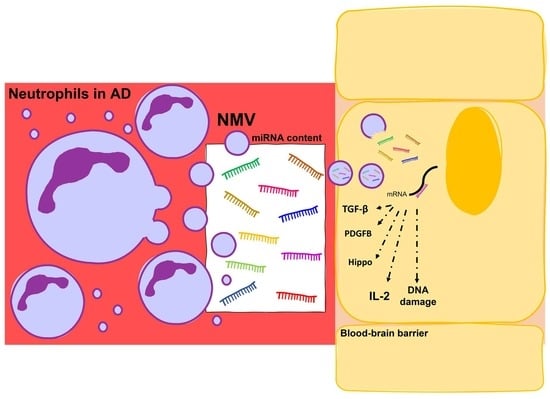
RNA-Seq Profiling of Neutrophil-Derived Microvesicles in Alzheimer’s Disease Patients Identifies a miRNA Signature That May Impact Blood–Brain Barrier Integrity
Abstract
:1. Introduction
2. Results
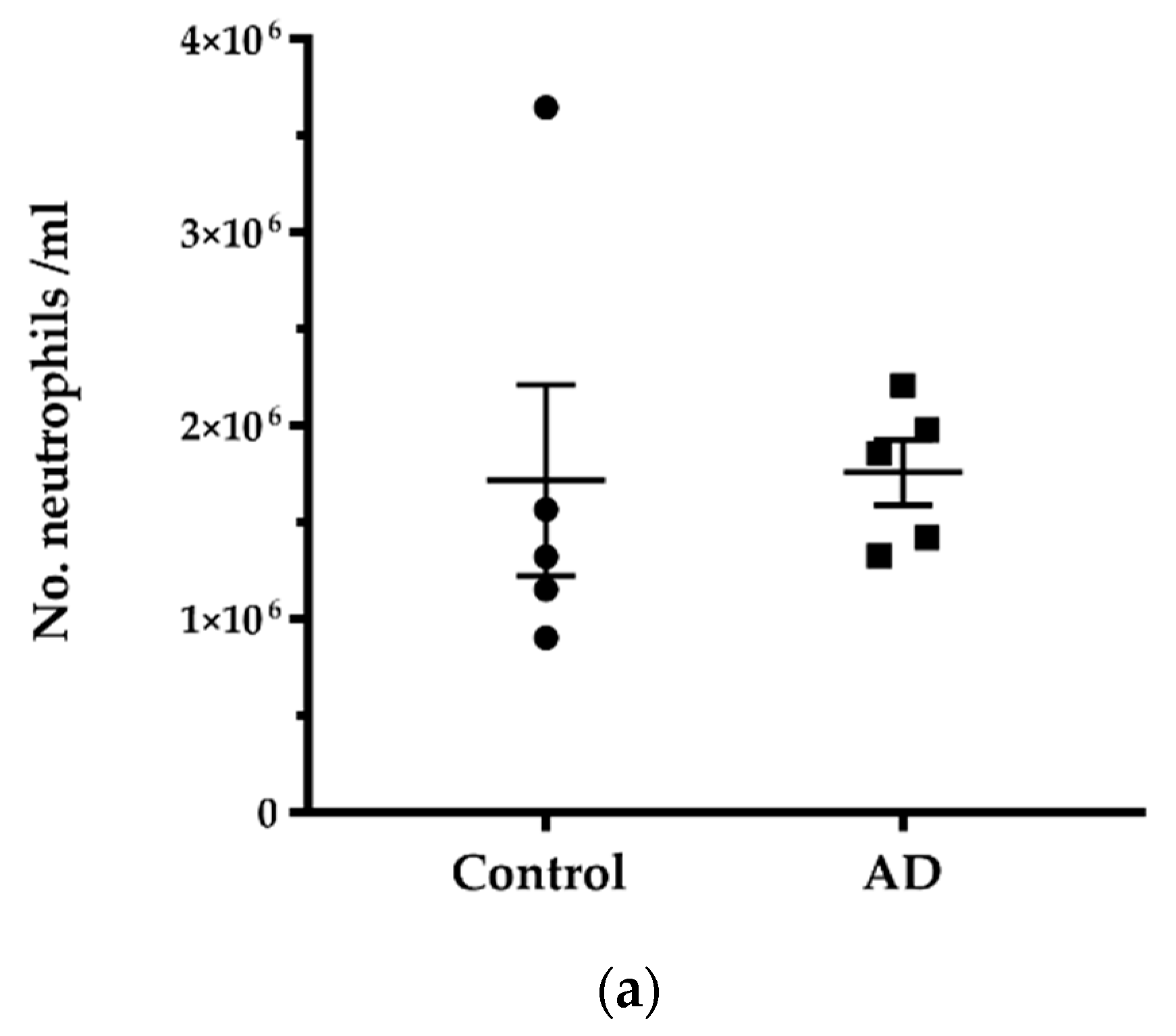
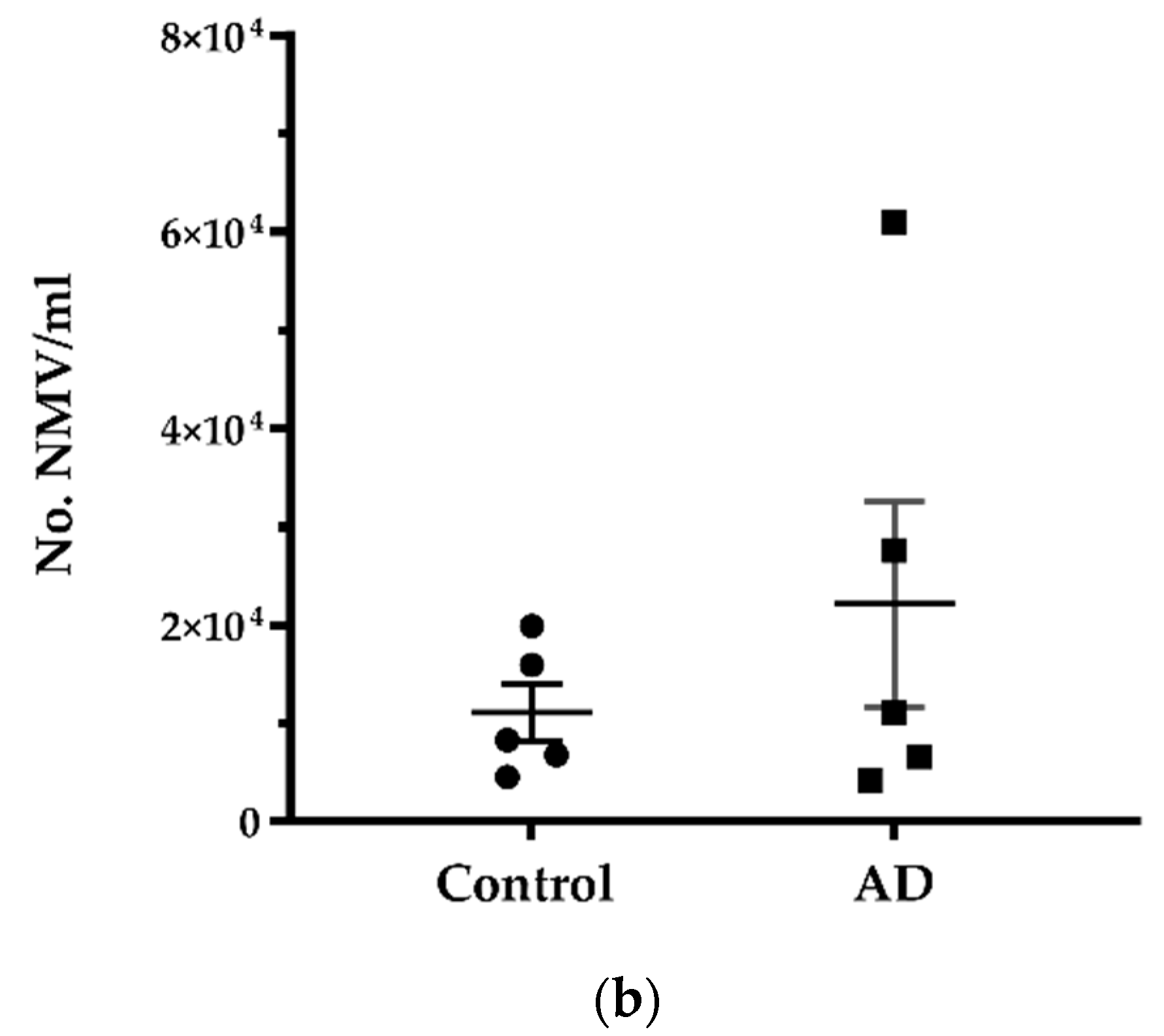
2.1. No Significant Difference in the Number of Neutrophils and NMV between AD and Control without Systemic Infection
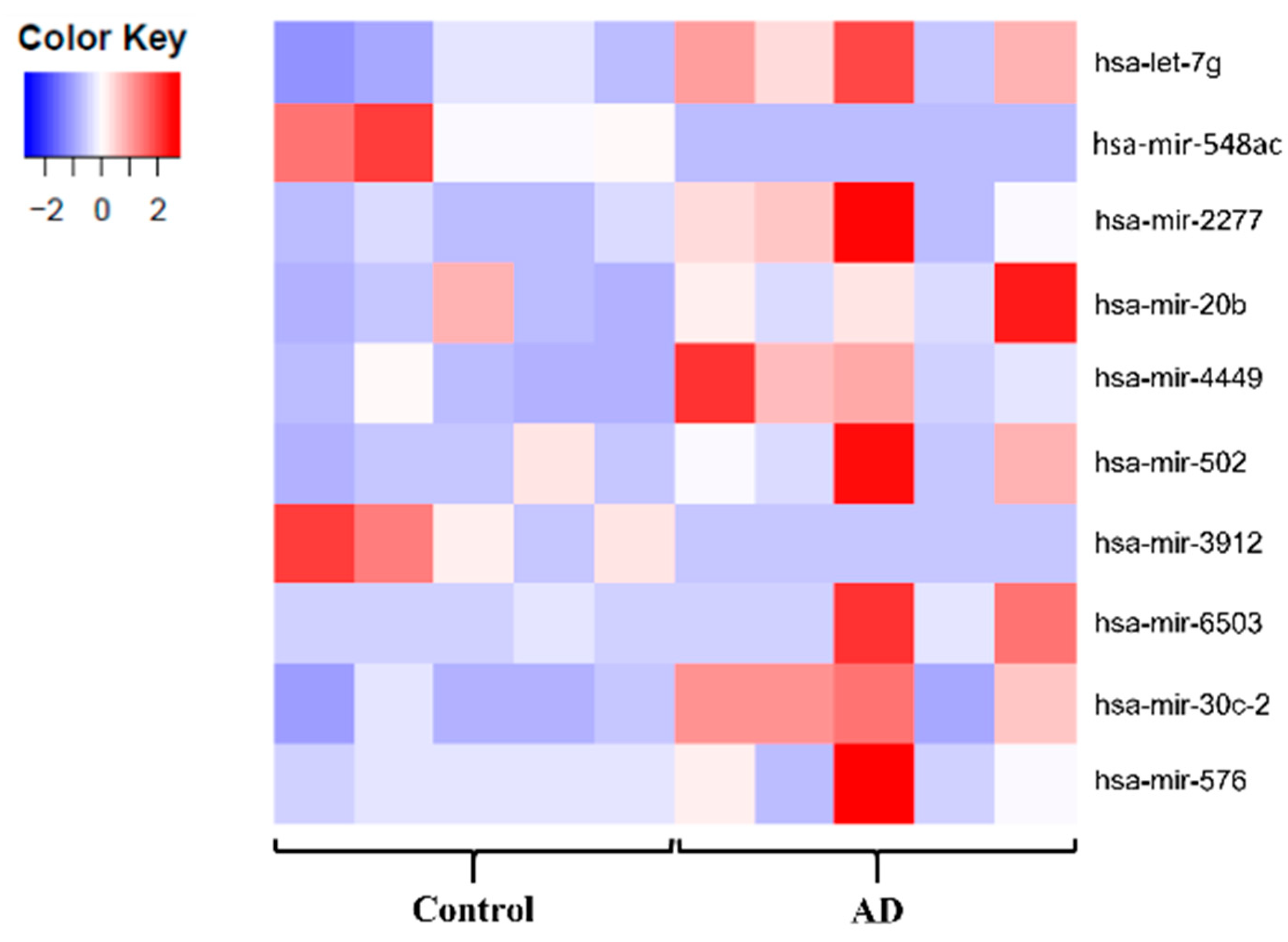
2.2. The miR Signature of NMV from AD Patients
2.2.1. Quality Control of Small RNA-seq Data
2.2.2. Detection of Dysregulated miRNA
2.2.3. Target and Pathway Analyses
3. Discussion
4. Materials and Methods
4.1. Neutrophil-Derived Microvesicle Isolation
4.2. Small RNA Purification, Library Preparation and Sequencing
4.3. Sequencing Data Analysis
4.4. Target and Pathway Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, C.; Dunn, N.; Mullee, M.; Perry, H. P2-251 Association between dementia and systemic infectious disease: Evidence from a case-control study. Neurobiol. Aging 2004, 25, S303–S304. [Google Scholar] [CrossRef]
- Cunningham, C.; Hennessy, E. Co-morbidity and systemic inflammation as drivers of cognitive decline: New experimental models adopting a broader paradigm in dementia research. Alzheimer’s Res. Ther. 2015, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asby, D.; Boche, D.; Allan, S.; Love, S.; Miners, J.S. Systemic infection exacerbates cerebrovascular dysfunction in Alzheimer’s disease. Brain 2021, 144, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Shad, K.F.; Aghazadeh, Y.; Ahmad, S.; Kress, B. Peripheral markers of Alzheimer’s disease: Surveillance of white blood cells. Synapse 2013, 67, 541–543. [Google Scholar] [CrossRef]
- Nachun, D.; Ramos, E.M.; Karydas, A.; Dokuru, D.; Gao, F.; Yang, Z.; Van Berlo, V.; Sears, R.; Kramer, J.; Boxer, A.L.; et al. Systems-level analysis of peripheral blood gene expression in dementia patients reveals an innate immune response shared across multiple disorders. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Nao, J.; Shi, J.; Zheng, D. Predictive Value of Routine Peripheral Blood Biomarkers in Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 332. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Lagarde, J.; Xicota, L.; Corne, H.; Chantran, Y.; Ms, T.C.; Crestani, B.; Bottlaender, M.; Potier, M.-C.; Aucouturier, P.; et al. Neutrophil hyperactivation correlates with Alzheimer’s disease progression. Ann. Neurol. 2018, 83, 387–405. [Google Scholar] [CrossRef]
- Bawa, K.K.; Initiative, F.T.A.D.N.; Krance, S.; Herrmann, N.; Cogo-Moreira, H.; Ouk, M.; Yu, D.; Wu, C.-Y.; Black, S.E.; Lanctôt, K.L.; et al. A peripheral neutrophil-related inflammatory factor predicts a decline in executive function in mild Alzheimer’s disease. J. Neuroinflamm. 2020, 17, 1–11. [Google Scholar] [CrossRef]
- Zenaro, E.; Pietronigro, E.; Della Bianca, V.; Piacentino, G.; Marongiu, L.; Budui, S.; Turano, E.; Rossi, B.; Angiari, S.; Dusi, S.; et al. Neutrophils promote Alzheimer’s disease–like pathology and cognitive decline via LFA-1 integrin. Nat. Med. 2015, 21, 880–886. [Google Scholar] [CrossRef]
- Gomez, I.; Ward, B.; Souilhol, C.; Recarti, C.; Ariaans, M.; Johnston, J.; Burnett, A.; Mahmoud, M.; Luong, L.A.; West, L.; et al. Neutrophil microvesicles drive atherosclerosis by delivering miR-155 to atheroprone endothelium. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.R.; Wang, X.N.; Sheng, C.; Li, Y.X.; Li, F.Z.T.; Sun, Y.; Han, Y. Extracellular vesicles as an emerging tool for the early detection of Alzheimer’s disease. Mech. Ageing Dev. 2019, 184, 111175. [Google Scholar] [CrossRef] [PubMed]
- Ajikumar, A.; Long, M.B.; Heath, P.R.; Wharton, S.B.; Ince, P.G.; Ridger, V.C.; Simpson, J.E. Neutrophil-Derived Microvesicle Induced Dysfunction of Brain Microvascular Endothelial Cells In Vitro. Int. J. Mol. Sci. 2019, 20, 5227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolonics, F.; Szeifert, V.; Timár, C.I.; Ligeti, E.; Lőrincz, Á.M. The Functional Heterogeneity of Neutrophil-Derived Extracellular Vesicles Reflects the Status of the Parent Cell. Cells 2020, 9, 2718. [Google Scholar] [CrossRef]
- Wei, W.; Wang, Z.Y.; Ma, L.N.; Zhang, T.T.; Cao, Y.; Li, H. MicroRNAs in Alzheimer’s Disease: Function and Potential Applications as Diagnostic Biomarkers. Front. Mol. Neurosci. 2020, 13, 160. [Google Scholar] [CrossRef]
- Toyama, K.; Spin, J.M.; Deng, A.C.; Huang, T.T.; Wei, K.; Wagenhäuser, M.U.; Yoshino, T.; Nguyen, H.; Mulorz, J.; Kundu, S.; et al. MicroRNA-mediated therapy modulating blood-brain barrier disruption improves vascular cognitive impairment. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1392–1406. [Google Scholar] [CrossRef] [Green Version]
- Harati, R.; Hammad, S.; Tlili, A.; Mahfood, M.; Mabondzo, A.; Hamoudi, R. miR-27a-3p regulates expression of intercellular junctions at the brain endothelium and controls the endothelial barrier permeability. PLoS ONE 2022, 17, e0262152. [Google Scholar] [CrossRef]
- Lin, M.; Zhu, L.; Wang, J.; Xue, Y.; Shang, X. miR-424–5p maybe regulate blood-brain barrier permeability in a model in vitro with Abeta incubated endothelial cells. Biochem. Biophys. Res. Commun. 2019, 517, 525–531. [Google Scholar] [CrossRef]
- Li, A.-D.; Tong, L.; Xu, N.; Ye, Y.; Nie, P.-Y.; Wang, Z.-Y.; Ji, L.-L. miR-124 regulates cerebromicrovascular function in APP/PS1 transgenic mice via C1ql3. Brain Res. Bull. 2019, 153, 214–222. [Google Scholar] [CrossRef]
- Sheng, Q.; Vickers, K.; Zhao, S.; Wang, J.; Samuels, D.; Koues, O.; Shyr, Y.; Guo, Y. Multi-perspective quality control of Illumina RNA sequencing data analysis. Brief. Funct. Genom. 2016, 16, 194–204. [Google Scholar] [CrossRef] [Green Version]
- Baik, S.H.; Cha, M.Y.; Hyun, Y.M.; Cho, H.; Hamza, B.; Kim, D.K.; Han, S.H.; Choi, H.; Kim, K.H.; Moon, M.; et al. Migration of neutrophils targeting amyloid plaques in Alzheimer’s disease mouse model. Neurobiol. Aging 2014, 35, 1286–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salinaro, A.T.; Pennisi, M.; di Paola, R.; Scuto, M.; Crupi, R.; Cambria, M.T.; Ontario, M.L.; Tomasello, M.; Uva, M.; Maiolino, L.; et al. Neuroinflammation and neurohormesis in the pathogenesis of Alzheimer’s disease and Alzheimer-linked pathologies: Modulation by nutritional mushrooms. Immun. Ageing 2018, 15, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.; Calabrese, E.J.; Mattson, M.P. Cellular Stress Responses, The Hormesis Paradigm, and Vitagenes: Novel Targets for Therapeutic Intervention in Neurodegenerative Disorders. Antioxid. Redox Signal. 2010, 13, 1763–1811. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Zhang, X.; Yin, K.J. MicroRNAs in central nervous system diseases: A prospective role in regulating blood-brain barrier integrity. Exp. Neurol. 2019, 323, 113094. [Google Scholar] [CrossRef] [PubMed]
- Henriques, A.D.; Machado-Silva, W.; Leite, R.E.; Suemoto, C.K.; Leite, K.R.; Srougi, M.; Pereira, A.C.; Jacob-Filho, W.; Nóbrega, O.T. Genome-wide profiling and predicted significance of post-mortem brain microRNA in Alzheimer’s disease. Mech. Ageing Dev. 2020, 191, 111352. [Google Scholar] [CrossRef]
- Satoh, J.I.; Kino, Y.; Niida, S. MicroRNA-Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer’s Disease from Public Data. Biomark Insights 2015, 10, 21. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, S.S.; Nygaard, A.-B.; Nielsen, M.-Y.; Jensen, K.; Christensen, T. miRNA Expression Profiles in Cerebrospinal Fluid and Blood of Patients with Acute Ischemic Stroke. Transl. Stroke Res. 2014, 5, 711–718. [Google Scholar] [CrossRef]
- Jury, D.; Daugaard, I.; Sanders, K.J.; Hansen, L.L.; Agalliu, D.; Pedersen, I.M. miR-151a enhances Slug dependent angiogenesis. Oncotarget 2020, 11, 2160–2171. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H., 3rd; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-related changes in microRNA levels in serum. Aging 2013, 5, 725–740. [Google Scholar] [CrossRef] [Green Version]
- Dobrowolny, G.; Martone, J.; Lepore, E.; Casola, I.; Petrucci, A.; Inghilleri, M.; Morlando, M.; Colantoni, A.; Scicchitano, B.M.; Calvo, A.; et al. A longitudinal study defined circulating microRNAs as reliable biomarkers for disease prognosis and progression in ALS human patients. Cell Death Discov. 2021, 7, 1–11. [Google Scholar] [CrossRef]
- He, S.; Huang, L.; Shao, C.; Nie, T.; Xia, L.; Cui, B.; Lu, F.; Zhu, L.; Chen, B.; Yang, Q. Several miRNAs derived from serum extracellular vesicles are potential biomarkers for early diagnosis and progression of Parkinson’s disease. Transl. Neurodegener. 2021, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-L.; Min, L.; Guo, Q.-D.; Zhang, J.-X.; Jiang, H.-L.; Shao, S.; Xing, J.-G.; Yin, L.-L.; Liu, J.-H.; Liu, R.; et al. Profiling microRNA from Brain by Microarray in a Transgenic Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sierksma, A.; Lu, A.; Salta, E.; Vanden Eynden, E.; Callaerts-Vegh, Z.; D’Hooge, R.; Blum, D.; Buée, L.; Fiers, M.; De Strooper, B. Deregulation of neuronal miRNAs induced by amyloid-β or TAU pathology 11 Medical and Health Sciences 1109 Neurosciences. Mol. Neurodegener. 2018, 13, 1–15. [Google Scholar]
- Kumar, P.; Dezso, Z.; MacKenzie, C.; Oestreicher, J.; Agoulnik, S.; Byrne, M.; Bernier, F.; Yanagimachi, M.; Aoshima, K.; Oda, Y. Circulating miRNA Biomarkers for Alzheimer’s Disease. PLoS ONE 2013, 8, e69807. [Google Scholar] [CrossRef]
- Leidinger, P.; Backes, C.; Deutscher, S.; Schmitt, K.; Mueller, S.C.; Frese, K.; Haas, J.; Ruprecht, K.; Paul, F.; Stähler, C.; et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013, 14, R78. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Yu, J.T.; Tan, M.S.; Liu, Q.Y.; Wang, H.F.; Zhang, W.; Jiang, T.; Tan, L. Genome-Wide Serum microRNA Expression Profiling Identifies Serum Biomarkers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 40, 1017–1027. [Google Scholar] [CrossRef]
- Dong, H.; Li, J.; Huang, L.; Chen, X.; Li, D.; Wang, T.; Hu, C.; Xu, J.; Zhang, C.; Zen, K.; et al. Serum MicroRNA Profiles Serve as Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Dis. Markers 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Bernstein, D.L.; Rom, S. Let-7g* and miR-98 Reduce Stroke-Induced Production of Proinflammatory Cytokines in Mouse Brain. Front. Cell Dev. Biol. 2020, 8, 632. [Google Scholar] [CrossRef]
- Rom, S.; Dykstra, H.; Zuluaga-Ramirez, V.; Reichenbach, N.L.; Persidsky, Y. miR-98 and let-7g* Protect the Blood-Brain Barrier Under Neuroinflammatory Conditions. J. Cereb. Blood Flow Metab. 2015, 35, 1957–1965. [Google Scholar] [CrossRef] [Green Version]
- Prabhakar, P.; Chandra, S.R.; Christopher, R. Circulating microRNAs as potential biomarkers for the identification of vascular dementia due to cerebral small vessel disease. Age Ageing 2017, 46, 861–864. [Google Scholar] [CrossRef] [Green Version]
- Grasso, M.; Piscopo, P.; Talarico, G.; Ricci, L.; Crestini, A.; Tosto, G.; Gasparini, M.; Bruno, G.; Denti, M.A.; Confaloni, A. Plasma microRNA profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiol. Aging 2019, 84, 240.e1–240.e12. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, K.; Hu, X.; Chen, H.; Li, F.; Yin, N.; Liu, A.L.; Shan, K.; Qin, Y.W.; Huang, X.; Chang, Q.; et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. eBioMedicine 2019, 49, 341–353. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Zhao, H.; Wang, Y.; Chen, Z. Exosomal shuttled miR-424-5p from ischemic preconditioned microglia mediates cerebral endothelial cell injury through negatively regulation of FGF2/STAT3 pathway. Exp. Neurol. 2020, 333, 113411. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Qu, M.; Li, Y.; Wang, L.; Zhang, L.; Wang, Y.; Tang, Y.; Tian, H.L.; Zhang, Z.; Yang, G.Y. MicroRNA-126-3p/-5p Overexpression Attenuates Blood-Brain Barrier Disruption in a Mouse Model of Middle Cerebral Artery Occlusion. Stroke 2020, 51, 619–627. [Google Scholar] [CrossRef]
- Fu, X.; Niu, T.; Li, X. MicroRNA-126-3p Attenuates Intracerebral Hemorrhage-Induced Blood-Brain Barrier Disruption by Regulating VCAM-1 Expression. Front. Neurosci. 2019, 13, 866. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Li, C.; Sun, A.; Wang, Y.; Zhou, S. Quantification of microRNA-210 in the cerebrospinal fluid and serum: Implications for Alzheimer’s disease. Exp. Ther. Med. 2015, 9, 1013–1017. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; He, X.; Wang, Y.; Tang, Y.; Zheng, C.; Cai, H.; Liu, J.; Wang, Y.; Fu, Y.; Yang, G.-Y. MicroRNA-210 overexpression induces angiogenesis and neurogenesis in the normal adult mouse brain. Gene Ther. 2014, 21, 37–43. [Google Scholar] [CrossRef]
- Huang, L.; Ma, Q.; Li, Y.; Li, B.; Zhang, L. Inhibition of microRNA-210 suppresses pro-inflammatory response and reduces acute brain injury of ischemic stroke in mice HHS Public Access. Exp. Neurol. 2018, 300, 41–50. [Google Scholar] [CrossRef]
- Ma, Q.; Dasgupta, C.; Li, Y.; Huang, L.; Zhang, L. MicroRNA-210 Suppresses Junction Proteins and Disrupts Blood-Brain Barrier Integrity in Neonatal Rat Hypoxic-Ischemic Brain Injury. Int. J. Mol. Sci. 2017, 18, 1356. [Google Scholar] [CrossRef] [Green Version]
- Goodall, E.F.; Leach, V.; Wang, C.; Cooper-Knock, J.; Heath, P.R.; Baker, D.; Drew, D.R.; Saffrey, M.J.; Simpson, J.E.; Romero, I.A.; et al. Age-Associated mRNA and miRNA Expression Changes in the Blood-Brain Barrier. Int. J. Mol. Sci. 2019, 20, 3097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marco, A.; MacPherson, J.I.; Ronshaugen, M.; Griffiths-Jones, S. MicroRNAs from the same precursor have different targeting properties. Silence 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Pi, J.; Zou, D.; Wang, X.; Xu, J.; Yu, S.; Zhang, T.; Li, F.; Zhang, X.; Zhao, H.; et al. microRNA arm-imbalance in part from complementary targets mediated decay promotes gastric cancer progression. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, X.; Li, W.; Huang, S.; Yin, Z.; Yang, M.; Han, Z.; Han, Z.; Chen, F.; Wang, H.; Lei, P.; et al. Increased miR-21-3p in Injured Brain Microvascular Endothelial Cells after Traumatic Brain Injury Aggravates Blood–Brain Barrier Damage by Promoting Cellular Apoptosis and Inflammation through Targeting MAT2B. J. Neurotrauma 2019, 36, 1291–1305. [Google Scholar] [CrossRef]
- Ge, X.; Han, Z.; Chen, F.; Wang, H.; Zhang, B.; Jiang, R.; Lei, P.; Zhang, J. miR-21 alleviates secondary blood–brain barrier damage after traumatic brain injury in rats. Brain Res. 2015, 1603, 150–157. [Google Scholar] [CrossRef]
- Yao, X.; Wang, Y.; Zhang, D. microRNA-21 Confers Neuroprotection Against Cerebral Ischemia-Reperfusion Injury and Alleviates Blood-Brain Barrier Disruption in Rats via the MAPK Signaling Pathway. J. Mol. Neurosci. 2018, 65, 43–53. [Google Scholar] [CrossRef]
- Varmazyar, R.; Noori-Zadeh, A.; Rajaei, F.; Darabi, S.; Bakhtiyari, S. 17 β-Estradiol Oxidative Stress Attenuation and Autophagy-Induced Dopaminergic Neuroprotection. Cell J. 2019, 21, 1. [Google Scholar]
- Town, T.; Laouar, Y.; Pittenger, C.; Mori, T.; Szekely, C.A.; Tan, J.; Duman, R.S.; Flavell, R.A. Blocking TGF-β–Smad2/3 innate immune signaling mitigates Alzheimer-like pathology. Nat. Med. 2008, 14, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Baumann, J.; Huang, S.F.; Gassmann, M.; Tsao, C.C.; Ogunshola, O.O. Furin inhibition prevents hypoxic and TGFβ-mediated blood-brain barrier disruption. Exp. Cell Res. 2019, 383, 111503. [Google Scholar] [CrossRef] [Green Version]
- Mehdipour, M.; Etienne, J.; Chen, C.C.; Gathwala, R.; Rehman, M.; Kato, C.; Liu, C.; Liu, Y.; Zuo, Y.; Conboy, M.J.; et al. Rejuvenation of brain, liver and muscle by simultaneous pharmacological modulation of two signaling determinants, that change in opposite directions with age. Aging 2019, 11, 5628–5645. [Google Scholar] [CrossRef]
- Tominaga, K.; Suzuki, H.I. TGF-β Signaling in Cellular Senescence and Aging-Related Pathology. Int. J. Mol. Sci. 2019, 20, 5002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.-Y.; Qin, L.; Li, G.; Wang, Z.; Dahlman, J.E.; Malagon-Lopez, J.; Gujja, S.; Cilfone, N.A.; Kauffman, K.J.; Sun, L.; et al. Endothelial TGF-β signalling drives vascular inflammation and atherosclerosis. Nat. Metab. 2019, 1, 912–926. [Google Scholar] [CrossRef] [PubMed]
- Diniz, L.P.; Matias, I.; Siqueira, M.; Stipursky, J.; Gomes, F.C.A. Astrocytes and the TGF-β1 Pathway in the Healthy and Diseased Brain: A Double-Edged Sword. Mol. Neurobiol. 2018, 56, 4653–4679. [Google Scholar] [CrossRef] [PubMed]
- Howe, M.D.; Furr, J.W.; Munshi, Y.; Roy-O’Reilly, M.A.; Maniskas, M.E.; Koellhoffer, E.C.; d’Aigle, J.; Sansing, L.H.; McCullough, L.D.; Urayama, A. Transforming growth factor-β promotes basement membrane fibrosis, alters perivascular cerebrospinal fluid distribution, and worsens neurological recovery in the aged brain after stroke. GeroScience 2019, 41, 543. [Google Scholar] [CrossRef] [PubMed]
- Tallquist, M.; Kazlauskas, A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004, 15, 205–213. [Google Scholar] [CrossRef]
- Sweeney, M.; Foldes, G. It Takes Two: Endothelial-Perivascular Cell Cross-Talk in Vascular Development and Disease. Front. Cardiovasc. Med. 2018, 5, 154. [Google Scholar] [CrossRef]
- Shen, J.; Ishii, Y.; Xu, G.; Dang, T.C.; Hamashima, T.; Matsushima, T.; Yamamoto, S.; Hattori, Y.; Takatsuru, Y.; Nabekura, J.; et al. PDGFR-β as a Positive Regulator of Tissue Repair in a Mouse Model of Focal Cerebral Ischemia. J. Cereb. Blood Flow Metab. 2011, 32, 353–367. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, R.; Kawahara, M.; Nakano-Doi, A.; Takahashi, A.; Tanaka, Y.; Narita, A.; Kuwahara-Otani, S.; Hayakawa, T.; Yagi, H.; Matsuyama, T.; et al. Brain pericytes serve as microglia-generating multipotent vascular stem cells following ischemic stroke. J. Neuroinflamm. 2016, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Klett, F.; Potas, J.R.; Hilpert, D.; Blazej, K.; Radke, J.; Huck, J.; Engel, O.; Stenzel, W.; Genové, G.; Priller, J. Early loss of pericytes and perivascular stromal cell-induced scar formation after stroke. J. Cereb. Blood Flow Metab. 2013, 33, 428–439. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Xu, G.; Zhu, R.; Yuan, J.; Ishii, Y.; Hamashima, T.; Matsushima, T.; Yamamoto, S.; Takatsuru, Y.; Nabekura, J.; et al. PDGFR-β restores blood-brain barrier functions in a mouse model of focal cerebral ischemia. J. Cereb. Blood Flow Metab. 2018, 39, 1501–1515. [Google Scholar] [CrossRef]
- Sagare, A.P.; Sweeney, M.D.; Makshanoff, J.; Zlokovic, B.V. Shedding of soluble platelet-derived growth factor receptor-β from human brain pericytes. Neurosci. Lett. 2015, 607, 97–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagare, A.P.; Bell, R.D.; Zhao, Z.; Ma, Q.; Winkler, E.A.; Ramanathan, A.; Zlokovic, B.V. Pericyte loss influences Alzheimer-like neurodegeneration in mice. Nat. Commun. 2013, 4, 2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.-J.; Zhang, H.; Park, H.; Choi, K.-S.; Lee, H.W.; Agrawal, V.; Kim, Y.-M.; Kwon, Y.-G. Yes-associated protein regulates endothelial cell contact-mediated expression of angiopoietin-2. Nat. Commun. 2015, 6, 6943. [Google Scholar] [CrossRef]
- Wang, L.; Luo, J.-Y.; Li, B.; Tian, X.Y.; Chen, L.-J.; Huang, Y.; Liu, J.; Deng, D.; Lau, C.W.; Wan, S.; et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 2016, 540, 579–582. [Google Scholar] [CrossRef]
- Wang, X.; Valls, A.F.; Schermann, G.; Shen, Y.; Moya, I.M.; Castro, L.; Urban, S.; Solecki, G.M.; Winkler, F.; Riedemann, L.; et al. YAP/TAZ Orchestrate VEGF Signaling during Developmental Angiogenesis. Dev. Cell 2017, 42, 462–478.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, P.; Zhang, Z.; Zou, C.; Tian, Q.; Chen, X.; Hong, M.; Liu, X.; Chen, Q.; Xu, Z.; Li, M.; et al. Hippo/YAP signaling pathway mitigates blood-brain barrier disruption after cerebral ischemia/reperfusion injury Tian participated in making the MCAO model and measuring neurologic deficit scores HHS Public Access. Behav. Brain Res. 2019, 356, 8–17. [Google Scholar] [CrossRef]
- Park, H.W.; Kim, Y.C.; Yu, B.; Moroishi, T.; Mo, J.S.; Plouffe, S.W.; Meng, Z.; Lin, K.C.; Yu, F.X.; Wang, Y.; et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell 2015, 162, 780–794. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Shen, C.; Lu, Y.; Huang, Z.; Li, L.; Rand, C.D.; Pan, J.; Sun, X.-D.; Tan, Z.; Wang, H.; et al. Muscle Yap Is a Regulator of Neuromuscular Junction Formation and Regeneration. J. Neurosci. 2017, 37, 3465–3477. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, N.J.; Menet, R.; Picard, K.; Parent, G.; Tremblay, M.; ElAli, A. Canonical Wnt Pathway Maintains Blood-Brain Barrier Integrity upon Ischemic Stroke and Its Activation Ameliorates Tissue Plasminogen Activator Therapy. Mol. Neurobiol. 2019, 56, 6521–6538. [Google Scholar] [CrossRef]
- Wylezinski, L.S.; Hawiger, J. Interleukin 2 Activates Brain Microvascular Endothelial Cells Resulting in Destabilization of Adherens Junctions. J. Biol. Chem. 2016, 291, 22913–22923. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Li, L.; Peng, C.; Bian, C.; Ocak, P.E.; Zhang, J.H.; Yang, Y.; Zhou, D.; Chen, G.; Luo, Y. Targeting oxidative stress and inflammatory response for blood-brain barrier protection in intracerebral hemorrhage. Antioxid. Redox Signal. 2022. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Champ, C.; Day, J.; Aarts, E.; Bahr, B.A.; Bakker, M.; Bánáti, D.; Calabrese, V.; Cederholm, T.; Cryan, J.; et al. Poor cognitive ageing: Vulnerabilities, mechanisms and the impact of nutritional interventions. Ageing Res. Rev. 2018, 42, 40–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Powell, D. Degust: Interactive RNA-seq Analysis; Drpowell/Degust 4.1.1 (4.1.1); Zenodo: Geneva, Switzerland, 2019. [Google Scholar] [CrossRef]
- Rahman, R.-U.; Gautam, A.; Bethune, J.; Sattar, A.; Fiosins, M.; Magruder, D.S.; Capece, V.; Shomroni, O.; Bonn, S. Oasis 2: Improved online analysis of small RNA-seq data. BMC Bioinform. 2018, 19, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Aparicio-Puerta, E.; Lebrón, R.; Rueda, A.; Gómez-Martín, C.; Giannoukakos, S.; Jáspez, D.; Medina, J.M.; Zubković, A.; Jurak, I.; Fromm, B.; et al. sRNAbench and sRNAtoolbox 2019: Intuitive fast small RNA profiling and differential expression. Nucleic Acids Res. 2019, 47, W530–W535. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
| Tool | Top 5 DE miRNA | p-Value | log2FC | Top 15 DE miRNA across Tools | |
|---|---|---|---|---|---|
| Up | Down | ||||
| DESeq2—sno/miRNA track | MIRLET7G | 0.00002 | 1.36 | ||
| MIR548AC | 0.00106 | −2.22 | |||
| MIR20B | 0.0014 | 1.58 | |||
| MIR502 | 0.0016 | 1.56 | |||
| MIR4449 | 0.00329 | 1.32 | |||
| DESeq2—NCBI RefSeq track | hsa-let-7g | 0.00037 | 1.2 | ||
| hsa-mir-548ac | 0.00055 | −1.9 | |||
| hsa-mir-2277 | 0.00262 | 1.65 | hsa-let-7a-3p | hsa-mir-151a | |
| hsa-mir-20b | 0.00415 | 1.31 | hsa-let-7g | hsa-mir-548ac | |
| hsa-mir-4449 | 0.00534 | 1.19 | miR-4449 | hsa-miR-4485-3p | |
| Degust—sno/miRNA track | hsa-mir-20b | 0.00062 | 1.98 | miR-652-5p | hsa-miR-136-3p |
| hsa-let-7g | 0.00139 | 1.37 | miR-16-1-3p | hsa-miR-584-5p | |
| hsa-mir-151a | 0.00199 | −1.66 | miR-20b-5p | ||
| hsa-mir-4449 | 0.00373 | 1.47 | miR-502 | ||
| hsa-mir-6503 | 0.0073 | 1.74 | miR-2277 | ||
| Degust—NCBI RefSeq track | microRNA 20b | 0.0003 | 2.05 | miR-4443 | |
| microRNA let-7g | 0.00032 | 1.43 | miR-6503 | ||
| microRNA 4449 | 0.00151 | 1.57 | |||
| microRNA 151a | 0.00277 | −1.51 | |||
| microRNA 6503 | 0.00391 | 1.7 | |||
| OASIS 2.0 | hsa-miR-4485-3p | 2 × 10−7 | −1.19 | ||
| hsa-miR-652-5p | 0.000001 | 2.82 | |||
| hsa-let-7a-3p | 0.000004 | 2.34 | |||
| hsa-miR-4443 | 0.00002 | 2 | |||
| hsa-miR-16-1-3p | 0.000027 | 3.52 | |||
| sRNAToolbox | hsa-miR-136-3p | 0.000074 | −1.49 | ||
| hsa-miR-584-5p | 0.0028 | −1.02 | |||
| hsa-miR-4485-3p | 0.01 | −1.75 | |||
| hsa-let-7a-3p | 0.02 | 3.14 | |||
| hsa-miR-20b-5p | 0.02 | 2.35 | |||
| Upregulated miRNA Targets | Downregulated miRNA Targets | |||||
|---|---|---|---|---|---|---|
| Term | p-Value | Adjusted p-Value | Term | p-Value | Adjusted p-Value | |
| Bioplanet 2019 | Axon guidance | 3.67 × 10−11 | 4.22 × 10−8 | Generic transcription pathway | 1.04 × 10−11 | 1.57 × 10−8 |
| Developmental biology | 5.59 × 10−11 | 4.22 × 10−8 | Wnt signalling pathway | 8.47 × 10−11 | 5.69 × 10−8 | |
| Interleukin-2 signalling pathway | 2.31 × 10−8 | 9.24 × 10−6 | Developmental biology | 1.13 × 10−10 | 5.69 × 10−8 | |
| Insulin signalling pathway | 2.50 × 10−8 | 9.24 × 10−6 | Pathways in cancer | 2.02 × 10−10 | 7.61 × 10−8 | |
| TGF-beta regulation of extracellular matrix | 3.28 × 10−8 | 9.24 × 10−6 | Interleukin-2 signalling pathway | 5.63 × 10−10 | 1.70 × 10−7 | |
| Wnt signalling pathway | 4.22 × 10−8 | 9.24 × 10−6 | Axon guidance | 4.35 × 10−9 | 1.10 × 10−6 | |
| Generic transcription pathway | 4.28 × 10−8 | 9.24 × 10−6 | PDGFB signalling pathway | 5.66 × 10−9 | 1.22 × 10−6 | |
| Pathways in cancer | 7.50 × 10−7 | 1.39 × 10−4 | TGF-beta regulation of extracellular matrix | 2.32 × 10−8 | 4.22 × 10−6 | |
| PDGFB signalling pathway | 8.27 × 10−7 | 1.39 × 10−4 | Pancreatic cancer | 2.51 × 10−8 | 4.22 × 10−6 | |
| TGF-beta signalling pathway | 3.39 × 10−6 | 4.85 × 10−4 | ATM-dependent DNA damage response | 3.03 × 10−8 | 4.57 × 10−6 | |
| KEGG 2021 Human | Proteoglycans in cancer | 5.13 × 10−7 | 9.85 × 10−5 | Endocytosis | 1.32 × 10−9 | 4.21 × 10−7 |
| Axon guidance | 6.16 × 10−7 | 9.85 × 10−5 | Pathways in cancer | 1.09 × 10−8 | 1.75 × 10−6 | |
| T cell receptor signalling pathway | 1.26 × 10−5 | 1.19 × 10−3 | AGE-RAGE signalling pathway in diabetic complications | 5.69 × 10−8 | 6.07 × 10−6 | |
| AGE-RAGE signalling pathway in diabetic complications | 1.94 × 10−5 | 1.19 × 10−3 | MAPK signalling pathway | 1.62 × 10−7 | 1.30 × 10−5 | |
| Focal adhesion | 2.15 × 10−5 | 1.19 × 10−3 | Signalling pathways regulating pluripotency of stem cells | 2.42 × 10−7 | 1.55 × 10−5 | |
| Hippo signalling pathway | 2.60 × 10−5 | 1.19 × 10−3 | Proteoglycans in cancer | 3.36 × 10−7 | 1.79 × 10−5 | |
| MAPK signalling pathway | 2.61 × 10−5 | 1.19 × 10−3 | Herpes simplex virus 1 infection | 9.16 × 10−7 | 4.19 × 10−5 | |
| Insulin signalling pathway | 4.85 × 10−5 | 1.57 × 10−3 | Pancreatic cancer | 1.42 × 10−6 | 5.68 × 10−5 | |
| Yersinia infection | 4.85 × 10−5 | 1.57 × 10−3 | Inositol phosphate metabolism | 2.78 × 10−6 | 8.99 × 10−5 | |
| Cellular senescence | 4.92 × 10−5 | 1.57 × 10−3 | Yersinia infection | 2.81 × 10−6 | 8.99 × 10−5 | |
| Group | Age | Sex | Temperature (°C) | CRP (mg/L) |
|---|---|---|---|---|
| Control | 56 | F | 36.8 | NA |
| 55 | F | 36.5 | 1.3 | |
| 67 | F | 36.2 | 1.0 | |
| 71 | F | 36.7 | 4.4 | |
| 66 | M | 37.1 | 0.7 | |
| AD | 63 | F | 36.7 | 8.2 |
| 62 | M | 36.9 | 1.2 | |
| 53 | M | 35.8 | 0.6 | |
| 70 | M | 35.4 | 0.4 | |
| 56 | M | 36.1 | 0.3 | |
| 61.9 ± 6.6 | 36.4 ± 0.53 | 1.1 ± 2.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Villaseñor, I.; Smith, C.I.; Thang, Y.J.R.; Heath, P.R.; Wharton, S.B.; Blackburn, D.J.; Ridger, V.C.; Simpson, J.E. RNA-Seq Profiling of Neutrophil-Derived Microvesicles in Alzheimer’s Disease Patients Identifies a miRNA Signature That May Impact Blood–Brain Barrier Integrity. Int. J. Mol. Sci. 2022, 23, 5913. https://doi.org/10.3390/ijms23115913
Vázquez-Villaseñor I, Smith CI, Thang YJR, Heath PR, Wharton SB, Blackburn DJ, Ridger VC, Simpson JE. RNA-Seq Profiling of Neutrophil-Derived Microvesicles in Alzheimer’s Disease Patients Identifies a miRNA Signature That May Impact Blood–Brain Barrier Integrity. International Journal of Molecular Sciences. 2022; 23(11):5913. https://doi.org/10.3390/ijms23115913
Chicago/Turabian StyleVázquez-Villaseñor, Irina, Cynthia I. Smith, Yung J. R. Thang, Paul R. Heath, Stephen B. Wharton, Daniel J. Blackburn, Victoria C. Ridger, and Julie E. Simpson. 2022. "RNA-Seq Profiling of Neutrophil-Derived Microvesicles in Alzheimer’s Disease Patients Identifies a miRNA Signature That May Impact Blood–Brain Barrier Integrity" International Journal of Molecular Sciences 23, no. 11: 5913. https://doi.org/10.3390/ijms23115913
APA StyleVázquez-Villaseñor, I., Smith, C. I., Thang, Y. J. R., Heath, P. R., Wharton, S. B., Blackburn, D. J., Ridger, V. C., & Simpson, J. E. (2022). RNA-Seq Profiling of Neutrophil-Derived Microvesicles in Alzheimer’s Disease Patients Identifies a miRNA Signature That May Impact Blood–Brain Barrier Integrity. International Journal of Molecular Sciences, 23(11), 5913. https://doi.org/10.3390/ijms23115913