Interaction between Bacteria and the Immune System for Cancer Immunotherapy: The α-GalCer Alliance
Abstract
:1. Introduction
2. Results
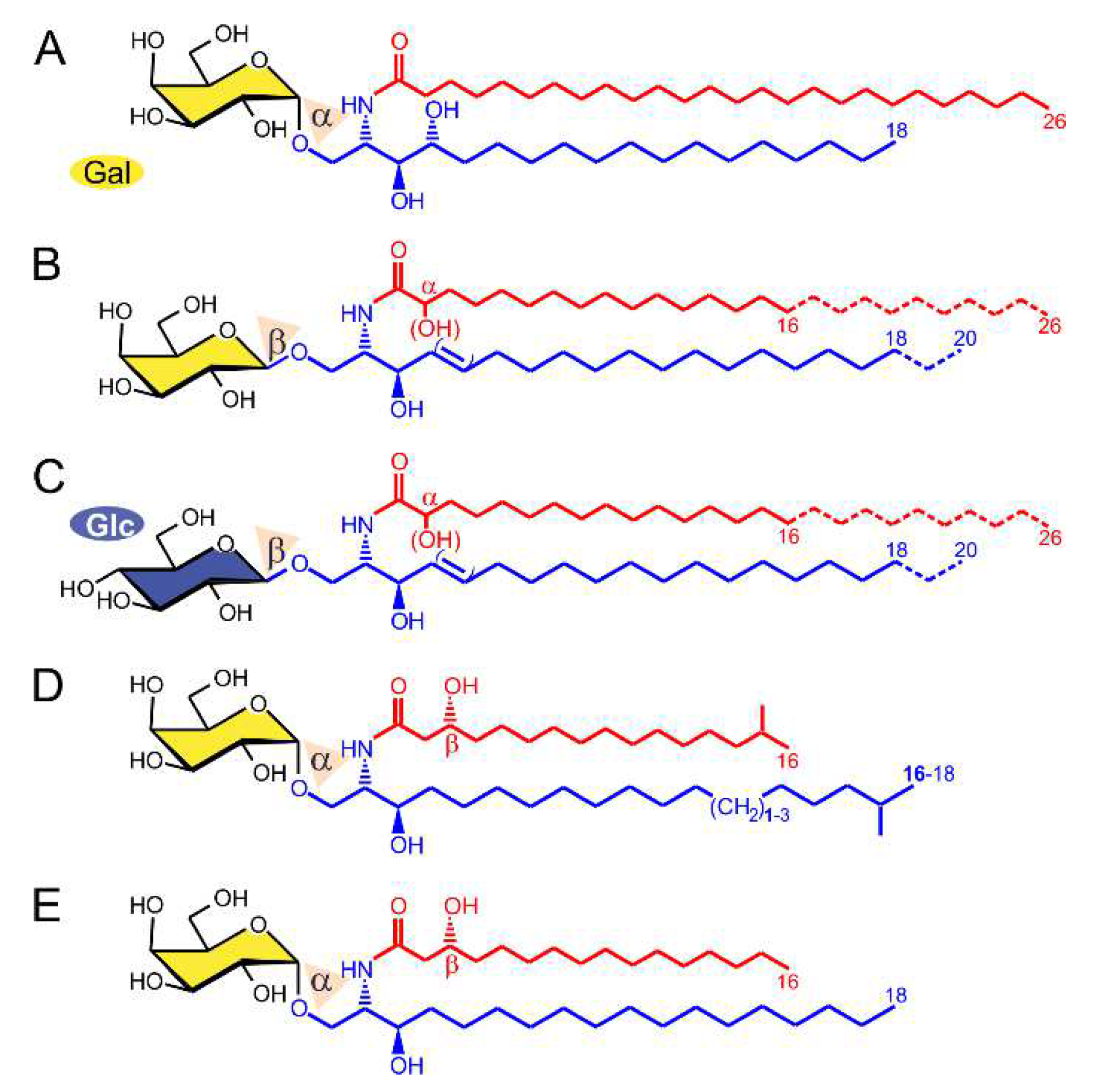
2.1. α-GalCer Is Produced by Human and Mouse Commensal Bacteria
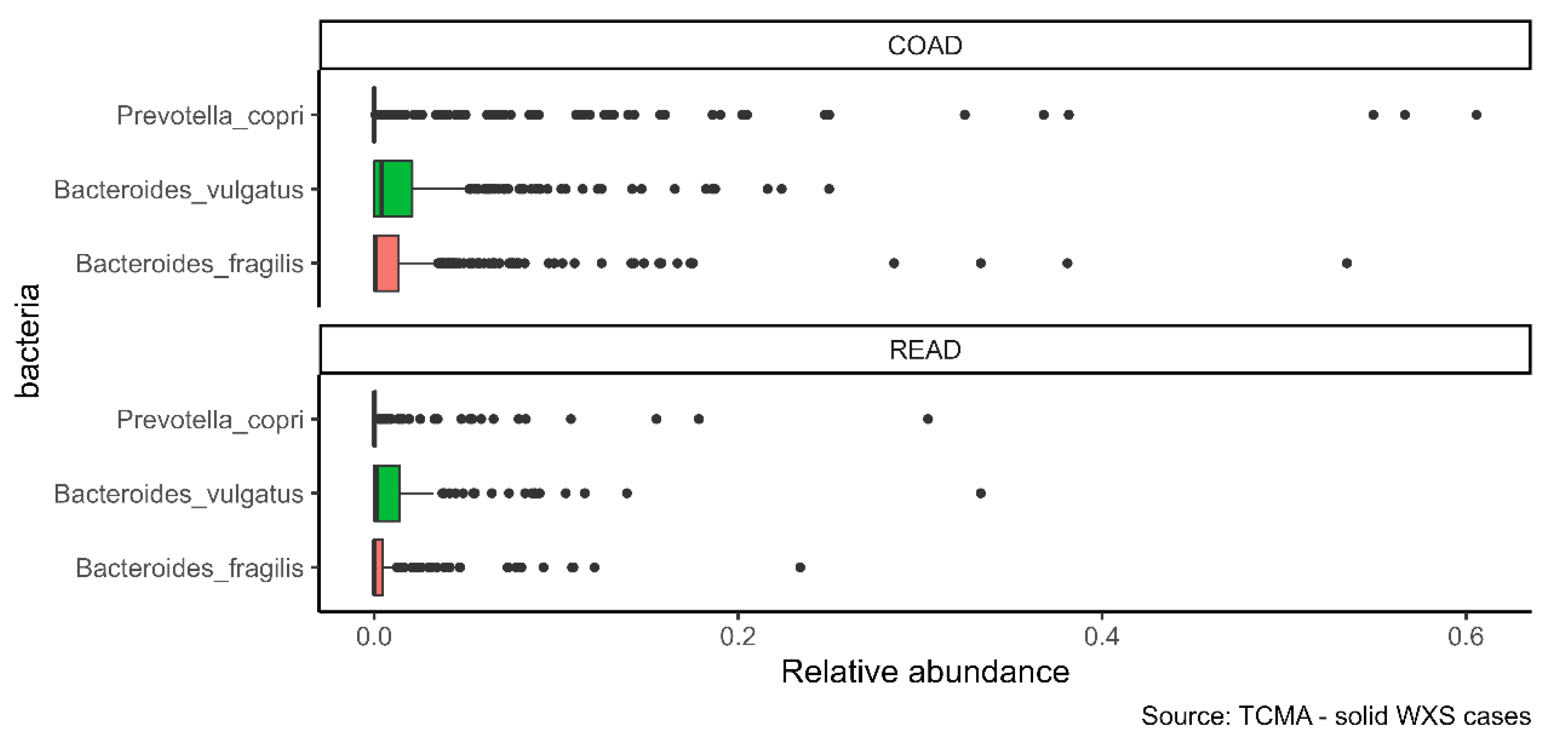
2.2. α-GalCer-Producing Bacteria Infiltrate Human Tumors
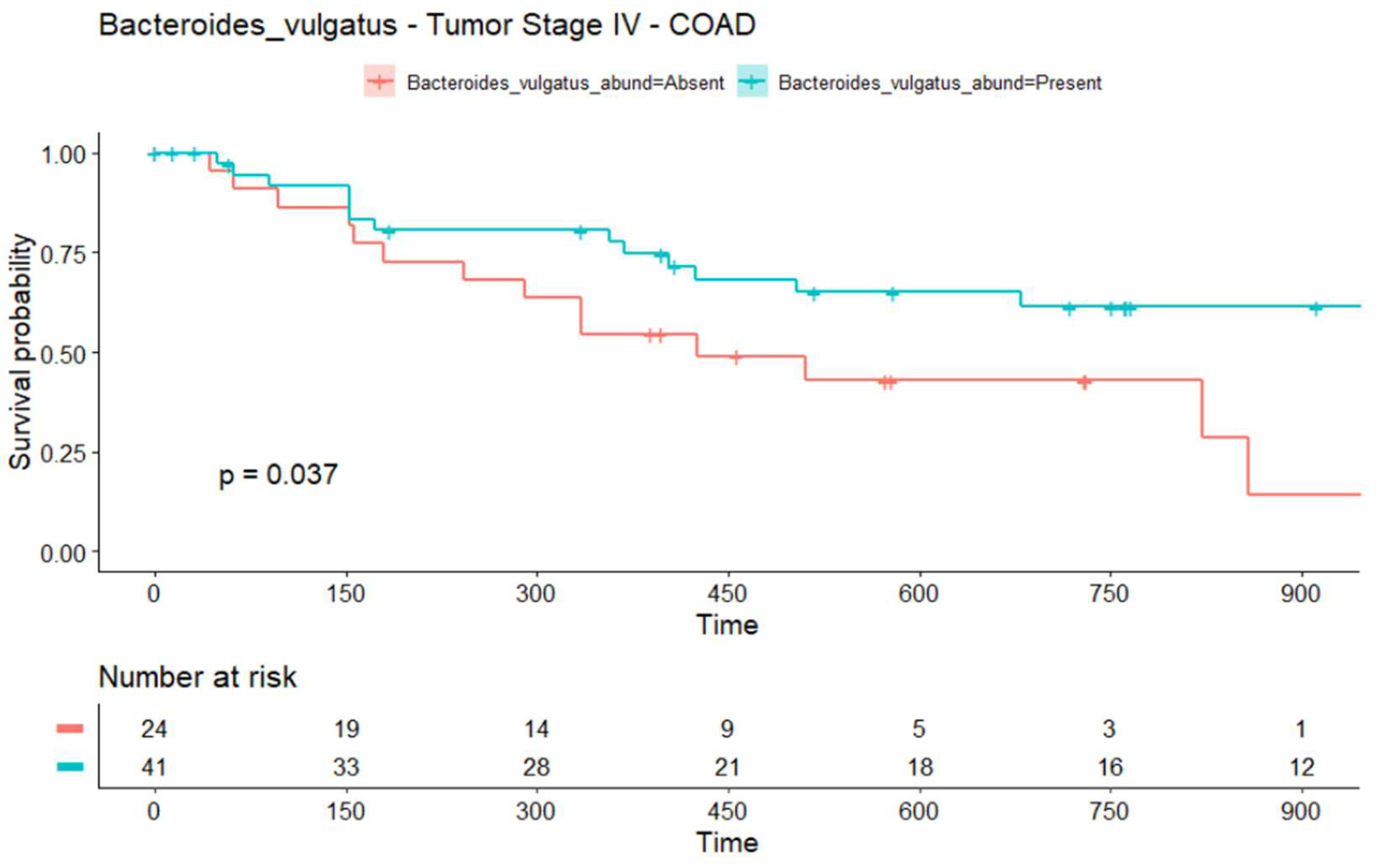
2.3. The Infiltration of Bacteroides vulgatus Is Associated with the Overall Survival (OS) in COAD
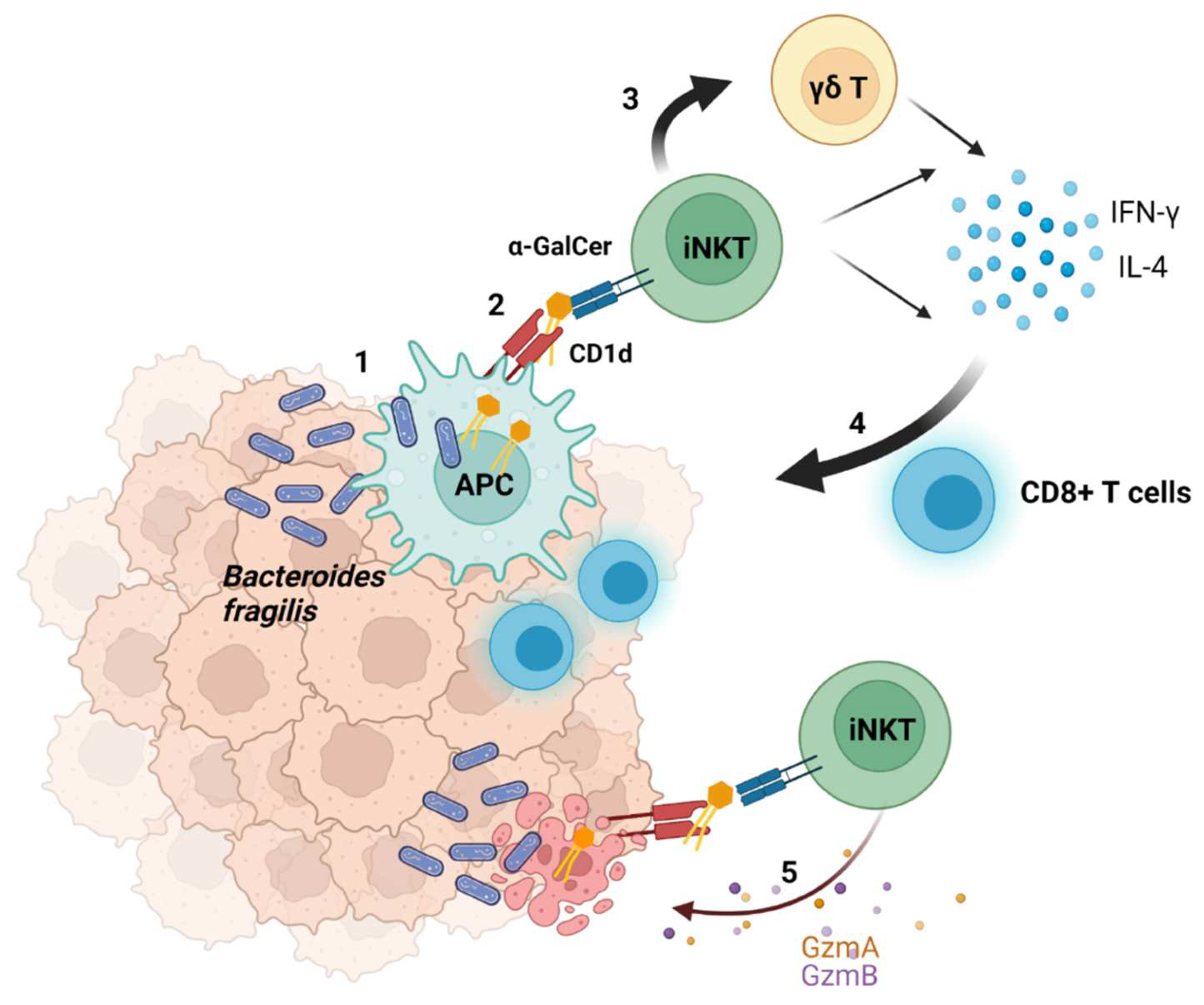
2.4. Suggested Model of the Interaction between Bacteria and Tumors
3. Discussion
4. Materials and Methods
4.1. Identification of α-GalCer-Producing Bacteria in Human Tissues
4.2. Treatment of Mice with Antibiotics
4.3. Lipid Extraction
4.4. Reversed-Phase LC-MS2 Analysis of Glycosylceramides (HexCer) Including Bacterial α-GalCer
4.5. Survival Analysis of the TCMA Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Godfrey, D.I.; Le Nours, J.; Andrews, D.M.; Uldrich, A.P.; Rossjohn, J. Unconventional T Cell Targets for Cancer Immunotherapy. Immunity 2018, 48, 453–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kain, L.; Webb, B.; Anderson, B.L.; Deng, S.; Holt, M.; Costanzo, A.; Zhao, M.; Self, K.; Teyton, A.; Everett, C.; et al. The Identification of the Endogenous Ligands of Natural Killer T Cells Reveals the Presence of Mammalian α-Linked Glycosylceramides. Immunity 2014, 41, 543–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kain, L.; Costanzo, A.; Webb, B.; Holt, M.; Bendelac, A.; Savage, P.B.; Teyton, L. Endogenous ligands of natural killer T cells are alpha-linked glycosylceramides. Mol. Immunol. 2015, 68, 94–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paget, C.; Chow, M.T.; Duret, H.; Mattarollo, S.R.; Smyth, M.J. Role of gammadelta T cells in al-pha-galactosylceramide-mediated immunity. J. Immunol. 2012, 188, 3928–3939. [Google Scholar] [CrossRef]
- Metelitsa, L.S. Anti-tumor potential of type-I NKT cells against CD1d-positive and CD1d-negative tumors in humans. Clin. Immunol. 2011, 140, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Dockry, E.; O’Leary, S.; Gleeson, L.E.; Lyons, J.; Keane, J.; Gray, S.G.; Doherty, D.G. Epigenetic induction of CD1d expression primes lung cancer cells for killing by invariant natural killer T cells. Oncoimmunology 2018, 7, e1428156. [Google Scholar] [CrossRef]
- Ni, C.; Wu, P.; Wu, X.; Zhang, T.; Zhang, T.; Wang, Z.; Zhang, S.; Qiu, F.; Huang, J. Thymosin alpha1 enhanced cytotoxicity of iNKT cells against colon cancer via upregulating CD1d expression. Cancer Lett. 2015, 356, 579–588. [Google Scholar] [CrossRef]
- Ling, Z.; Shao, L.; Liu, X.; Cheng, Y.; Yan, C.; Mei, Y.; Ji, F.; Liu, X. Regulatory T Cells and Plasmacytoid Dendritic Cells Within the Tumor Microenvironment in Gastric Cancer Are Correlated With Gastric Microbiota Dysbiosis: A Preliminary Study. Front. Immunol. 2019, 10, 533. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Su, H.; Johnson, C.H.; Khan, S.A.; Kluger, H.; Lu, L. Intratumour microbiome associated with the infiltration of cytotoxic CD8+ T cells and patient survival in cutaneous melanoma. Eur. J. Cancer 2021, 151, 25–34. [Google Scholar] [CrossRef]
- Ma, J.; Huang, L.; Hu, D.; Zeng, S.; Han, Y.; Shen, H. The role of the tumor microbe microenvironment in the tumor immune microenvironment: Bystander, activator, or inhibitor? J. Exp. Clin. Cancer Res. 2021, 40, 327. [Google Scholar] [CrossRef]
- Pinato, D.J.; Gramenitskaya, D.; Altmann, D.M.; Boyton, R.J.; Mullish, B.H.; Marchesi, J.R.; Bower, M. Antibiotic therapy and outcome from immune-checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 287. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohlman, A.B.; Arguijo Mendoza, D.; Ding, S.; Gao, M.; Dressman, H.; Iliev, I.D.; Lipkin, S.M.; Shen, X. The cancer microbiome atlas: A pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe 2021, 29, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Wong-Rolle, A.; Wei, H.K.; Zhao, C.; Jin, C. Unexpected guests in the tumor microenvironment: Microbiome in cancer. Protein Cell 2021, 12, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, K.; Zhang, P.; Zheng, J.; Min, C.; Li, X. Research Progress of Pancreas-Related Microorganisms and Pancreatic Cancer. Front. Oncol. 2020, 10, 604531. [Google Scholar] [CrossRef]
- Kazim, S.F.; Martinez, E.; Hough, T.J.; Spangler, B.Q.; Bowers, C.A.; Chohan, M.O. The Survival Benefit of Postoperative Bacterial Infections in Patients With Glioblastoma Multiforme: Myth or Reality? Front. Neurol. 2021, 12. [Google Scholar] [CrossRef]
- Morita, M.; Motoki, K.; Akimoto, K.; Natori, T.; Sakai, T.; Sawa, E.; Yamaji, K.; Koezuka, Y.; Kobayashi, E.; Fukushima, H. Structure-activity relationship of alpha-galactosylceramides against B16-bearing mice. J. Med. Chem 1995, 38, 2176–2187. [Google Scholar] [CrossRef]
- Sandhoff, R.; Sandhoff, K. Emerging concepts of ganglioside metabolism. FEBS Lett. 2018, 592, 3835–3864. [Google Scholar] [CrossRef] [Green Version]
- Wieland Brown, L.C.; Penaranda, C.; Kashyap, P.C.; Williams, B.B.; Clardy, J.; Kronenberg, M.; Sonnenburg, J.L.; Comstock, L.E.; Bluestone, J.A.; Fischbach, M.A. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013, 11, e1001610. [Google Scholar] [CrossRef] [Green Version]
- von Gerichten, J.; Schlosser, K.; Lamprecht, D.; Morace, I.; Eckhardt, M.; Wachten, D.; Jennemann, R.; Grone, H.J.; Mack, M.; Sandhoff, R. Diastereomer-specific quantification of bioactive hexosylceramides from bacteria and mammals. J. Lipid Res. 2017, 58, 1247–1258. [Google Scholar] [CrossRef] [Green Version]
- von Gerichten, J.; Lamprecht, D.; Opalka, L.; Soulard, D.; Marsching, C.; Pilz, R.; Sencio, V.; Herzer, S.; Galy, B.; Nordstrom, V.; et al. Bacterial immunogenic alpha-galactosylceramide identified in the murine large intestine: Dependency on diet and inflammation. J. Lipid Res. 2019, 60, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Bendelac, A.; Savage, P.B.; Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef] [Green Version]
- Kalaora, S.; Nagler, A.; Nejman, D.; Alon, M.; Barbolin, C.; Barnea, E.; Ketelaars, S.L.C.; Cheng, K.; Vervier, K.; Shental, N.; et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021, 592, 138–143. [Google Scholar] [CrossRef]
- Lehmann, N.; Paret, C.; El Malki, K.; Russo, A.; Neu, M.A.; Wingerter, A.; Seidmann, L.; Foersch, S.; Ziegler, N.; Roth, L.; et al. Tumor Lipids of Pediatric Papillary Renal Cell Carcinoma Stimulate Unconventional T Cells. Front. Immunol. 2020, 11, 1819. [Google Scholar] [CrossRef] [PubMed]
- Debesa-Tur, G.; Pérez-Brocal, V.; Ruiz-Ruiz, S.; Castillejo, A.; Latorre, A.; Soto, J.L.; Moya, A. Metagenomic analysis of formalin-fixed paraffin-embedded tumor and normal mucosa reveals differences in the microbiome of colorectal cancer patients. Sci. Rep. 2021, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Xu, E.; Ai, D. Inferring Bacterial Infiltration in Primary Colorectal Tumors From Host Whole Genome Sequencing Data. Front. Genet. 2019, 10, 213. [Google Scholar] [CrossRef] [Green Version]
- Ooki, A.; Shinozaki, E.; Yamaguchi, K. Immunotherapy in Colorectal Cancer: Current and Future Strategies. J. Anus. Rectum. Colon. 2021, 5, 11–24. [Google Scholar] [CrossRef]
- Diez-Alonso, M.; Mendoza-Moreno, F.; Jimenez-Alvarez, L.; Nunez, O.; Blazquez-Martin, A.; Sanchez-Gollarte, A.; Matias-Garcia, B.; Molina, R.; San-Juan, A.; Gutierrez-Calvo, A. Prognostic factors of survival in stage IV colorectal cancer with synchronous liver metastasis: Negative effect of the KRAS mutation. Mol. Clin. Oncol. 2021, 14, 93. [Google Scholar] [CrossRef]
- Yokota, T.; Ura, T.; Shibata, N.; Takahari, D.; Shitara, K.; Nomura, M.; Kondo, C.; Mizota, A.; Utsunomiya, S.; Muro, K.; et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br. J. Cancer 2011, 104, 856–862. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, S.H.; Harrison, T.A.; Phipps, A.I.; Steinfelder, R.; Trinh, Q.M.; Qu, C.; Banbury, B.L.; Georgeson, P.; Grasso, C.S.; Giannakis, M.; et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat. Commun. 2020, 11, 3644. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Kawamura, H.; Honda, M.; Takano, Y.; Kinuta, S.; Kamiga, T.; Yamazaki, S.; Muto, A.; Shiraso, S.; Yamashita, N.; et al. Impact of histological subtype on prognosis in stage IV colorectal cancer: A population-based cohort study. PLoS ONE 2022, 17, e0264652. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Song, I.H.; Lee, A.; Kang, J.; Lee, Y.S.; Lee, I.K.; Song, Y.S.; Lee, S.H. Enhancing the landscape of colorectal cancer using targeted deep sequencing. Sci. Rep. 2021, 11, 8154. [Google Scholar] [CrossRef] [PubMed]
- Chasov, V.; Zaripov, M.; Mirgayazova, R.; Khadiullina, R.; Zmievskaya, E.; Ganeeva, I.; Valiullina, A.; Rizvanov, A.; Bulatov, E. Promising New Tools for Targeting p53 Mutant Cancers: Humoral and Cell-Based Immunotherapies. Front. Immunol. 2021, 12, 707734. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, T.; Onodera, H.; Tsuruyama, T.; Mori, A.; Nagayama, S.; Hiai, H.; Imamura, M. Increased intratumor Valpha24-positive natural killer T cells: A prognostic factor for primary colorectal carcinomas. Clin. Cancer Res. 2005, 11, 7322–7327. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.K.; Mehrabian, P.; Boyajian, S.; Wu, W.L.; Selicha, J.; Vonderfecht, S.; Mazmanian, S.K. The Protective Role of Bacteroides fragilis in a Murine Model of Colitis-Associated Colorectal Cancer. mSphere 2018, 3, e00587-18. [Google Scholar] [CrossRef] [Green Version]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated with Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients with NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Huang, J.; Liu, D.; Wang, Y.; Liu, L.; Li, J.; Yuan, J.; Jiang, Z.; Jiang, Z.; Hsiao, W.W.; Liu, H.; et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 2022, 71, 734–745. [Google Scholar] [CrossRef]
- Ouaknine Krief, J.; Helly de Tauriers, P.; Dumenil, C.; Neveux, N.; Dumoulin, J.; Giraud, V.; Labrune, S.; Tisserand, J.; Julie, C.; Emile, J.F.; et al. Role of antibiotic use, plasma citrulline and blood microbiome in advanced non-small cell lung cancer patients treated with nivolumab. J. Immunother. Cancer 2019, 7, 176. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T. A Package for Survival Analysis in R. R Package Version 3.3-1. Available online: https://CRAN.R-project.org/package=survival (accessed on 23 May 2022).
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Package ‘survminer’. Drawing Survival Curves Using ‘ggplot2’ (R package version 0.4.9). Available online: https://CRAN.R-project.org/package=survminer (accessed on 23 May 2022).

| Tissue | Samples | Bacteroides vulgatus (%) | Bacteroides fragilis (%) | Prevotella copri (%) |
|---|---|---|---|---|
| Bone (T) | 39 | 0 | 0 | 10.26 |
| Breast (N-FA) | 28 | 0 | 0 | 0 |
| Breast (N) | 51 | 1.96 | 3.92 | 3.92 |
| Breast (NAT) | 168 | 1.19 | 0.6 | 5.36 |
| Breast (T) | 338 | 1.48 | 1.48 | 9.47 |
| Colon (NAT) | 22 | 45.45 | 31.82 | 13.64 |
| Colon (T) | 22 | 40.91 | 45.45 | 27.27 |
| Extraction control | 432 | 0.23 | 1.62 | 3.94 |
| GBM (T) | 40 | 0 | 0 | 10 |
| Lung (NAT) | 231 | 0.87 | 1.73 | 3.03 |
| Lung (T) | 243 | 0.41 | 2.88 | 6.58 |
| Melanoma (T) | 197 | 3.55 | 4.57 | 2.54 |
| NTC | 204 | 0 | 1.96 | 2.45 |
| Ovary (N- fallop) | 17 | 5.88 | 17.65 | 23.53 |
| Ovary (NAT) | 12 | 8.33 | 0 | 0 |
| Ovary (T) | 56 | 1.79 | 1.79 | 8.93 |
| Pancreas (T) | 67 | 5.97 | 5.97 | 5.97 |
| Paraffin control | 165 | 0.61 | 0.61 | 3.64 |
| Project | Samples | Bacteroides vulgatus (%) | Bacteroides fragilis (%) | Prevotella copri (%) |
|---|---|---|---|---|
| COAD | 439 | 65.83 | 55.35 | 28.47 |
| READ | 159 | 54.09 | 37.74 | 27.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ustjanzew, A.; Sencio, V.; Trottein, F.; Faber, J.; Sandhoff, R.; Paret, C. Interaction between Bacteria and the Immune System for Cancer Immunotherapy: The α-GalCer Alliance. Int. J. Mol. Sci. 2022, 23, 5896. https://doi.org/10.3390/ijms23115896
Ustjanzew A, Sencio V, Trottein F, Faber J, Sandhoff R, Paret C. Interaction between Bacteria and the Immune System for Cancer Immunotherapy: The α-GalCer Alliance. International Journal of Molecular Sciences. 2022; 23(11):5896. https://doi.org/10.3390/ijms23115896
Chicago/Turabian StyleUstjanzew, Arsenij, Valentin Sencio, François Trottein, Jörg Faber, Roger Sandhoff, and Claudia Paret. 2022. "Interaction between Bacteria and the Immune System for Cancer Immunotherapy: The α-GalCer Alliance" International Journal of Molecular Sciences 23, no. 11: 5896. https://doi.org/10.3390/ijms23115896
APA StyleUstjanzew, A., Sencio, V., Trottein, F., Faber, J., Sandhoff, R., & Paret, C. (2022). Interaction between Bacteria and the Immune System for Cancer Immunotherapy: The α-GalCer Alliance. International Journal of Molecular Sciences, 23(11), 5896. https://doi.org/10.3390/ijms23115896







