Ancient Origins of Cytoskeletal Crosstalk: Spectraplakin-like Proteins Precede the Emergence of Cortical Microtubule Stabilization Complexes as Crosslinkers
Abstract
1. Introduction
2. Results and Discussion
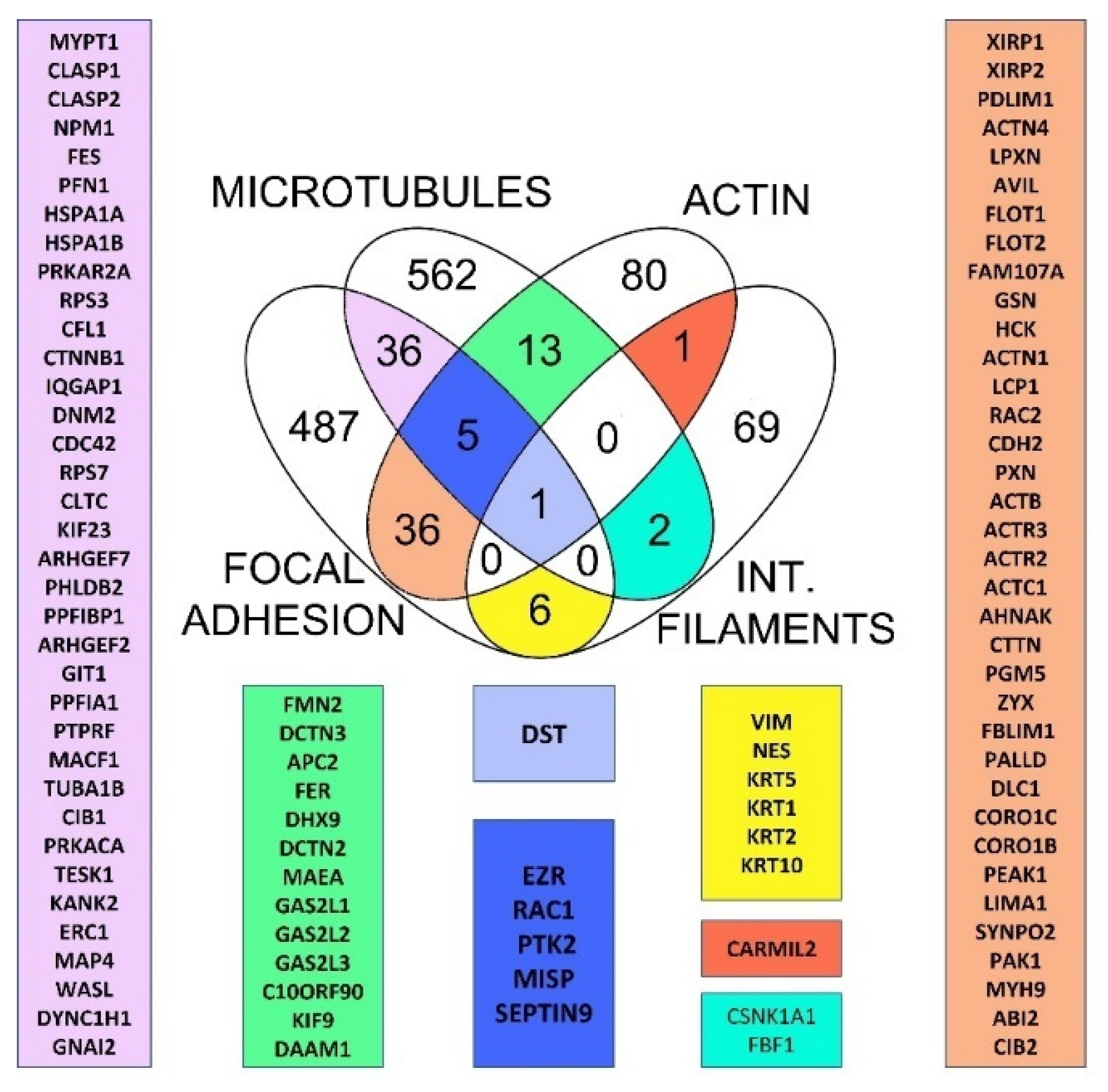
2.1. Retrieving of Cytoskeleton Crosstalk Proteins from Databases Pointed Dystonin as Major Regulator of Cytoskeletal Crosstalk
2.2. Evolutionary Conservation of Cytoskeletal Crosstalk Proteins
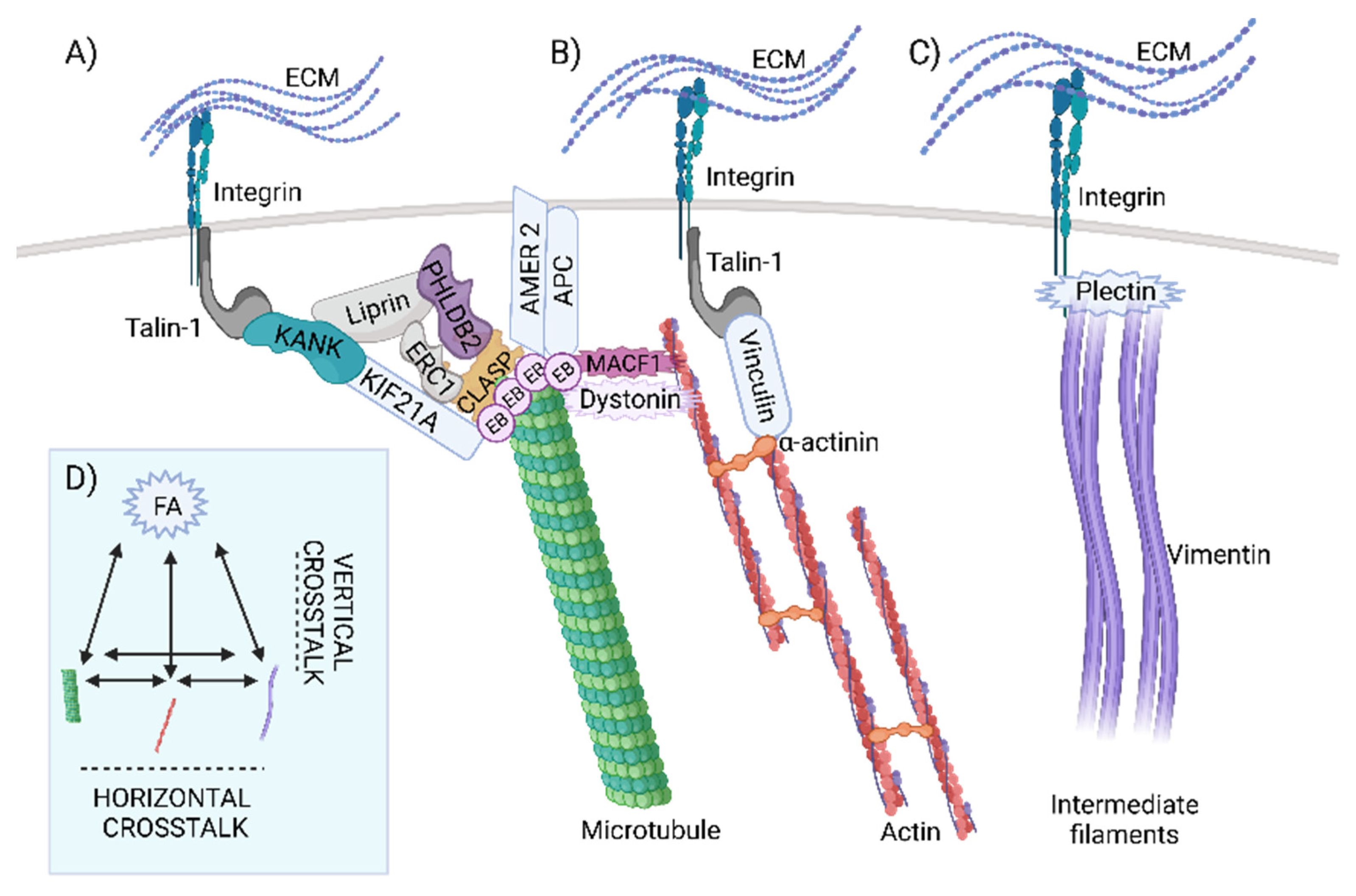
2.3. Evolution of Horizontal Crosstalk Interaction Hot Spots
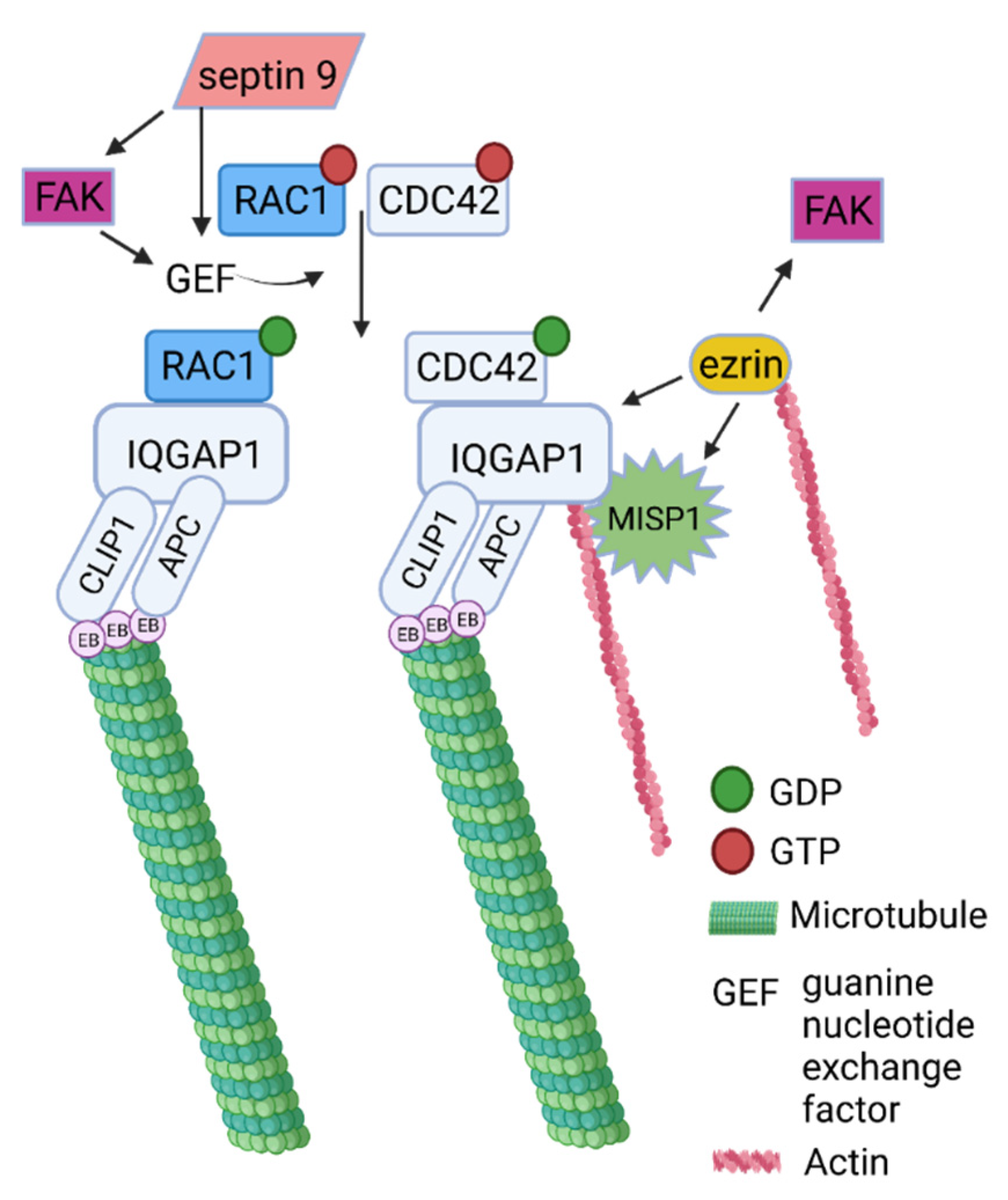
2.4. Dataset Analysis Revealed MISP as a New Player in Human Cytoskeletal Crosstalk
2.5. The Evolution of Signal Transduction to Microtubules through CMSC and the KANK-KIF21 Axis Revealed Its Origin in Metazoa
3. Conclusions
4. Methods
4.1. Protein Dataset Assessment—Updating a List of Proteins Belonging to Human Cytoskeleton
4.2. Clustering of Human Cytoskeleton Proteins
4.3. Model Organism Dataset
4.4. Data Collection
4.5. Identification and Analysis of Protein Conserved Domains
4.6. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaalouk, D.E.; Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009, 10, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Llort Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed]
- Samanta, D.; Almo, S.C. Nectin family of cell-adhesion molecules: Structural and molecular aspects of function and specificity. Cell. Mol. Life Sci. 2015, 72, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: A family of cell surface receptors. Cell 1987, 48, 549–554. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, Allosteric Signaling Machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Menter, D.G.; Dubois, R.N. Prostaglandins in cancer cell adhesion, migration, and invasion. Int. J. Cell Biol. 2012, 2012, 723419. [Google Scholar] [CrossRef]
- Davies, P.F.; Robotewskyj, A.; Griem, M.L. Quantitative studies of endothelial cell adhesion. Directional remodeling of focal adhesion sites in response to flow forces. J. Clin. Investig. 1994, 93, 2031–2038. [Google Scholar] [CrossRef]
- Yue, J.; Zhang, Y.; Liang, W.G.; Gou, X.; Lee, P.; Liu, H.; Lyu, W.; Tang, W.-J.; Chen, S.-Y.; Yang, F.; et al. In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat. Commun. 2016, 7, 11692. [Google Scholar] [CrossRef]
- Buehler, M.J. Mechanical players-The role of intermediate filaments in cell mechanics and organization. Biophys. J. 2013, 105, 1733–1734. [Google Scholar] [CrossRef][Green Version]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef]
- Pollard, T.D.; Goldman, R.D. Overview of the Cytoskeleton from an Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a030288. [Google Scholar] [CrossRef] [PubMed]
- Jékely, G. Origin and Evolution of the Self-Organizing Cytoskeleton in the Network of Eukaryotic Organelles. Cold Spring Harb. Perspect. Biol. 2014, 6, a016030. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peter, A.; Stick, R. Evolutionary aspects in intermediate filament proteins. Curr. Opin. Cell Biol. 2015, 32, 48–55. [Google Scholar] [CrossRef]
- Tekle, Y.I.; Williams, J.R. Cytoskeletal architecture and its evolutionary significance in amoeboid eukaryotes and their mode of locomotion. R. Soc. Open Sci. 2016, 3, 160283. [Google Scholar] [CrossRef] [PubMed]
- Manich, M.; Hernandez-Cuevas, N.; Ospina-Villa, J.D.; Syan, S.; Marchat, L.A.; Olivo-Marin, J.-C.; Guillén, N. Morphodynamics of the Actin-Rich Cytoskeleton in Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2018, 8, 179. [Google Scholar] [CrossRef]
- Kang, S.; Tice, A.K.; Stairs, C.W.; Jones, R.E.; Lahr, D.J.G.; Brown, M.W. The integrin-mediated adhesive complex in the ancestor of animals, fungi, and amoebae. Curr. Biol. 2021, 31, 3073–3085.e3073. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Roger, A.J.; Lang, F.B.; King, N.; Ruiz-Trillo, I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. USA 2010, 107, 10142–10147. [Google Scholar] [CrossRef]
- Heiss, A.A.; Walker, G.; Simpson, A.G. The microtubular cytoskeleton of the apusomonad Thecamonas, a sister lineage to the opisthokonts. Protist 2013, 164, 598–621. [Google Scholar] [CrossRef]
- Parra-Acero, H.; Harcet, M.; Sánchez-Pons, N.; Casacuberta, E.; Brown, N.H.; Dudin, O.; Ruiz-Trillo, I. Integrin-Mediated Adhesion in the Unicellular Holozoan Capsaspora owczarzaki. Curr. Biol. 2020, 30, 4270–4275.e4. [Google Scholar] [CrossRef]
- Kożyczkowska, A.; Najle, S.R.; Ocaña-Pallarès, E.; Aresté, C.; Shabardina, V.; Ara, P.S.; Ruiz-Trillo, I.; Casacuberta, E. Stable transfection in protist Corallochytrium limacisporum identifies novel cellular features among unicellular animals relatives. Curr. Biol. 2021, 31, 4104–4110.e4. [Google Scholar] [CrossRef]
- Mitchell, J.M.; Nichols, S.A. Diverse cell junctions with unique molecular composition in tissues of a sponge (Porifera). EvoDevo 2019, 10, 26. [Google Scholar] [CrossRef]
- Miller, P.W.; Pokutta, S.; Mitchell, J.M.; Chodaparambil, J.V.; Clarke, D.N.; Nelson, W.J.; Weis, W.I.; Nichols, S.A. Analysis of a vinculin homolog in a sponge (phylum Porifera) reveals that vertebrate-like cell adhesions emerged early in animal evolution. J. Biol. Chem. 2018, 293, 11674–11686. [Google Scholar] [CrossRef] [PubMed]
- DuBuc, T.Q.; Ryan, J.F.; Martindale, M.Q. “Dorsal–Ventral” Genes Are Part of an Ancient Axial Patterning System: Evidence from Trichoplax adhaerens (Placozoa). Mol. Biol. Evol. 2019, 36, 966–973. [Google Scholar] [CrossRef]
- Srivastava, M.; Begovic, E.; Chapman, J.; Putnam, N.H.; Hellsten, U.; Kawashima, T.; Kuo, A.; Mitros, T.; Salamov, A.; Carpenter, M.L.; et al. The Trichoplax genome and the nature of placozoans. Nature 2008, 454, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Kollmar, M. Polyphyly of nuclear lamin genes indicates an early eukaryotic origin of the metazoan-type intermediate filament proteins. Sci. Rep. 2015, 5, 10652. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Cusseddu, R.; Robert, A.; Côté, J.-F. Strength Through Unity: The Power of the Mega-Scaffold MACF1. Front. Cell Dev. Biol. 2021, 9, 577. [Google Scholar] [CrossRef]
- Künzli, K.; Favre, B.; Chofflon, M.; Borradori, L. One gene but different proteins and diseases: The complexity of dystonin and bullous pemphigoid antigen 1. Exp. Dermatol. 2016, 25, 10–16. [Google Scholar] [CrossRef]
- Stroud, M.J.; Nazgiewicz, A.; McKenzie, E.A.; Wang, Y.; Kammerer, R.A.; Ballestrem, C. GAS2-like proteins mediate communication between microtubules and actin through interactions with end-binding proteins. J. Cell Sci. 2014, 127, 2672–2682. [Google Scholar] [CrossRef]
- Ortega, E.; Manso, J.A.; Buey, R.M.; Carballido, A.M.; Carabias, A.; Sonnenberg, A.; de Pereda, J.M. The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape. J. Biol. Chem. 2016, 291, 18643–18662. [Google Scholar] [CrossRef]
- Fogl, C.; Mohammed, F.; Al-Jassar, C.; Jeeves, M.; Knowles, T.J.; Rodriguez-Zamora, P.; White, S.A.; Odintsova, E.; Overduin, M.; Chidgey, M. Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin. Nat. Commun. 2016, 7, 10827. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, F.; Trieber, C.; Overduin, M.; Chidgey, M. Molecular mechanism of intermediate filament recognition by plakin proteins. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2020, 1867, 118801. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.B.; Guerreiro, P.S.; Canato, S.; Janody, F. The spectraplakin Dystonin antagonizes YAP activity and suppresses tumourigenesis. Sci. Rep. 2019, 9, 19843. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.R. Cell adhesion molecules as targets of autoantibodies in pemphigus and pemphigoid, bullous diseases due to defective epidermal cell adhesion. Adv. Immunol. 1993, 53, 291–325. [Google Scholar] [CrossRef]
- Poliakova, K.; Adebola, A.; Leung, C.L.; Favre, B.; Liem, R.K.; Schepens, I.; Borradori, L. BPAG1a and b associate with EB1 and EB3 and modulate vesicular transport, Golgi apparatus structure, and cell migration in C2.7 myoblasts. PLoS ONE 2014, 9, e107535. [Google Scholar] [CrossRef] [PubMed]
- Tomar, A.; Schlaepfer, D.D. Focal adhesion kinase: Switching between GAPs and GEFs in the regulation of cell motility. Curr. Opin. Cell Biol. 2009, 21, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Oyang, L.; Rao, S.; Han, Y.; Luo, X.; Yi, P.; Lin, J.; Xia, L.; Hu, J.; Tan, S.; et al. Rac1, A Potential Target for Tumor Therapy. Front. Oncol. 2021, 11, 674426. [Google Scholar] [CrossRef]
- Nagata, K.; Kawajiri, A.; Matsui, S.; Takagishi, M.; Shiromizu, T.; Saitoh, N.; Izawa, I.; Kiyono, T.; Itoh, T.J.; Hotani, H.; et al. Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. J. Biol. Chem. 2003, 278, 18538–18543. [Google Scholar] [CrossRef]
- Nakos, K.; Rosenberg, M.; Spiliotis, E.T. Regulation of microtubule plus end dynamics by septin 9. Cytoskeleton 2019, 76, 83–91. [Google Scholar] [CrossRef]
- Kschonsak, Y.T.; Hoffmann, I. Activated ezrin controls MISP levels to ensure correct NuMA polarization and spindle orientation. J. Cell Sci. 2018, 131, jcs214544. [Google Scholar] [CrossRef]
- Fontao, L.; Geerts, D.; Kuikman, I.; Koster, J.; Kramer, D.; Sonnenberg, A. The interaction of plectin with actin: Evidence for cross-linking of actin filaments by dimerization of the actin-binding domain of plectin. J. Cell Sci. 2001, 114, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Leube, R.E.; Moch, M.; Windoffer, R. Intermediate filaments and the regulation of focal adhesion. Curr. Opin. Cell Biol. 2015, 32, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Osmanagic-Myers, S.; Rus, S.; Wolfram, M.; Brunner, D.; Goldmann, W.H.; Bonakdar, N.; Fischer, I.; Reipert, S.; Zuzuarregui, A.; Walko, G.; et al. Plectin reinforces vascular integrity by mediating crosstalk between the vimentin and the actin networks. J. Cell Sci. 2015, 128, 4138–4150. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Knifley, T.; Chen, M.; O’Connor, K.L. Integrin α6β4 requires plectin and vimentin for adhesion complex distribution and invasive growth. J. Cell Sci. 2022, 135, jcs258471. [Google Scholar] [CrossRef]
- Laly, A.C.; Sliogeryte, K.; Pundel, O.J.; Ross, R.; Keeling, M.C.; Avisetti, D.; Waseem, A.; Gavara, N.; Connelly, J.T. The keratin network of intermediate filaments regulates keratinocyte rigidity sensing and nuclear mechanotransduction. Sci. Adv. 2021, 7, eabd6187. [Google Scholar] [CrossRef]
- Preisner, H.; Habicht, J.; Garg, S.G.; Gould, S.B. Intermediate filament protein evolution and protists. Cytoskeleton 2018, 75, 231–243. [Google Scholar] [CrossRef]
- Baines, A.J. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem. Soc. Trans. 2009, 37, 796–803. [Google Scholar] [CrossRef]
- Broderick, M.J.F.; Winder, S.J. Spectrin, α-Actinin, and Dystrophin. In Advances in Protein Chemistry; Academic Press: Cambridge, MA, USA, 2005; Volume 70, pp. 203–246. [Google Scholar]
- Virel, A.; Backman, L. A Comparative and Phylogenetic Analysis of the α-Actinin Rod Domain. Mol. Biol. Evol. 2007, 24, 2254–2265. [Google Scholar] [CrossRef]
- Pérez-Munive, C.; Moreno Díaz de la Espina, S. Nuclear spectrin-like proteins are structural actin-binding proteins in plants. Biol. Cell 2011, 103, 145–157. [Google Scholar] [CrossRef]
- Virel, A.; Backman, L. Molecular Evolution and Structure of α-Actinin. Mol. Biol. Evol. 2004, 21, 1024–1031. [Google Scholar] [CrossRef]
- Baines, A.J. Evolution of the spectrin-based membrane skeleton. Transfus. Clin. Biol. 2010, 17, 95–103. [Google Scholar] [CrossRef]
- Persson, K.; Backman, L. Structural and functional characterization of a plant alpha-actinin. FEBS Open Bio 2021, 11, 2198–2210. [Google Scholar] [CrossRef]
- Stroud, M.J.; Kammerer, R.A.; Ballestrem, C. Characterization of G2L3 (GAS2-like 3), a new microtubule- and actin-binding protein related to spectraplakins. J. Biol. Chem. 2011, 286, 24987–24995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yue, J.; Wu, X. Spectraplakin family proteins—Cytoskeletal crosslinkers with versatile roles. J. Cell Sci. 2017, 130, 2447–2457. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.R.; Fuchs, E.; Slep, K.C. Structure of the ACF7 EF-Hand-GAR Module and Delineation of Microtubule Binding Determinants. Structure 2017, 25, 1130–1138.e1136. [Google Scholar] [CrossRef] [PubMed]
- Liem, R.K. Cytoskeletal Integrators: The Spectrin Superfamily. Cold Spring Harb. Perspect. Biol. 2016, 8, a018259. [Google Scholar] [CrossRef] [PubMed]
- Kurochkina, N.; Guha, U. SH3 domains: Modules of protein-protein interactions. Biophys. Rev. 2013, 5, 29–39. [Google Scholar] [CrossRef]
- Dionne, U.; Bourgault, É.; Dubé, A.K.; Bradley, D.; Chartier, F.J.M.; Dandage, R.; Dibyachintan, S.; Després, P.C.; Gish, G.D.; Pham, N.T.H.; et al. Protein context shapes the specificity of SH3 domain-mediated interactions in vivo. Nat. Commun. 2021, 12, 1597. [Google Scholar] [CrossRef]
- Gushchina, L.V.; Gabdulkhakov, A.G.; Nikonov, S.V.; Filimonov, V.V. High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide. J. Biomol. Struct. Dyn. 2011, 29, 485–495. [Google Scholar] [CrossRef]
- Koster, J.; Geerts, D.; Favre, B.; Borradori, L.; Sonnenberg, A. Analysis of the interactions between BP180, BP230, plectin and the integrin α6β4 important for hemidesmosome assembly. J. Cell Sci. 2003, 116, 387–399. [Google Scholar] [CrossRef]
- Hijikata, T.; Nakamura, A.; Isokawa, K.; Imamura, M.; Yuasa, K.; Ishikawa, R.; Kohama, K.; Takeda, S.; Yorifuji, H. Plectin 1 links intermediate filaments to costameric sarcolemma through β-synemin, α-dystrobrevin and actin. J. Cell Sci. 2008, 121, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Koster, J.; van Wilpe, S.; Kuikman, I.; Litjens, S.H.M.; Sonnenberg, A. Role of Binding of Plectin to the Integrin β4 Subunit in the Assembly of Hemidesmosomes. Mol. Biol. Cell 2003, 15, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Ortega, E.; Buey, R.M.; Sonnenberg, A.; de Pereda, J.M. The Structure of the Plakin Domain of Plectin Reveals a Non-canonical SH3 Domain Interacting with Its Fourth Spectrin Repeat. J. Biol. Chem. 2011, 286, 12429–12438. [Google Scholar] [CrossRef] [PubMed]
- Janda, L.; Damborský, J.; Rezniczek, G.A.; Wiche, G. Plectin repeats and modules: Strategic cysteines and their presumed impact on cytolinker functions. BioEssays News Rev. Mol. Cell. Dev. Biol. 2001, 23, 1064–1069. [Google Scholar] [CrossRef]
- Favre, B.; Schneider, Y.; Lingasamy, P.; Bouameur, J.E.; Begré, N.; Gontier, Y.; Steiner-Champliaud, M.F.; Frias, M.A.; Borradori, L.; Fontao, L. Plectin interacts with the rod domain of type III intermediate filament proteins desmin and vimentin. Eur. J. Cell Biol. 2011, 90, 390–400. [Google Scholar] [CrossRef]
- Nikolic, B.; Mac Nulty, E.; Mir, B.; Wiche, G. Basic amino acid residue cluster within nuclear targeting sequence motif is essential for cytoplasmic plectin-vimentin network junctions. J. Cell Biol. 1996, 134, 1455–1467. [Google Scholar] [CrossRef]
- Karashima, T.; Tsuruta, D.; Hamada, T.; Ishii, N.; Ono, F.; Hashikawa, K.; Ohyama, B.; Natsuaki, Y.; Fukuda, S.; Koga, H.; et al. Interaction of plectin and intermediate filaments. J. Dermatol. Sci. 2012, 66, 44–50. [Google Scholar] [CrossRef]
- Gullmets, J.; Torvaldson, E.; Lindqvist, J.; Imanishi, S.Y.; Taimen, P.; Meinander, A.; Eriksson, J.E. Internal epithelia in Drosophila display rudimentary competence to form cytoplasmic networks of transgenic human vimentin. FASEB J. 2017, 31, 5332–5341. [Google Scholar] [CrossRef]
- Filić, V.; Mijanović, L.; Putar, D.; Talajić, A.; Ćetković, H.; Weber, I. Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom. Cells 2021, 10, 1592. [Google Scholar] [CrossRef]
- Zeng, Y.; Cao, Y.; Liu, L.; Zhao, J.; Zhang, T.; Xiao, L.; Jia, M.; Tian, Q.; Yu, H.; Chen, S.; et al. SEPT9_i1 regulates human breast cancer cell motility through cytoskeletal and RhoA/FAK signaling pathway regulation. Cell Death Dis. 2019, 10, 720. [Google Scholar] [CrossRef]
- Cao, L.; Yu, W.; Wu, Y.; Yu, L. The evolution, complex structures and function of septin proteins. Cell. Mol. Life Sci. 2009, 66, 3309. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Settele, F.; Kotak, S.; Sanchez-Pulido, L.; Ehret, L.; Ponting, C.P.; Gönczy, P.; Hoffmann, I. MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. J. Cell Biol. 2013, 200, 773–787. [Google Scholar] [CrossRef]
- Lakämper, S.; Meyhöfer, E. Back on track—On the role of the microtubule for kinesin motility and cellular function. J. Muscle Res. Cell Motil. 2006, 27, 161. [Google Scholar] [CrossRef]
- Sun, Z.; Tseng, H.-Y.; Tan, S.; Senger, F.; Kurzawa, L.; Dedden, D.; Mizuno, N.; Wasik, A.A.; Thery, M.; Dunn, A.R.; et al. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 2016, 18, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Bouchet, B.P.; Gough, R.E.; Ammon, Y.-C.; van de Willige, D.; Post, H.; Jacquemet, G.; Altelaar, A.M.; Heck, A., Jr.; Goult, B.T.; Akhmanova, A. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife 2016, 5, e18124. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liao, S.; Zhu, Z.; Li, Y.; Li, F.; Xu, C. Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins. J. Biol. Chem. 2018, 293, 557–566. [Google Scholar] [CrossRef]
- van der Vaart, B.; van Riel, W.E.; Doodhi, H.; Kevenaar, J.T.; Katrukha, E.A.; Gumy, L.; Bouchet, B.P.; Grigoriev, I.; Spangler, S.A.; Yu, K.L.; et al. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell 2013, 27, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Mengos, A.E.; Mandarino, L.J.; Sekulic, A. Association of liprin β-1 with kank proteins in melanoma. Exp. Dermatol. 2016, 25, 321–323. [Google Scholar] [CrossRef]
- Lansbergen, G.; Grigoriev, I.; Mimori-Kiyosue, Y.; Ohtsuka, T.; Higa, S.; Kitajima, I.; Demmers, J.; Galjart, N.; Houtsmuller, A.B.; Grosveld, F.; et al. CLASPs Attach Microtubule Plus Ends to the Cell Cortex through a Complex with LL5β. Dev. Cell 2006, 11, 21–32. [Google Scholar] [CrossRef]
- Hensley, M.R.; Cui, Z.; Chua, R.F.M.; Simpson, S.; Shammas, N.L.; Yang, J.-Y.; Leung, Y.F.; Zhang, G. Evolutionary and developmental analysis reveals KANK genes were co-opted for vertebrate vascular development. Sci. Rep. 2016, 6, 27816. [Google Scholar] [CrossRef]
- Liang, M.; Jin, G.; Xie, X.; Zhang, W.; Li, K.; Niu, F.; Yu, C.; Wei, Z. Oligomerized liprin-α promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins. Cell Rep. 2021, 34, 108901. [Google Scholar] [CrossRef] [PubMed]
- Weng, Z.; Shang, Y.; Yao, D.; Zhu, J.; Zhang, R. Structural analyses of key features in the KANK1·KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex. J. Biol. Chem. 2018, 293, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Kakinuma, N.; Wang, Y.; Kiyama, R. Kank proteins: A new family of ankyrin-repeat domain-containing proteins. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2008, 1780, 128–133. [Google Scholar] [CrossRef]
- LaFlamme, S.E.; Mathew-Steiner, S.; Singh, N.; Colello-Borges, D.; Nieves, B. Integrin and microtubule crosstalk in the regulation of cellular processes. Cell. Mol. Life Sci. 2018, 75, 4177–4185. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.H.J.; Humphries, J.D.; Byron, A.; Millon-Frémillon, A.; Humphries, M.J. Microtubule-Dependent Modulation of Adhesion Complex Composition. PLoS ONE 2014, 9, e115213. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Microtubules at focal adhesions—A double-edged sword. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Nammalwar, R.C.; Heil, A.; Gerke, V. Ezrin interacts with the scaffold protein IQGAP1 and affects its cortical localization. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2015, 1853, 2086–2094. [Google Scholar] [CrossRef]
- Vodicska, B.; Cerikan, B.; Schiebel, E.; Hoffmann, I. MISP regulates the IQGAP1/Cdc42 complex to collectively orchestrate spindle orientation and mitotic progression. Sci. Rep. 2018, 8, 6330. [Google Scholar] [CrossRef]
- Watanabe, T.; Wang, S.; Noritake, J.; Sato, K.; Fukata, M.; Takefuji, M.; Nakagawa, M.; Izumi, N.; Akiyama, T.; Kaibuchi, K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell 2004, 7, 871–883. [Google Scholar] [CrossRef]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. [Google Scholar] [CrossRef]
- Komarova, Y.; Lansbergen, G.; Galjart, N.; Grosveld, F.; Borisy, G.G.; Akhmanova, A. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell 2005, 16, 5334–5345. [Google Scholar] [CrossRef] [PubMed]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef]
- Szeverenyi, I.; Cassidy, A.J.; Chung, C.W.; Lee, B.T.; Common, J.E.; Ogg, S.C.; Chen, H.; Sim, S.Y.; Goh, W.L.; Ng, K.W.; et al. The Human Intermediate Filament Database: Comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 2008, 29, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Noordstra, I.; Akhmanova, A. Linking cortical microtubule attachment and exocytosis. F1000Research 2017, 6, 469. [Google Scholar] [CrossRef] [PubMed]
- Garcin, C.; Straube, A. Microtubules in cell migration. Essays Biochem. 2019, 63, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Winograd-Katz, S.E.; Fässler, R.; Geiger, B.; Legate, K.R. The integrin adhesome: From genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 273–288. [Google Scholar] [CrossRef]
- Horton, E.R.; Byron, A.; Askari, J.A.; Ng, D.H.J.; Millon-Fremillon, A.; Robertson, J.; Koper, E.J.; Paul, N.R.; Warwood, S.; Knight, D.; et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 2015, 17, 1577–1587. [Google Scholar] [CrossRef]
- Paradžik, M.; Humphries, J.D.; Stojanović, N.; Nestić, D.; Majhen, D.; Dekanić, A.; Samaržija, I.; Sedda, D.; Weber, I.; Humphries, M.J.; et al. KANK2 Links αVβ5 Focal Adhesions to Microtubules and Regulates Sensitivity to Microtubule Poisons and Cell Migration. Front. Cell Dev. Biol. 2020, 8, 125. [Google Scholar] [CrossRef]
- Lock, J.G.; Jones, M.C.; Askari, J.A.; Gong, X.; Oddone, A.; Olofsson, H.; Göransson, S.; Lakadamyali, M.; Humphries, M.J.; Strömblad, S. Reticular adhesions are a distinct class of cell-matrix adhesions that mediate attachment during mitosis. Nat. Cell Biol. 2018, 20, 1290–1302. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: bioinfogp.cnb.csic.es/tools/venny/index.html] (accessed on 8 January 2022).
- Frickey, T.; Lupas, A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The New Tree of Eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Shabardina, V.; Kashima, Y.; Suzuki, Y.; Makalowski, W. Emergence and Evolution of ERM Proteins and Merlin in Metazoans. Genome Biol. Evol. 2020, 12, 3710–3724. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Gabaldón, T. Gene gain and loss across the metazoan tree of life. Nat. Ecol. Evol. 2020, 4, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, D136–D143. [Google Scholar] [CrossRef]
- Ciccarelli, F.D.; Doerks, T.; von Mering, C.; Creevey, C.J.; Snel, B.; Bork, P. Toward automatic reconstruction of a highly resolved tree of life. Science 2006, 311, 1283–1287. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Morpheus by Broad Institute (RRID:SCR_017386). Available online: software.broadinstitute.org/morpheus (accessed on 11 May 2022).
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Biđin, S.; Vujaklija, I.; Paradžik, T.; Bielen, A.; Vujaklija, D. Leitmotif: Protein motif scanning 2.0. Bioinformatics 2020, 36, 3566–3567. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Geneious. Available online: geneious.com (accessed on 10 May 2022).
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I. phyloT: Phylogenetic Tree Generator. Available online: phylot.biobyte.de (accessed on 1 December 2021).
- Biorender. Available online: biorender.com (accessed on 5 May 2022).




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paradžik, T.; Podgorski, I.I.; Vojvoda Zeljko, T.; Paradžik, M. Ancient Origins of Cytoskeletal Crosstalk: Spectraplakin-like Proteins Precede the Emergence of Cortical Microtubule Stabilization Complexes as Crosslinkers. Int. J. Mol. Sci. 2022, 23, 5594. https://doi.org/10.3390/ijms23105594
Paradžik T, Podgorski II, Vojvoda Zeljko T, Paradžik M. Ancient Origins of Cytoskeletal Crosstalk: Spectraplakin-like Proteins Precede the Emergence of Cortical Microtubule Stabilization Complexes as Crosslinkers. International Journal of Molecular Sciences. 2022; 23(10):5594. https://doi.org/10.3390/ijms23105594
Chicago/Turabian StyleParadžik, Tina, Iva I. Podgorski, Tanja Vojvoda Zeljko, and Mladen Paradžik. 2022. "Ancient Origins of Cytoskeletal Crosstalk: Spectraplakin-like Proteins Precede the Emergence of Cortical Microtubule Stabilization Complexes as Crosslinkers" International Journal of Molecular Sciences 23, no. 10: 5594. https://doi.org/10.3390/ijms23105594
APA StyleParadžik, T., Podgorski, I. I., Vojvoda Zeljko, T., & Paradžik, M. (2022). Ancient Origins of Cytoskeletal Crosstalk: Spectraplakin-like Proteins Precede the Emergence of Cortical Microtubule Stabilization Complexes as Crosslinkers. International Journal of Molecular Sciences, 23(10), 5594. https://doi.org/10.3390/ijms23105594





