Sequence Analysis and Functional Verification of the Effects of Three Key Structural Genes, PdTHC2’GT, PdCHS and PdCHI, on the Isosalipurposide Synthesis Pathway in Paeonia delavayi var. lutea
Abstract
:1. Introduction
2. Results
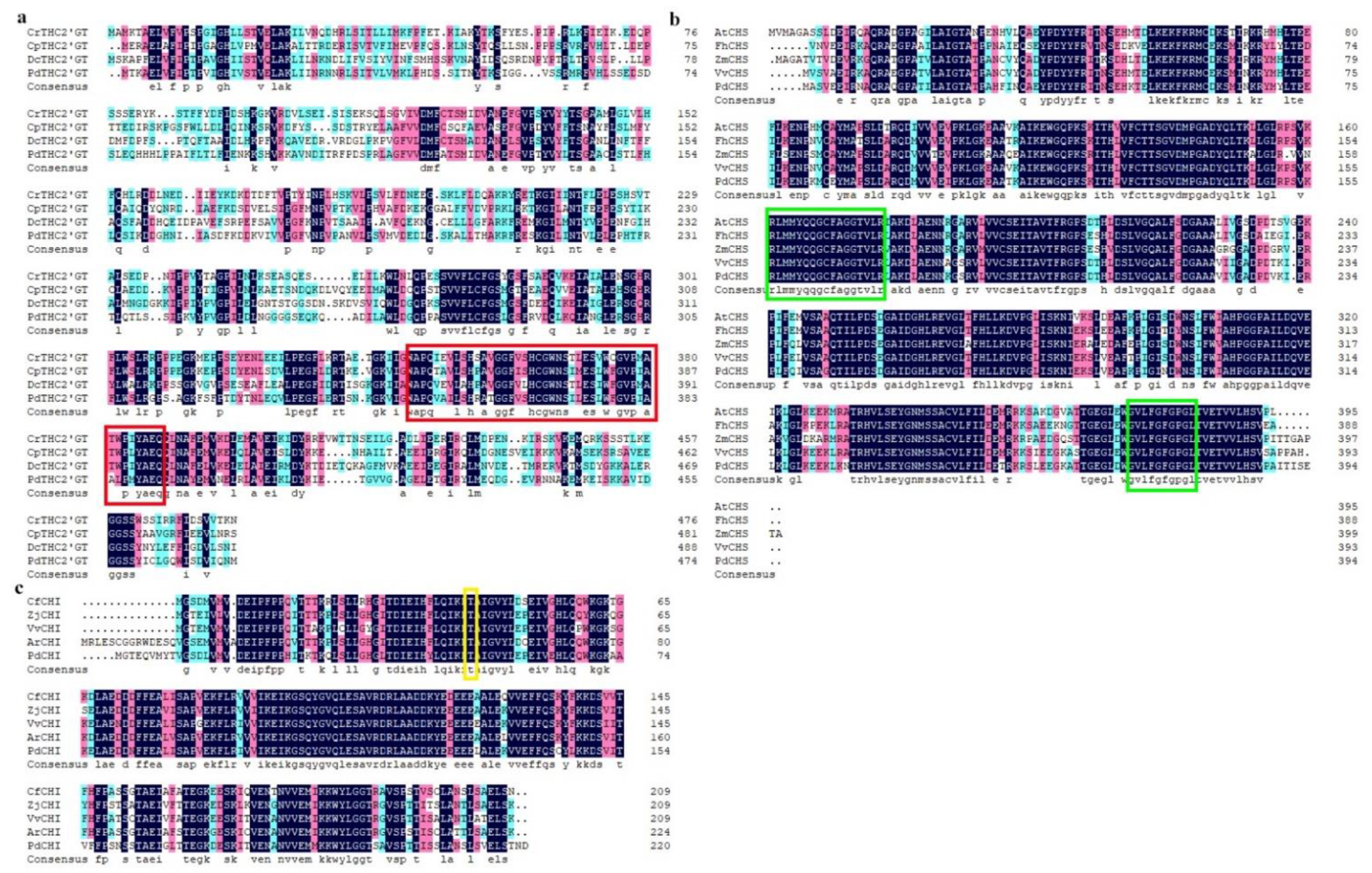
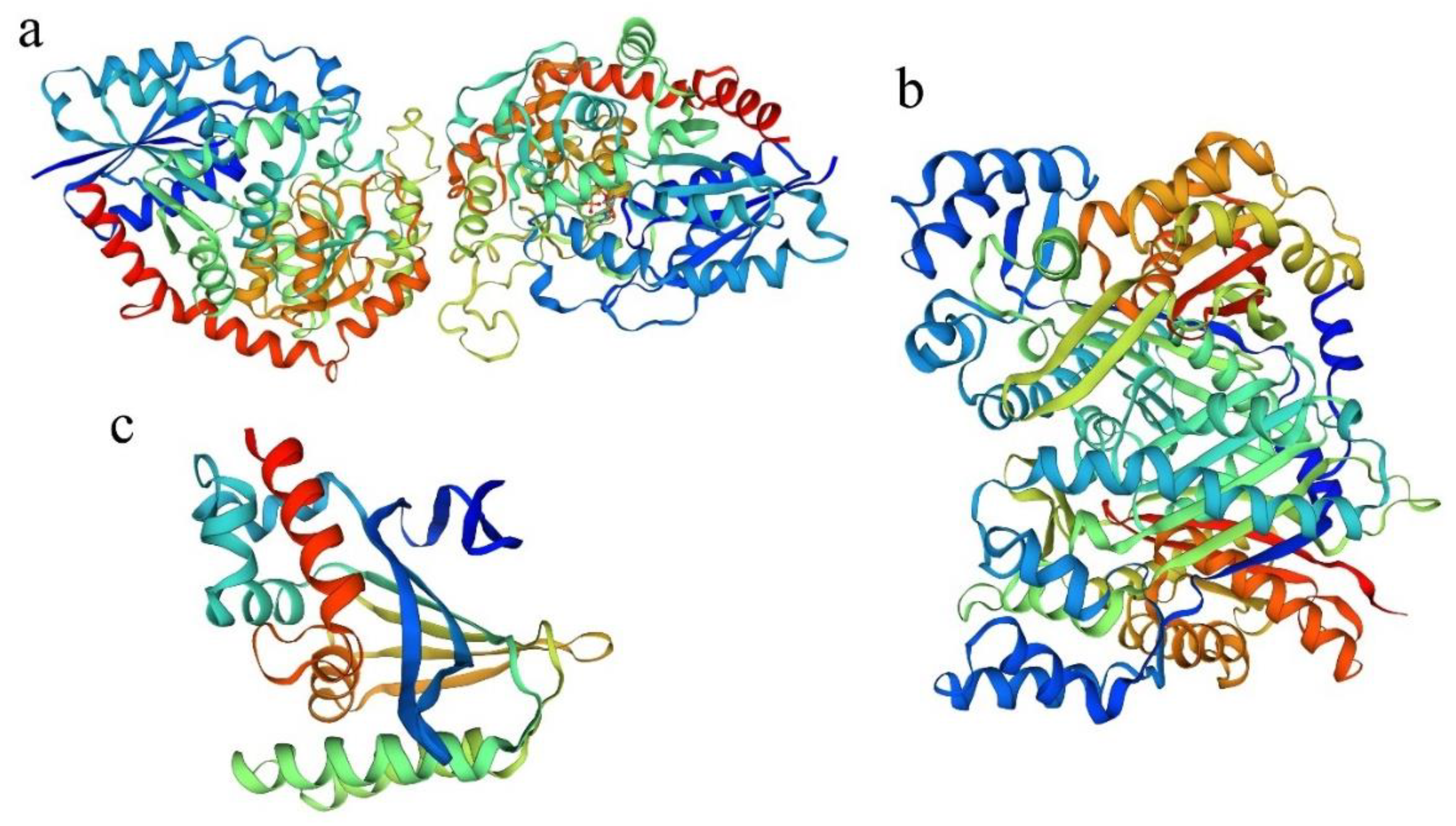
2.1. Cloning and Sequence Analysis of PdTHC2′GT, PdCHS, and PdCHI
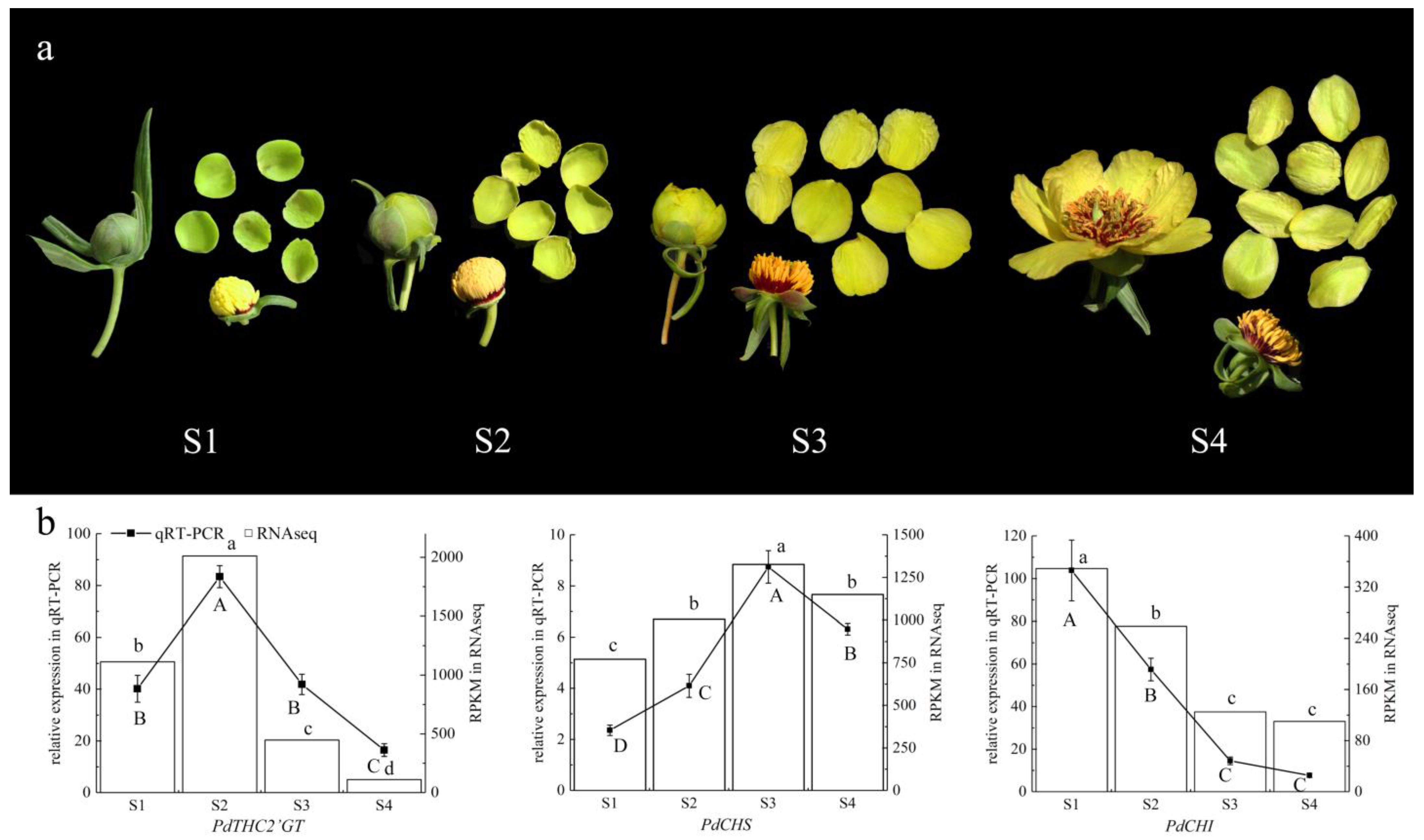
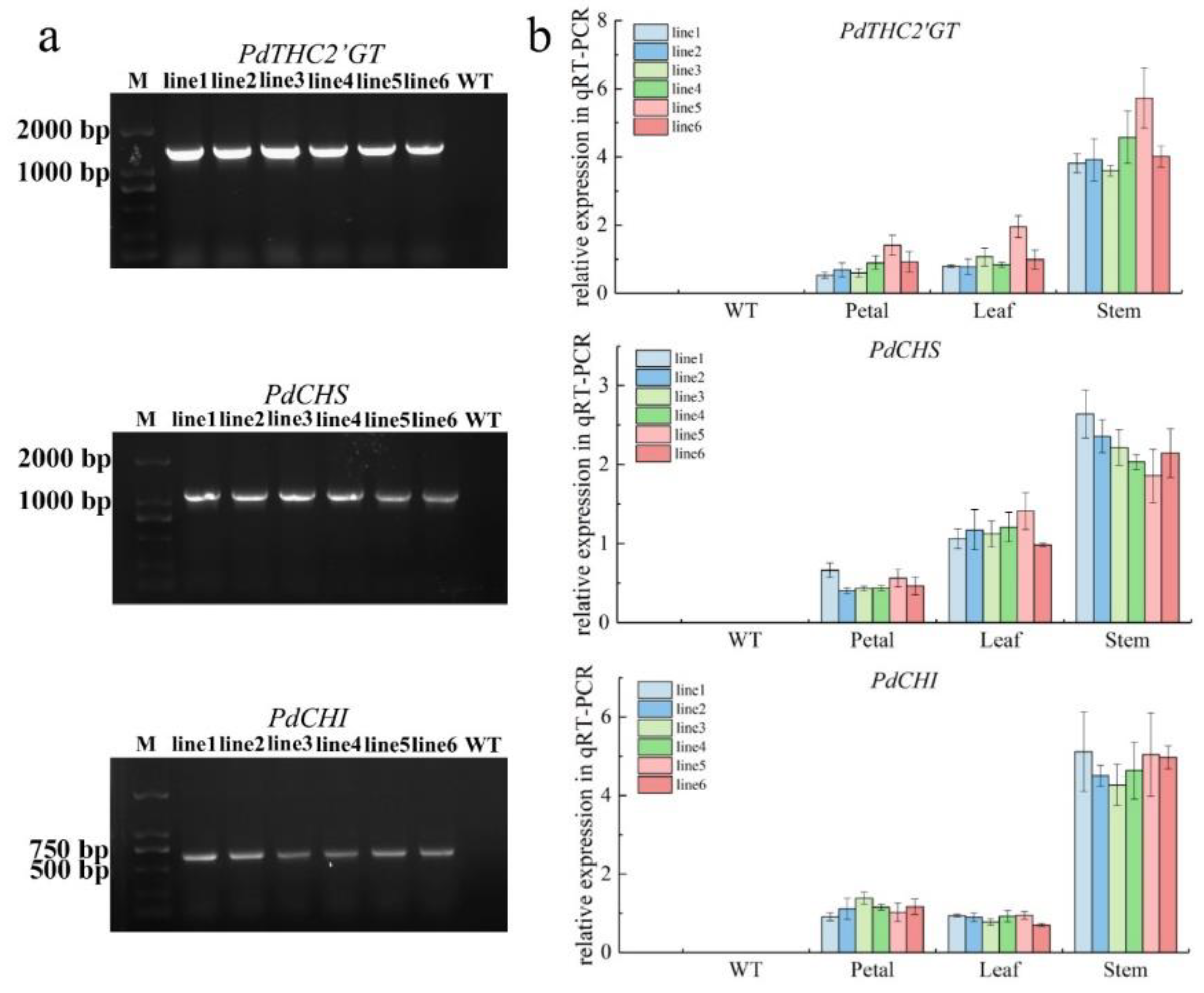
2.2. The Expression Pattern of PdTHC2′GT, PdCHS, and PdCHI
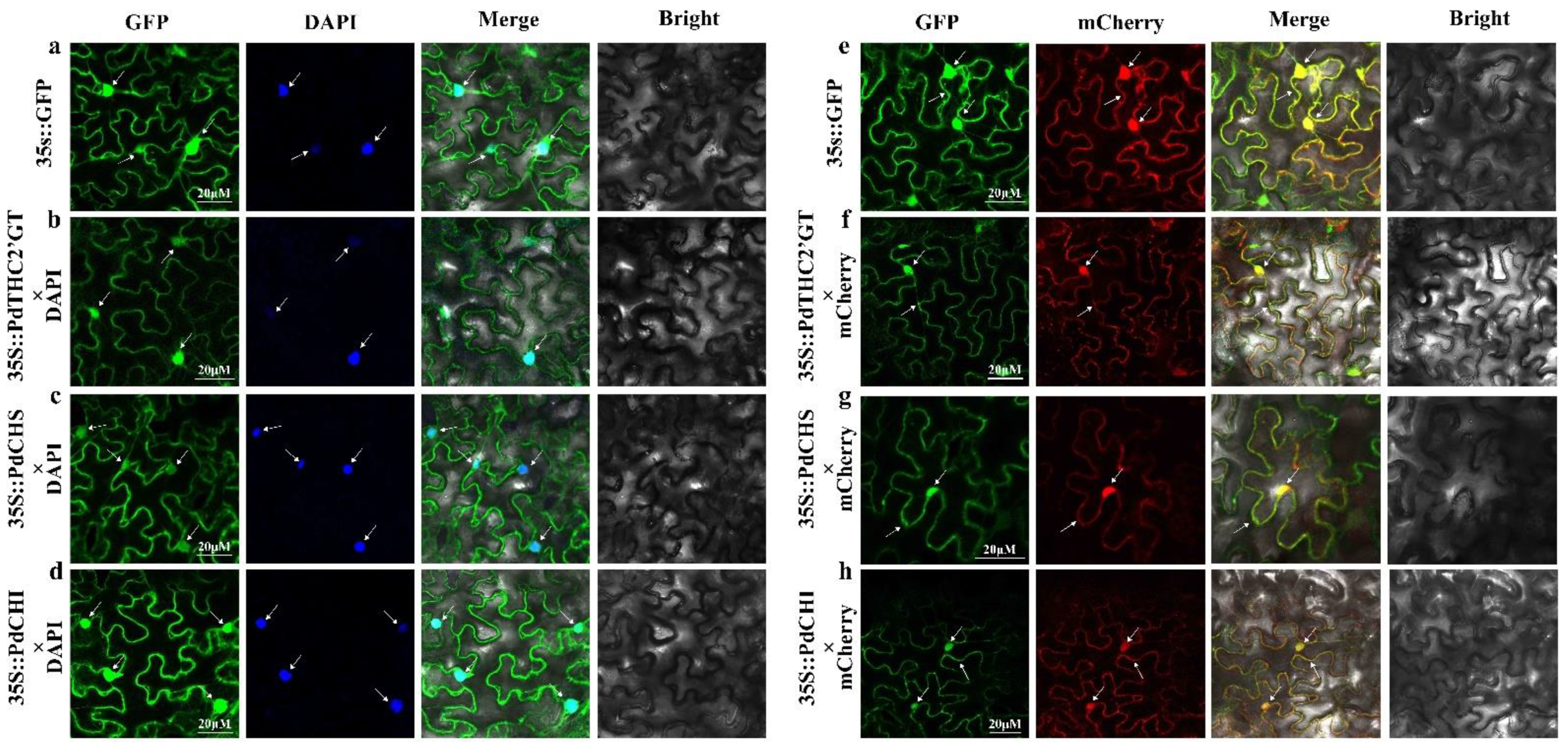
2.3. Subcellular Localization of PdTHC2′GT, PdCHS, and PdCHI
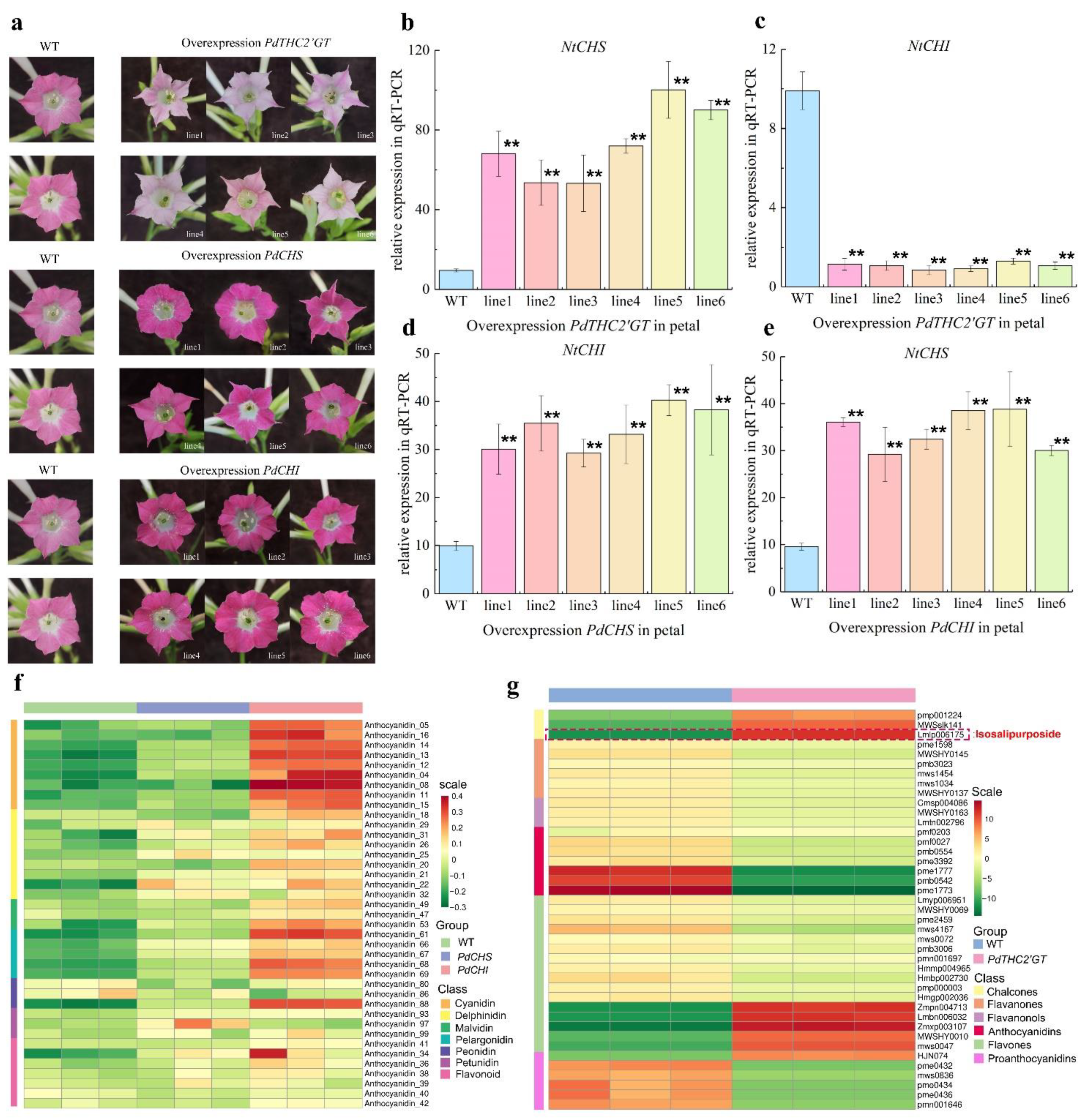
2.4. Overexpression of PdTHC2′GT, PdCHS, and PdCHI in Tobacco
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Gene Cloning and Sequence Analysis
4.3. Gene Expression Analysis
4.4. Subcellular Localization Analyses
4.5. Tobacco Stable Transformation
4.6. Extraction and Detection of Anthocyanins
4.7. Extraction and Detection of Flavonoids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, K.; Wang, Y.; Zheng, B.; Zhu, X.; Wu, H.; Shi, Q. Pollen morphology of 40 Paeonia delavayi (Paeoniaceae) populations. J. Beijing For. Univ. 2011, 33, 94–103. (In Chinese) [Google Scholar]
- Li, C.; Du, H.; Wang, L.; Shu, Q.; Zheng, Y.; Xu, Y.; Zhang, J.; Yang, R.; Ge, Y. Flavonoid composition and antioxidant activity of tree peony (Paeonia Section, Moutan) yellow flowers. J. Agric. Food Chem. 2009, 57, 8496–8503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Y.; Lv, C.; Peng, Z. Identification of components of flower pigments in petals of Paeonia lutea wild population in Yunnan. J. Northeast. For. Univ. 2011, 39, 52–54. [Google Scholar] [CrossRef]
- Itoh, Y.; Higeta, D.; Suzuki, A.; Yoshida, H.; Ozeki, Y. Excision of transposable elements from the chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation (Dianthus caryophyllus). Plant Cell Physiol. 2002, 43, 578–585. [Google Scholar] [CrossRef] [Green Version]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef] [Green Version]
- Schijlen, E.; Ric de Vos, C.H.; Jonker, H.; Broeck, H.; Molthoff, J.; Tunee, A.; Martens, S.; Bovy, A. Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol. J. 2006, 4, 433–444. [Google Scholar] [CrossRef]
- Sharma, S.B.; Dixon, R.A. Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J. 2005, 44, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Itoh, Y.; Ozeki, Y.; Iwashina, T.; Yamaguchi, M. Variation in chalcononaringenin 2′-O-glucoside content in the petals of carnations (Dianthus caryophyllus) bearing yellow flowers. Sci. Hortic. 2004, 99, 175–186. [Google Scholar] [CrossRef]
- Harborne, J.B. Comparative Biochemistry of flavonoids—I.: Distribution of chalcone and aurone pigments in plants. Phytochemistry 1966, 5, 111–115. [Google Scholar] [CrossRef]
- Brkljaca, R.; White, J.; Urban, S. Phytochemical investigation of the constituents derived from the australian plant Macropidia fuliginosa. J. Nat. Prod. 2015, 78, 1600–1608. [Google Scholar] [CrossRef]
- Togami, J.; Okuhara, H.; Nakamura, N.; Ishiguro, K.; Hirose, C.; Ochiai, M.; Fukui, Y.; Yamaguchi, M.; Tanaka, Y. Isolation of cDNAs encoding tetrahydroxychalcone 2′-glucosyltransferase activity from carnation, cyclamen, and catharanthus. Plant Biotechnol. 2011, 28, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.; Ji, X.; Jiao, Z.; Luo, Y.; Zhang, G.; Tao, S.; Lei, Z.; Zhang, J.; Wang, Y.; Liu, Z.; et al. Functional analysis of a novel C-glycosyltransferase in the orchid Dendrobium catenatum. Hortic. Res. 2020, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Reimold, U.; Kröger, M.; Kreuzaler, F.; Hahlbrock, K. Coding and 3′ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J. 1983, 2, 1801–1805. [Google Scholar] [CrossRef]
- Sommer, H.; Saedler, H. Structure of the chalcone synthase gene of Antirrhinum majus. Mol. Genet. Genom. 1986, 202, 429–434. [Google Scholar] [CrossRef]
- Saslowsky, D.E.; Dana, C.D.; Winkel-Shirley, B. An allelic series for the chalcone synthase locus in Arabidopsis. Gene 2000, 255, 127–138. [Google Scholar] [CrossRef]
- Lo, C.; Coolbaugh, R.C.; Nicholson, R.L. Molecular characterization and in silico expression analysis of a chalcone synthase gene family in Sorghum bicolor. Physiol. Mol. Plant Pathol. 2002, 61, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Fukusaki, E.I.; Kawasaki, K.; Kajiyama, S.; An, C.; Suzuki, K.; Tanaka, Y.; Kobayashi, A. Flower color modulations of Torenia hybrida by downregulation of chalcone synthase genes with RNA interference. J. Biotechnol. 2004, 111, 229–240. [Google Scholar] [CrossRef]
- Li, M.; Cao, Y.; Ye, S.; Irshad, M.; Pan, T.; Qiu, D. Isolation of CHS Gene from Brunfelsia acuminata flowers and its regulation in anthocyanin biosysthesis. Molecules 2016, 22, 44. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S.; Hori, W.; Hosokawa, M.; Tatsuzawa, F.; Doi, M. Post-transcriptional silencing of chalcone synthase, is involved in phenotypic lability in petals and leaves of bicolor dahlia (Dahlia variabilis) ‘Yuino’. Planta 2018, 247, 413–428. [Google Scholar] [CrossRef] [Green Version]
- Ohta, Y.; Atsumi, G.; Yoshida, C.; Takahashi, S.; Shimizu, M.; Nishihara, M.; Nakatsuka, T. Post-transcriptional gene silencing of the chalcone synthase gene CHS causes corolla lobe-specific whiting of Japanese gentian. Planta 2022, 255, 29. [Google Scholar] [CrossRef]
- Wang, Y.; Dou, Y.; Wang, R.; Guan, X.; Hu, Z.; Zheng, J. Molecular characterization and functional analysis of chalcone synthase from Syringa oblata Lindl. in the flavonoid biosynthetic pathway. Gene 2017, 635, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Zhu, J.; Hao, Q.; Yuan, Y.W.; Duan, Y.W.; Men, S.; Wang, Q.; Hou, Q.; Liu, Z.A.; Shu, Q.; et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 2018, 60, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Saito, N.; Tatsuzawa, F.; Hoshino, A.; Abe, Y.; Ichimura, M.; Yokoi, M.; Toki, K.; Morita, Y.; Iida, S.; Honda, T. Anthocyanin pigmentation controlled by speckled and c-1 mutations of japanese morning glory. J. Jpn. Soc. Hortic. Sci. 2011, 80, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Forkmann, G.; Dangelmayr, B. Genetic control of chalcone isomerase activity in flowers of Dianthus caryophyllus. Biochem. Genet. 1980, 18, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.D.; Lee, S.; Choi, W.; Yim, W.C.; Cushman, J.C. Laying the foundation for crassulacean acid metabolism (CAM) Biodesign: Expression of the C4 metabolism cycle genes of CAM in Arabidopsis. Front. Plant Sci. 2019, 10, 101. [Google Scholar] [CrossRef]
- Saslowsky, D.E.; Warek, U.; Winkel, B.S.J. Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 2005, 280, 23735–23740. [Google Scholar] [CrossRef] [Green Version]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, ncell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, N.; Kwon, Y.; Gotoh, M.; Narimatsu, H. Comparison of glycosyltransferase families using the profile hidden Markov model. Biochem. Biophys. Res. Commun. 2003, 310, 574–579. [Google Scholar] [CrossRef]
- Yu, X.; Chen, M.; Liu, C. Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso) flavones in Medicago truncatula. Plant J. 2008, 55, 382–396. [Google Scholar] [CrossRef]
- Winkel, B.S.J. When an enzyme isn’t just an enzyme anymore. J. Exp. Bot. 2017, 68, 1387–1389. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.M.; Ahmad, I.; Ansari, M.Y.; Haqqi, T.M. Wogonin, a natural flavonoid, intercalates with genomic DNA and exhibits protective effects in IL-1β stimulated osteoarthritis chondrocytes. Chem. Biol. Interact. 2017, 274, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Tu, R.; Wang, H.; Liu, Q.; Wang, D.; Zhou, X.; Xu, P.; Zhang, Y.; Wu, W.; Chen, D.; Gao, L.; et al. Characterization and genetic analysis of the oshpl3 rice lesion mimic mutant showing spontaneous cell death and enhanced bacterial blight resistance. Plant Physiol. Biochem. 2020, 154, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Romina, D.D.P.; José, S.; Ana, M.G.; Julián, C.R. Liquid chromatographic-mass spectrometric analysis of anthocyanin composition of dark blue bee pollen from Echium plantagineum. J. Chromatogr. A 2004, 1054, 205–210. [Google Scholar] [CrossRef]
- Ferrars, R.M.D.; Czank, C.; Saha, S.; Needs, P.W.; Zhang, Q.; Raheem, K.S.; Botting, N.P.; Kroon, P.A.; Kay, C.D. Methods for isolating, identifying, and quantifying anthocyanin metabolites in clinical samples. Anal. Chem. 2014, 86, 10052–10058. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, L. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
- Zhu, Z.; Andrew, W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, H.; Han, L.; Yuan, M.; Zhang, M.; Zhou, L.; Wang, Y. Sequence Analysis and Functional Verification of the Effects of Three Key Structural Genes, PdTHC2’GT, PdCHS and PdCHI, on the Isosalipurposide Synthesis Pathway in Paeonia delavayi var. lutea. Int. J. Mol. Sci. 2022, 23, 5696. https://doi.org/10.3390/ijms23105696
Zou H, Han L, Yuan M, Zhang M, Zhou L, Wang Y. Sequence Analysis and Functional Verification of the Effects of Three Key Structural Genes, PdTHC2’GT, PdCHS and PdCHI, on the Isosalipurposide Synthesis Pathway in Paeonia delavayi var. lutea. International Journal of Molecular Sciences. 2022; 23(10):5696. https://doi.org/10.3390/ijms23105696
Chicago/Turabian StyleZou, Hongzhu, Lulu Han, Meng Yuan, Mengjie Zhang, Lin Zhou, and Yan Wang. 2022. "Sequence Analysis and Functional Verification of the Effects of Three Key Structural Genes, PdTHC2’GT, PdCHS and PdCHI, on the Isosalipurposide Synthesis Pathway in Paeonia delavayi var. lutea" International Journal of Molecular Sciences 23, no. 10: 5696. https://doi.org/10.3390/ijms23105696





