Lifestyles Shape the Cytochrome P450 Repertoire of the Bacterial Phylum Proteobacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Deltaproteobacterial Species Have the Highest P450 Diversity
2.2. Proteobacterial Species Have Highly Diverse P450s
2.3. More P450s Are Involved in Secondary Metabolism in Delta-, Compared to Alpha-, Gamma-, and Beta-Proteobacterial Species
2.4. P450 Repertoire of Proteobacterial Species Shaped by Their Lifestyle
3. Materials and Methods
3.1. Species and Their Genome Database Information
3.2. Genome Data Mining and Annotation of P450s
3.3. Phylogenetic Analysis of P450s
3.4. Generation of P450 Profile Heat-Maps
3.5. Identification of P450s Part of smBGCs
3.6. P450 Key Features Analysis
3.7. Comparative Analysis of P450s and smBGCs Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, H. Fifty Years of Cytochrome P450 Research; Springer: Tokyo, Japan, 2014; p. IX, 409. [Google Scholar]
- Podust, L.M.; Sherman, D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012, 29, 1251–1266. [Google Scholar] [CrossRef] [Green Version]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef] [Green Version]
- Klingenberg, M. Pigments of rat liver microsomes. Arch. Biochem. Biophys. 1958, 75, 376–386. [Google Scholar] [CrossRef]
- Garfinkel, D. Studies on pig liver microsomes. I. Enzymic and pigment composition of different microsomal fractions. Arch. Biochem. Biophys. 1958, 77, 493–509. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962, 237, 1375–1376. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar] [CrossRef]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R.; et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar] [CrossRef] [Green Version]
- Mnguni, F.C.; Padayachee, T.; Chen, W.; Gront, D.; Yu, J.-H.; Nelson, D.R.; Syed, K. More P450s are involved in secondary metabolite biosynthesis in Streptomyces compared to Bacillus, Cyanobacteria and Mycobacterium. Int. J. Mol. Sci. 2020, 21, 4814. [Google Scholar] [CrossRef]
- Msomi, N.N.; Padayachee, T.; Nzuza, N.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. In silico analysis of P450s and their role in secondary metabolism in the bacterial class Gammaproteobacteria. Molecules 2021, 26, 1538. [Google Scholar] [CrossRef]
- Padayachee, T.; Nzuza, N.; Chen, W.; Nelson, D.R.; Syed, K. Impact of lifestyle on cytochrome P450 monooxygenase repertoire is clearly evident in the bacterial phylum Firmicutes. Sci. Rep. 2020, 10, 13982. [Google Scholar] [CrossRef]
- Khumalo, M.J.; Nzuza, N.; Padayachee, T.; Chen, W.; Yu, J.-H.; Nelson, D.; Syed, K. Comprehensive analyses of cytochrome P450 monoxygenases and secondary metabolite biosynthetic gene clusters in Cyanobacteria. Int. J. Mol. Sci. 2020, 21, 656. [Google Scholar] [CrossRef] [Green Version]
- Nzuza, N.; Padayachee, T.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Ancient Bacterial Class Alphaproteobacteria Cytochrome P450 Monooxygenases Can Be Found in Other Bacterial Species. Int. J. Mol. Sci. 2021, 22, 5542. [Google Scholar] [CrossRef]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef] [Green Version]
- Syed, P.R.; Chen, W.; Nelson, D.R.; Kappo, A.P.; Yu, J.H.; Karpoormath, R.; Syed, K. Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. Int. J. Mol. Sci. 2019, 20, 2690. [Google Scholar] [CrossRef] [Green Version]
- van Wyk, R.; van Wyk, M.; Mashele, S.S.; Nelson, D.R.; Syed, K. Comprehensive comparative analysis of cholesterol catabolic genes/proteins in mycobacterial species. Int. J. Mol. Sci. 2019, 20, 1032. [Google Scholar] [CrossRef] [Green Version]
- Berman, J.J. Taxonomic Guide to Infectious Diseases: Understanding the Biologic Classes of Pathogenic Organisms; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Kersters, K.; De Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. Introduction to the Proteobacteria. In The Prokaryotes: A Handbook on the Biology of Bacteria; Springer: Berlin/Heidelberg, Germany, 2006; Volume 5, pp. 3–37. [Google Scholar]
- Williams, K.P.; Kelly, D.P. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013, 63, 2901–2906. [Google Scholar] [CrossRef]
- Phung, N.T.; Lee, J.; Kang, K.H.; Chang, I.S.; Gadd, G.M.; Kim, B.H. Analysis of microbial diversity in oligotrophic microbial fuel cells using 16S rDNA sequences. FEMS Microbiol. Lett. 2004, 233, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Tang, K.; Zhang, L.; Zhao, Z.; Xie, X.; Chen, C.T.A.; Wang, D.; Jiao, N.; Zhang, Y. Coupled carbon, sulfur, and nitrogen cycles mediated by microorganisms in the water column of a shallow-water hydrothermal ecosystem. Front. Microbiol. 2018, 9, 2178. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wu, M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 2015, 5, 7949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, M.; Falkow, S.; Rosenberg, E.; Schleifer, K.-H.; Stackebrandt, E. The Prokaryotes: Volume 5: Proteobacteria: Alpha and Beta Subclasses; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Bhat, M.A.; Mishra, A.K.; Bhat, M.A.; Banday, M.I.; Bashir, O.; Rather, I.A.; Rahman, S.; Shah, A.A.; Jan, A.T. Myxobacteria as a Source of New Bioactive Compounds: A Perspective Study. Pharmaceutics 2021, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998, 107, 15–24. [Google Scholar]
- Lamb, D.C.; Hargrove, T.Y.; Zhao, B.; Wawrzak, Z.; Goldstone, J.V.; Nes, W.D.; Kelly, S.L.; Waterman, M.R.; Stegeman, J.J.; Lepesheva, G.I. Concerning P450 Evolution: Structural Analyses Support Bacterial Origin of Sterol 14α-Demethylases. Mol. Biol. Evol. 2021, 38, 952–967. [Google Scholar] [CrossRef]
- Marin, J.; Battistuzzi, F.U.; Brown, A.C.; Hedges, S.B. The timetree of prokaryotes: New insights into their evolution and speciation. Mol. Biol. Evol. 2016, 34, 437–446. [Google Scholar] [CrossRef]
- Battistuzzi, F.U.; Hedges, S.B. A major clade of prokaryotes with ancient adaptations to life on land. Mol. Biol. Evol. 2009, 26, 335–343. [Google Scholar] [CrossRef]
- Kgosiemang, I.K.R.; Syed, K.; Mashele, S.S. Comparative genomics and evolutionary analysis of cytochrome P450 monooxygenases in fungal subphylum Saccharomycotina. J. Pure Appl. Microbiol. 2014, 8, 12. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, S.E.; Peterson, J.A. How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 1999, 369, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [Green Version]
- Kryś, J.D.; Gront, D. VisuaLife: Library for interactive visualization in rich web applications. Bioinformatics 2021, 37, 3662–3663. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Pascal Andreu, V.; de Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
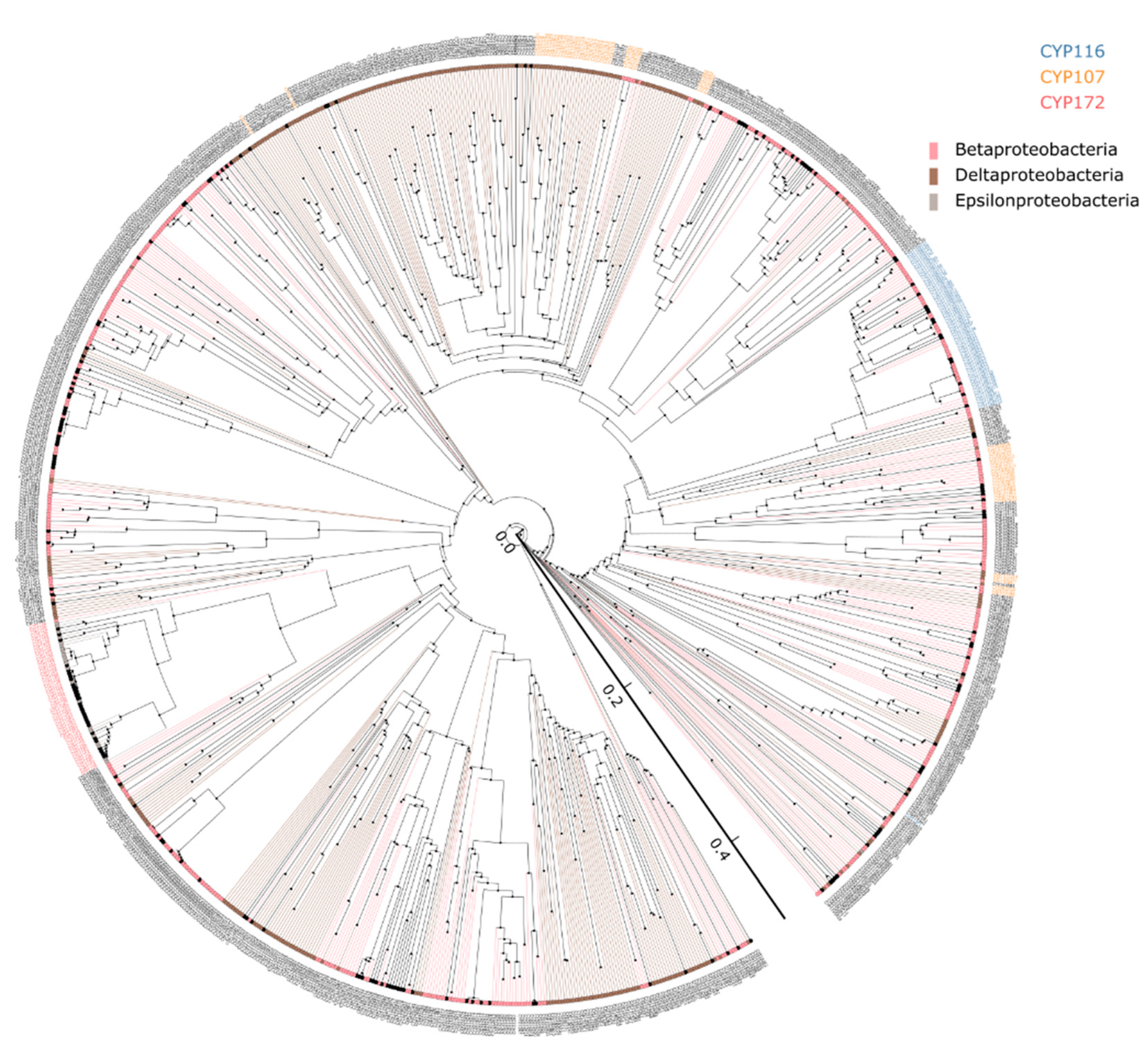
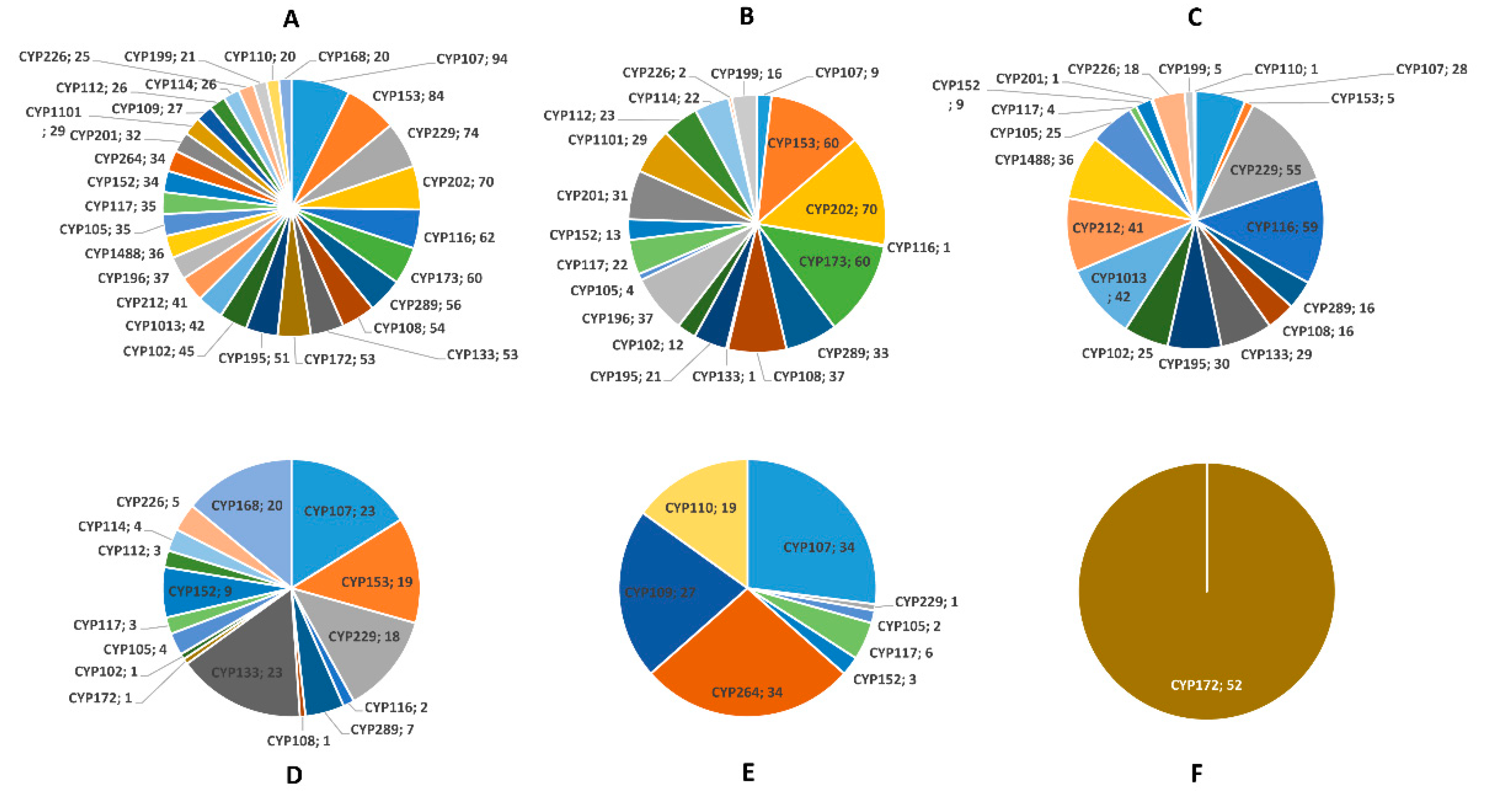

| Category | Alphaproteobacteria | Betaproteobacteria | Gammaproteobacteria | Deltaproteobacteria | Epsilonproteobacteria |
|---|---|---|---|---|---|
| Species analyzed | 599 | 513 | 1261 | 107 | 216 |
| Species without P450s | 370 | 223 | 1091 | 84 | 163 |
| Species with P450s | 229 | 290 | 169 | 23 | 53 |
| Percentage of species with P450s | 38 | 57 | 13 | 21 | 25 |
| No. of P450s | 873 | 603 | 277 | 333 | 53 |
| No. of families | 143 | 79 | 81 | 74 | 2 |
| No. of subfamilies | 214 | 119 | 102 | 171 | 2 |
| Dominant P450 family | CYP202 | CYP116 | CYP133 & CYP107 | CYP107 | CYP172 |
| Average no. of P450s | 4 | 2 | 2 | 14 | 1 |
| P450 diversity percentage | 0,07 | 0,05 | 0,17 | 0,97 | 0,07 |
| No. of P450s part of BGCs | 21 | 107 | 49 | 69 | 0 |
| No. of P450 families part of BGCs | 16 | 18 | 22 | 37 | 0 |
| Percentage of P450s part of BGCs | 2 | 18 | 18 | 21 | 0 |
| Reference | [15] | This study | [12] | This study | This study |
| Alphaproteobacteria | Betaproteobacteria | Gammaproteobacteria | Deltaproteobacteria | ||||
|---|---|---|---|---|---|---|---|
| P450 Family | Count | P450 Family | Count | P450 Family | Count | P450 Family | Count |
| CYP206 | 5 | CYP1013 | 39 | CYP107 | 18 | CYP107 | 6 |
| CYP1101 | 2 | CYP1488 | 37 | CYP1465 | 4 | CYP1011 | 4 |
| CYP2334 | 1 | CYP107 | 4 | CYP105 | 3 | CYP262 | 4 |
| CYP199 | 1 | CYP116 | 4 | CYP126 | 3 | CYP264 | 4 |
| CYP173 | 1 | CYP117 | 4 | CYP134 | 2 | CYP110 | 3 |
| CYP153 | 1 | CYP133 | 4 | CYP153 | 2 | CYP120 | 3 |
| CYP152 | 1 | CYP1486 | 3 | CYP159 | 2 | CYP253 | 3 |
| CYP1302 | 1 | CYP1464 | 2 | CYP116 | 1 | CYP263 | 3 |
| CYP127 | 1 | CYP1104 | 1 | CYP1200 | 1 | CYP1069 | 2 |
| CYP1246 | 1 | CYP1200 | 1 | CYP1247 | 1 | CYP109 | 2 |
| CYP1138 | 1 | CYP1246 | 1 | CYP1278 | 1 | CYP126 | 2 |
| CYP1104 | 1 | CYP1318 | 1 | CYP1414 | 1 | CYP1329 | 2 |
| CYP1138 | 1 | CYP1481 | 1 | CYP1475 | 1 | CYP1486 | 2 |
| CYP108 | 1 | CYP1686 | 1 | CYP163 | 1 | CYP183 | 2 |
| CYP107 | 1 | CYP183 | 1 | CYP289 | 1 | CYP251 | 2 |
| CYP1326 | 1 | CYP2308 | 1 | CYP1201 | 1 | CYP51 | 2 |
| CYP261 | 1 | CYP1468 | 1 | CYP105 | 1 | ||
| CYP267 | 1 | CYP1469 | 1 | CYP1224 | 1 | ||
| CYP1472 | 1 | CYP1298 | 1 | ||||
| CYP1477 | 1 | CYP1347 | 1 | ||||
| CYP1779 | 1 | CYP1448 | 1 | ||||
| CYP2242 | 1 | CYP147 | 1 | ||||
| CYP1489 | 1 | ||||||
| CYP1490 | 1 | ||||||
| CYP1491 | 1 | ||||||
| CYP1494 | 1 | ||||||
| CYP1497 | 1 | ||||||
| CYP1498 | 1 | ||||||
| CYP1499 | 1 | ||||||
| CYP1503 | 1 | ||||||
| CYP1504 | 1 | ||||||
| CYP152 | 1 | ||||||
| CYP167 | 1 | ||||||
| CYP209 | 1 | ||||||
| CYP229 | 1 | ||||||
| CYP242 | 1 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Msweli, S.; Chonco, A.; Msweli, L.; Syed, P.R.; Karpoormath, R.; Chen, W.; Gront, D.; Nkosi, B.V.Z.; Nelson, D.R.; Syed, K. Lifestyles Shape the Cytochrome P450 Repertoire of the Bacterial Phylum Proteobacteria. Int. J. Mol. Sci. 2022, 23, 5821. https://doi.org/10.3390/ijms23105821
Msweli S, Chonco A, Msweli L, Syed PR, Karpoormath R, Chen W, Gront D, Nkosi BVZ, Nelson DR, Syed K. Lifestyles Shape the Cytochrome P450 Repertoire of the Bacterial Phylum Proteobacteria. International Journal of Molecular Sciences. 2022; 23(10):5821. https://doi.org/10.3390/ijms23105821
Chicago/Turabian StyleMsweli, Siphesihle, Andiswa Chonco, Lihle Msweli, Puleng Rosinah Syed, Rajshekhar Karpoormath, Wanping Chen, Dominik Gront, Bridget Valeria Zinhle Nkosi, David R. Nelson, and Khajamohiddin Syed. 2022. "Lifestyles Shape the Cytochrome P450 Repertoire of the Bacterial Phylum Proteobacteria" International Journal of Molecular Sciences 23, no. 10: 5821. https://doi.org/10.3390/ijms23105821
APA StyleMsweli, S., Chonco, A., Msweli, L., Syed, P. R., Karpoormath, R., Chen, W., Gront, D., Nkosi, B. V. Z., Nelson, D. R., & Syed, K. (2022). Lifestyles Shape the Cytochrome P450 Repertoire of the Bacterial Phylum Proteobacteria. International Journal of Molecular Sciences, 23(10), 5821. https://doi.org/10.3390/ijms23105821







