Proteomics Analysis
4.7. LC-MS/MS Analysis
4.8. Analysis of the PulA Gene Region
4.9. Analysis of Publicly Available Vaginal Metagenomes
4.10. Amylopullulanase Sequence Variant Detection
4.11. Identification of PulA Genes with Transposase Insertions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Ethnic Group | Number of Metagenomes | Number of Metagenomes with PulA | Number of Metagenomes with Defective PulA |
|---|---|---|---|
| Total | 1507 | 270 | 62 |
| Black or African American | 781 | 102 | 12 |
| White or Caucasian | 163 | 55 | 19 |
| Chinese | 76 | 4 | 2 |
| Unknown | 472 | 108 | 28 |
| Hispanic, mixed or other | 15 | 1 | 1 |
References
- O’Hanlon, D.E.; Moench, T.R.; Cone, R.A. Vaginal pH and Microbicidal Lactic Acid When Lactobacilli Dominate the Microbiota. PLoS ONE 2013, 8, e80074. [Google Scholar] [CrossRef] [PubMed]
- Mirmonsef, P.; Hotton, A.L.; Gilbert, D.; Burgad, D.; Landay, A.; Weber, K.M.; Cohen, M.; Ravel, J.; Spear, G.T. Free Glycogen in Vaginal Fluids is Associated with Lactobacillus Colonization and Low Vaginal pH. PLoS ONE 2014, 9, e102467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal Microbiome of Reproductive-Age Women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef] [Green Version]
- Dols, J.A.M.; Molenaar, D.; van der Helm, J.J.; Caspers, M.P.M.; de Kat Angelino-Bart, A.; Schuren, F.H.J.; Speksnijder, A.G.C.L.; Westerhoff, H.V.; Richardus, J.H.; Boon, M.E.; et al. Molecular Assessment of Bacterial Vaginosis by Lactobacillus Abundance and Species Diversity. BMC Infect. Dis. 2016, 16, 180. [Google Scholar] [CrossRef] [Green Version]
- Gosmann, C.; Anahtar, M.N.; Handley, S.A.; Farcasanu, M.; Abu-Ali, G.; Bowman, B.A.; Padavattan, N.; Desai, C.; Droit, L.; Moodley, A.; et al. Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated with Increased HIV Acquisition in Young South African Women. Immunity 2017, 46, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Seta, F.; Lonnee-Hoffmann, R.; Campisciano, G.; Comar, M.; Verstraelen, H.; Vieira-Baptista, P.; Ventolini, G.; Lev-Sagie, A. The Vaginal Microbiome: III. The Vaginal Microbiome in Various Urogenital Disorders. J. Low. Genit. Tract Dis. 2022, 26, 85–92. [Google Scholar] [CrossRef]
- Ceccarani, C.; Foschi, C.; Parolin, C.; D’Antuono, A.; Gaspari, V.; Consolandi, C.; Laghi, L.; Camboni, T.; Vitali, B.; Severgnini, M.; et al. Diversity of Vaginal Microbiome and Metabolome during Genital Infections. Sci. Rep. 2019, 9, 14095. [Google Scholar] [CrossRef] [Green Version]
- Koedooder, R.; Singer, M.; Schoenmakers, S.; Savelkoul, P.H.M.; Morré, S.A.; de Jonge, J.D.; Poort, L.; Cuypers, W.; Beckers, N.G.M.; Broekmans, F.J.M.; et al. The Vaginal Microbiome as a Predictor for Outcome of in Vitro Fertilization with or without Intracytoplasmic Sperm Injection: A Prospective Study. Hum. Reprod. 2019, 34, 1042–1054. [Google Scholar] [CrossRef]
- Vergaro, P.; Tiscornia, G.; Barragán, M.; García, D.; Rodriguez, A.; Santaló, J.; Vassena, R. Vaginal Microbiota Profile at the Time of Embryo Transfer Does Not Affect Live Birth Rate in IVF Cycles with Donated Oocytes. Reprod. Biomed. Online 2019, 38, 883–891. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The Vaginal Microbiome and Preterm Birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef] [Green Version]
- Usyk, M.; Schlecht, N.F.; Pickering, S.; Williams, L.; Sollecito, C.C.; Gradissimo, A.; Porras, C.; Safaeian, M.; Pinto, L.; Herrero, R.; et al. MolBV Reveals Immune Landscape of Bacterial Vaginosis and Predicts Human Papillomavirus Infection Natural History. Nat. Commun. 2022, 13, 233. [Google Scholar] [CrossRef] [PubMed]
- Dols, J.A.M.; Reid, G.; Kort, R.; Schuren, F.H.J.; Tempelman, H.; Bontekoe, T.R.; Korporaal, H.; Van der Veer, E.M.; Smit, P.W.; Boon, M.E. PCR-Based Identification of Eight Lactobacillus Species and 18 Hr-HPV Genotypes in Fixed Cervical Samples of South African Women at Risk of HIV and BV. Diagn. Cytopathol. 2012, 40, 472–477. [Google Scholar] [CrossRef] [PubMed]
- van der Veer, C.; Hertzberger, R.Y.; Bruisten, S.M.; Tytgat, H.L.P.; Swanenburg, J.; de Kat Angelino-Bart, A.; Schuren, F.; Molenaar, D.; Reid, G.; de Vries, H.; et al. Comparative Genomics of Human Lactobacillus crispatus Isolates Reveals Genes for Glycosylation and Glycogen Degradation: Implications for in Vivo Dominance of the Vaginal Microbiota. Microbiome 2019, 7, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolston, B.M.; Jenkins, D.J.; Hood-Pishchany, M.I.; Nahoum, S.R.; Balskus, E.P. Characterization of Vaginal Microbial Enzymes Identifies Amylopullulanases That Support Growth of Lactobacillus crispatus on Glycogen. bioRxiv 2021, 2021.07.19.452977. [Google Scholar] [CrossRef]
- Pan, M.; Hidalgo-Cantabrana, C.; Barrangou, R. Host and Body Site-Specific Adaptation of Lactobacillus crispatus Genomes. NAR Genom. Bioinform. 2020, 2, lqaa001. [Google Scholar] [CrossRef] [Green Version]
- Nasioudis, D.; Beghini, J.; Bongiovanni, A.M.; Giraldo, P.C.; Linhares, I.M.; Witkin, S.S. Alpha-Amylase in Vaginal Fluid: Association With Conditions Favorable to Dominance of Lactobacillus. Reprod. Sci. 2015, 22, 1393–1398. [Google Scholar] [CrossRef]
- Spear, G.T.; French, A.L.; Gilbert, D.; Zariffard, M.R.; Mirmonsef, P.; Sullivan, T.H.; Spear, W.W.; Landay, A.; Micci, S.; Lee, B.H.; et al. Human Alpha-Amylase Present in Lower-Genital-Tract Mucosal Fluid Processes Glycogen to Support Vaginal Colonization by Lactobacillus. J. Infect. Dis. 2014, 210, 1019–1028. [Google Scholar] [CrossRef] [Green Version]
- Kenetta, L.N.; Geremy, C.C.; Joshua, N.A.; Engbrecht, K.; Fillmore, T.; Forney, L.J.; Vincent, B.Y. Amylases in the Human Vagina. mSphere 2020, 5, e00943-20. [Google Scholar] [CrossRef]
- Spear, G.T.; McKenna, M.; Landay, A.L.; Makinde, H.; Hamaker, B.; French, A.L.; Lee, B.H. Effect of PH on Cleavage of Glycogen by Vaginal Enzymes. PLoS ONE 2015, 10, e0132646. [Google Scholar] [CrossRef] [Green Version]
- Collins, S.L.; McMillan, A.; Seney, S.; van der Veer, C.; Kort, R.; Sumarah, M.W.; Reid, G. Promising Prebiotic Candidate Established by Evaluation of Lactitol, Lactulose, Raffinose, and Oligofructose for Maintenance of a Lactobacillus-Dominated Vaginal Microbiota. Appl. Environ. Microbiol. 2018, 84, e02200-17. [Google Scholar] [CrossRef] [Green Version]
- Møller, M.S.; Goh, Y.J.; Rasmussen, K.B.; Cypryk, W.; Celebioglu, H.U.; Klaenhammer, T.R.; Svensson, B.; Hachem, M.A. An Extracellular Cell-Attached Pullulanase Confers Branched α-Glucan Utilization in Human Gut Lactobacillus Acidophilus. Appl. Environ. Microbiol. 2017, 83, e00402-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muscariello, L.; Marasco, R.; De Felice, M.; Sacco, M. The Functional CcpA Gene Is Required for Carbon Catabolite Repression in Lactobacillus Plantarum. Appl. Environ. Microbiol. 2001, 67, 2903–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahr, K.; Hillen, W.; Titgemeyer, F. Carbon Catabolite Repression in Lactobacillus Pentosus: Analysis of the CcpA Region. Appl. Environ. Microbiol. 2000, 66, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Azcarate-Peril, M.A.; Duong, T.; Conners, S.B.; Kelly, R.M.; Klaenhammer, T.R. Global Analysis of Carbohydrate Utilization by Lactobacillus Acidophilus Using CDNA Microarrays. Proc. Natl. Acad. Sci. USA 2006, 103, 3816–3821. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.; Turner, K.W.; Thomas, T.D. Catabolite Inhibition and Sequential Metabolism of Sugars by Streptococcus Lactis. J. Bacteriol. 1978, 133, 1163–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, N.; Stanley, G.A.; Bunte, A.; Rdstrm, P. Product Formation and Phosphoglucomutase Activities in Lactococcus Lactis: Cloning and Characterization of a Novel Phosphoglucomutase Gene. Microbiology 1997, 143 Pt 3, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Åvall-Jääskeläinen, S.; Palva, A. Lactobacillus Surface Layers and Their Applications. FEMS Microbiol. Rev. 2005, 29, 511–529. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; France, M.T.; Crabtree, J.; Holm, J.B.; Humphrys, M.S.; Brotman, R.M.; Ravel, J. A Comprehensive Non-Redundant Gene Catalog Reveals Extensive within-Community Intraspecies Diversity in the Human Vagina. Nat. Commun. 2020, 11, 940. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, H.; Molenaar, D.; Kleerebezem, M.; van Hylckama Vlieg, J.E. High Local Substrate Availability Stabilizes a Cooperative Trait. ISME J. 2011, 5, 929–932. [Google Scholar] [CrossRef]
- Brandt, K.; Barrangou, R. Adaptive Response to Iterative Passages of Five Lactobacillus Species in Simulated Vaginal Fluid. BMC Microbiol. 2020, 20, 339. [Google Scholar] [CrossRef]
- Lithgow, K.V.; Cochinamogulos, A.; Muirhead, K.; Konschuh, S.; Oluoch, L.; Mugo, N.R.; Roxby, A.C.; Sycuro, L.K. Resolution of glycogen and glycogen-degrading activities reveals correlates of Lactobacillus crispatus dominance in a cohort of young African women. bioRxiv 2022, 2022.03.29.486257. [Google Scholar] [CrossRef]
- Srinivasan, S.; Morgan, M.T.; Fiedler, T.L.; Djukovic, D.; Hoffman, N.G.; Raftery, D.; Marrazzo, J.M.; Fredricks, D.N. Metabolic Signatures of Bacterial Vaginosis. mBio 2015, 6, e00204-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregoire, A.T. Carbohydrates of Human Vaginal Tissue. Nature 1963, 198, 996. [Google Scholar] [CrossRef] [PubMed]
- MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification|Nature Biotechnology. Available online: https://www.nature.com/articles/nbt.1511 (accessed on 15 April 2022).
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences 3 MATERIAL AND METHODS Learning Parameters and Prediction of Protein-Coding Genes. Available online: https://www.semanticscholar.org/paper/Automatic-Annotation-of-Microbial-Genomes-and-3-AND-Solovyev-Salamov/88cd5fdbfb2dba16a6b23987d30ad9437ff0c805 (accessed on 15 April 2022).
- Lu, Y.; Song, S.; Tian, H.; Yu, H.; Zhao, J.; Chen, C. Functional Analysis of the Role of CcpA in Lactobacillus Plantarum Grown on Fructooligosaccharides or Glucose: A Transcriptomic Perspective. Microb. Cell Factories 2018, 17, 201. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. A Framework for Human Microbiome Research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Chen, C.; Wei, W.; Wang, Z.; Dai, J.; Hao, L.; Song, L.; Zhang, X.; Zeng, L.; Du, H.; et al. The Metagenome of the Female Upper Reproductive Tract. GigaScience 2018, 7, giy107. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating Species Abundance in Metagenomics Data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: A Bioconductor Package for Handling and Analysis of High-Throughput Phylogenetic Sequence Data. In Pacific Symposium on Biocomputing; World Scientific: Singapore, 2012; pp. 235–246. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obenchain, V.; Lawrence, M.; Carey, V.; Gogarten, S.; Shannon, P.; Morgan, M. VariantAnnotation: A Bioconductor Package for Exploration and Annotation of Genetic Variants. Bioinformatics 2014, 30, 2076–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]


| ACC NO | Description | PEP | RL09 GAL | RL10 GAL | RL10 GLU | RL10 GLY | 2logR1 | 2logR2 |
|---|---|---|---|---|---|---|---|---|
| A0A4Q0LMV5 | Amylopullulanase | 64 | 2.8 × 105 | 2.5 × 107 | 7.5 × 105 | 1.8 × 107 | 6.5 | 4.6 |
| A0A135Z5X6 | Cell surface protein | 11 | 1.5 × 106 | 3.2 ×106 | 1.2 × 106 | 3.2 × 106 | 1.1 | 1.4 |
| D5GXS8 | Lysin (Glycoside Hydrolase Family) | 10 | 2.9 × 105 | 3.6 × 105 | 4.0 × 105 | 9.7 × 105 | 0.3 | 1.3 |
| D5H1Q3 | Aggregation promoting factor | 4 | ND | ND | 1.5 × 105 | 2.2 × 105 | ND | 0.6 |
| A0A135ZFI7 | N-acetylmuramidase | 17 | 1.3 × 106 | 2.0 × 106 | 4.0 × 106 | 4.1 × 106 | 0.6 | 0.04 |
| K1MET7 | Uncharacterized protein | 29 | 6.4 × 106 | 1.0 × 107 | 7.6 × 106 | 2.3 × 106 | 0.7 | 0.04 |
| A0A6M1GK10 | S-layer protein (Fragment) | 32 | 4.2 × 105 | 6.9 × 105 | 3.0 × 105 | 3.0 × 105 | 0.7 | −0.02 |
| C7XI68 | SlpA domain-containing protein | 14 | 3.2 × 105 | 7.3 × 105 | 6.4 × 105 | 6.1 × 105 | 1.2 | −0.07 |
| A0A6B8SPR2 | SlpA domain-containing protein | 15 | 3.1 × 107 | 2.2 × 107 | 1.3 × 107 | 1.1 × 107 | −0.5 | −0.2 |
| A0A135ZF64 | Amidase | 10 | 5.7 × 105 | 1.7 × 106 | 1.7 × 106 | 1.4 × 106 | 1.6 | −0.2 |
| A0A135ZH73 | Lysin | 11 | 4.5 × 106 | 8.8 × 106 | 6.6 × 106 | 5.6 × 106 | 1.0 | −0.2 |
| A0A7V8KTK5 | Putative surface layer protein | 48 | 1.6 × 107 | 2.8 × 106 | 2.7 × 107 | 2.2 × 107 | 0.8 | −0.3 |
| E3R4X6 | Uncharacterized protein | 9 | 2.6 × 106 | 2.8 × 106 | 2.7 × 106 | 2.3 × 106 | 0.1 | −0.3 |
| K1MJV3 | Uncharacterized protein | 22 | 1.6 × 107 | 2.9 × 107 | 2.0 × 107 | 1.6 × 107 | 0.8 | −0.3 |
| V5EI62 | Lysin (Glycoside Hydrolase Family) | 15 | 2.4 × 106 | 4.6 × 106 | 1.1 × 107 | 7.6 × 106 | 0.9 | −0.5 |
| V5EF53 | SlpA domain-containing protein | 12 | 2.1 × 107 | 3.0 × 107 | 6.1 × 107 | 4.3 × 107 | 0.5 | −0.5 |
| A0A4Q0LUE2 | Uncharacterized protein | 24 | 2.7 × 107 | 1.8 × 107 | 3.3 × 107 | 2.3 × 107 | −0.6 | −0.5 |
| A0A4R6CSS1 | Cell surface protein (SlpA) | 21 | 2.4 × 106 | 5.8 × 106 | 6.6 × 106 | 4.6 × 106 | 1.3 | −0.5 |
| D0DJ09 | Bacterial surface layer protein | 45 | 3.6 × 108 | 4.8 × 108 | 5.1 × 108 | 3.5 × 108 | 0.4 | −0.5 |
| K1M186 | Uncharacterized protein | 12 | 8.0 × 106 | 9.7 × 106 | 9.5 × 106 | 6.1 × 106 | 0.3 | −0.6 |
| A0A135YZ62 | Bacterial surface layer protein (SlpA) | 32 | 2.0 × 107 | 3.3 × 107 | 8.7 × 107 | 5.5 × 107 | 0.8 | −0.6 |
| D0DIV9 | Uncharacterized protein | 36 | 5.3 × 107 | 7.6 × 107 | 1.3 × 108 | 7.9 × 107 | 0.5 | −0.7 |
| V5GA31 | Lactocepin s-layer protein | 10 | 1.8 × 106 | 2.7 × 106 | 8.4 × 106 | 3.2 × 106 | 0.6 | −1.4 |
| A0A120DHV3 | Uncharacterized protein | 16 | 6.0 × 106 | 1.1 × 107 | 2.1 × 107 | 7.7 × 106 | 0.8 | −1.4 |
| K1N369 | Uncharacterized protein | 8 | 9.8 × 104 | 3.7 × 104 | 1.4 × 105 | 3.0 × 104 | −1.4 | −2.2 |
| A0A125P6L8 | S-layer protein (SlpA) | 13 | 1.1 × 104 | 1.3 × 105 | 8.7 × 105 | 4.2 × 103 | 3.5 | −7.7 |
| A0A7T1TNA6 | Bacterial surface layer protein | 30 | 4.7 × 106 | 5.7 × 106 | 6.1 × 107 | 2.5 × 105 | 0.3 | −7.9 |
| A0A1C2D5C0 | Cell surface protein | 5 | ND | 1.5 × 106 | 8.1 × 105 | ND | ND | ND |
| Fraction of total detected proteins (iBAQ) | 0.47 | 0.46 | 0.22 | 0.40 | ||||
| Strain | PulA Variant |
|---|---|
| RL03, RL08, RL10, RL11, RL13, RL14, RL15, RL16, RL17, RL20, RL21, RL22, RL23, RL24, RL25, RL27, RL28, RL29, RL30, RL33 | Wild type |
| RL02 and RL09 | Two nucleotide deletion |
| RL06 and RL07 | N-terminal insertion of mobile element (variant 1) |
| RL19 and RL26 | N-terminal insertion of mobile element (variant 2) |
| RL05 | Large deletion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hertzberger, R.; May, A.; Kramer, G.; van Vondelen, I.; Molenaar, D.; Kort, R. Genetic Elements Orchestrating Lactobacillus crispatus Glycogen Metabolism in the Vagina. Int. J. Mol. Sci. 2022, 23, 5590. https://doi.org/10.3390/ijms23105590
Hertzberger R, May A, Kramer G, van Vondelen I, Molenaar D, Kort R. Genetic Elements Orchestrating Lactobacillus crispatus Glycogen Metabolism in the Vagina. International Journal of Molecular Sciences. 2022; 23(10):5590. https://doi.org/10.3390/ijms23105590
Chicago/Turabian StyleHertzberger, Rosanne, Ali May, Gertjan Kramer, Isabelle van Vondelen, Douwe Molenaar, and Remco Kort. 2022. "Genetic Elements Orchestrating Lactobacillus crispatus Glycogen Metabolism in the Vagina" International Journal of Molecular Sciences 23, no. 10: 5590. https://doi.org/10.3390/ijms23105590
APA StyleHertzberger, R., May, A., Kramer, G., van Vondelen, I., Molenaar, D., & Kort, R. (2022). Genetic Elements Orchestrating Lactobacillus crispatus Glycogen Metabolism in the Vagina. International Journal of Molecular Sciences, 23(10), 5590. https://doi.org/10.3390/ijms23105590






