Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Results
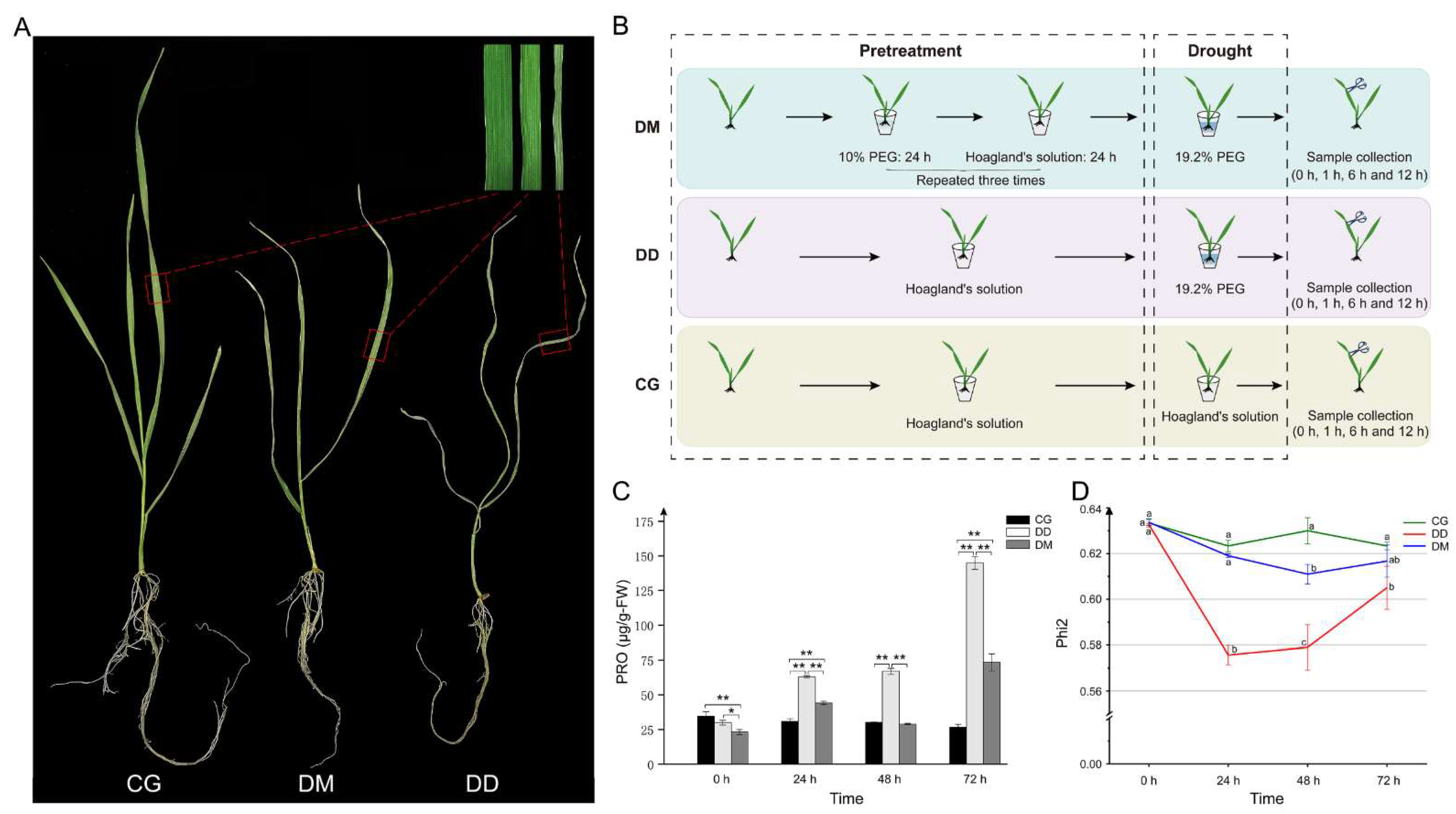
2.1. The Influence of Drought Memory to Plant Drought Resistance
2.2. High-Throughput Sequencing of miRNAs
2.3. Identification of Conserved, Known and Novel miRNAs
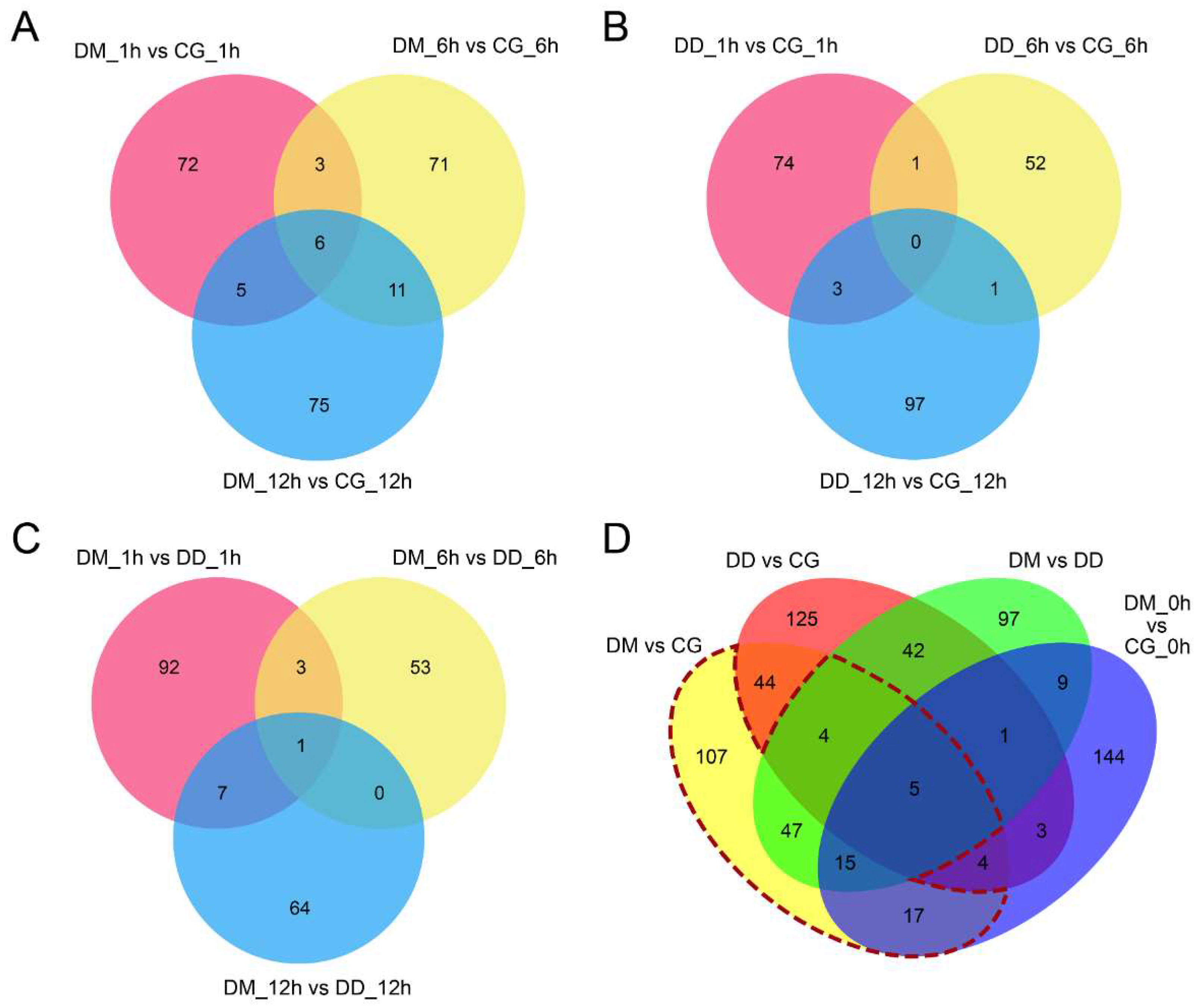
2.4. Identification of Drought Memory-Related miRNAs
2.5. Validation of Target Genes of Drought Memory-Related miRNAs in Wheat Using Degradome Sequencing
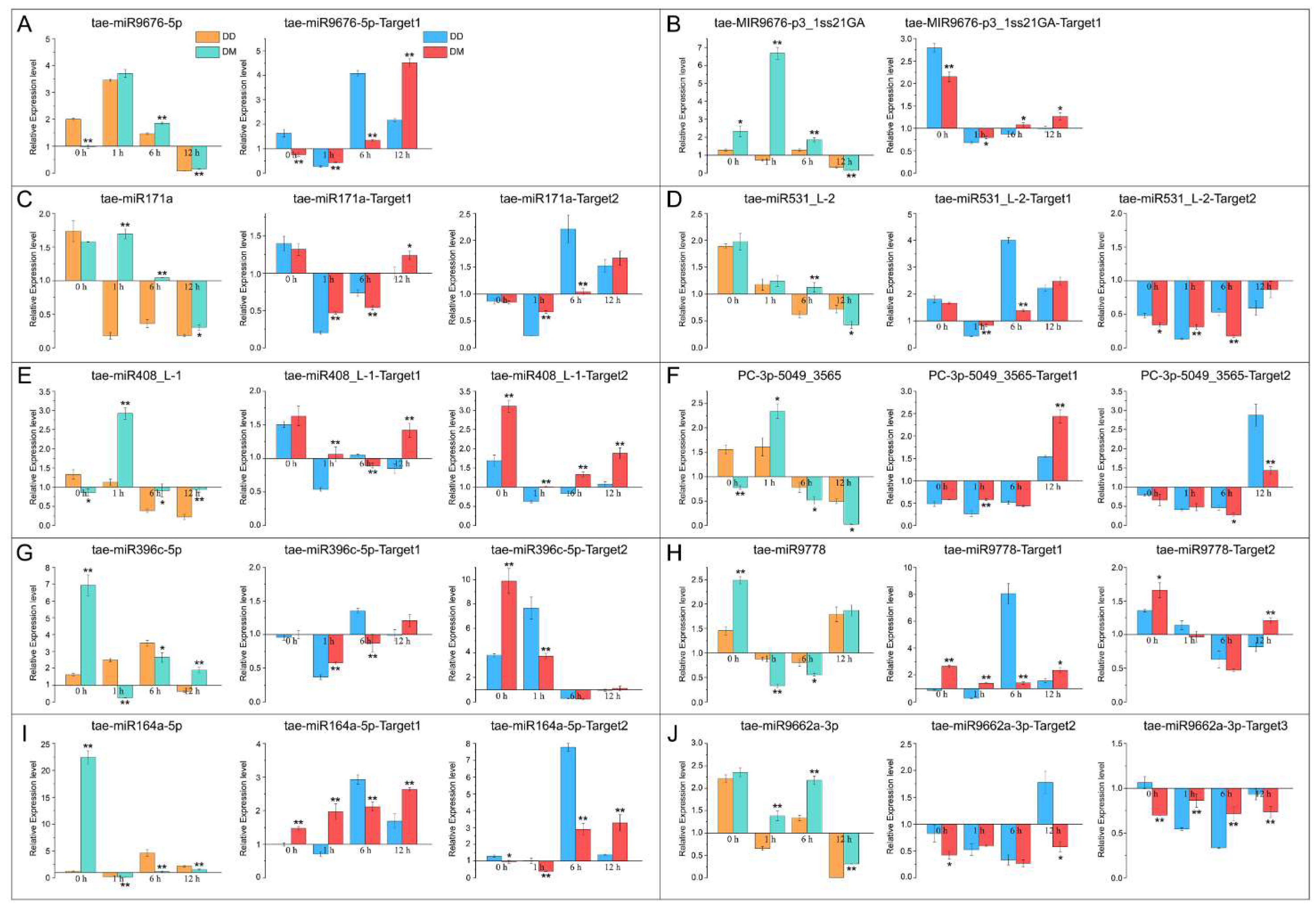
2.6. Validation of Drought Memory-Related miRNAs and Their Target Genes by RT-qPCR
2.7. Overexpression of the tae-miR531_L-2 Precursor in Arabidopsis thaliana
3. Discussion
3.1. Identification of Drought Memory-Related miRNAs and Their Target Genes in Wheat
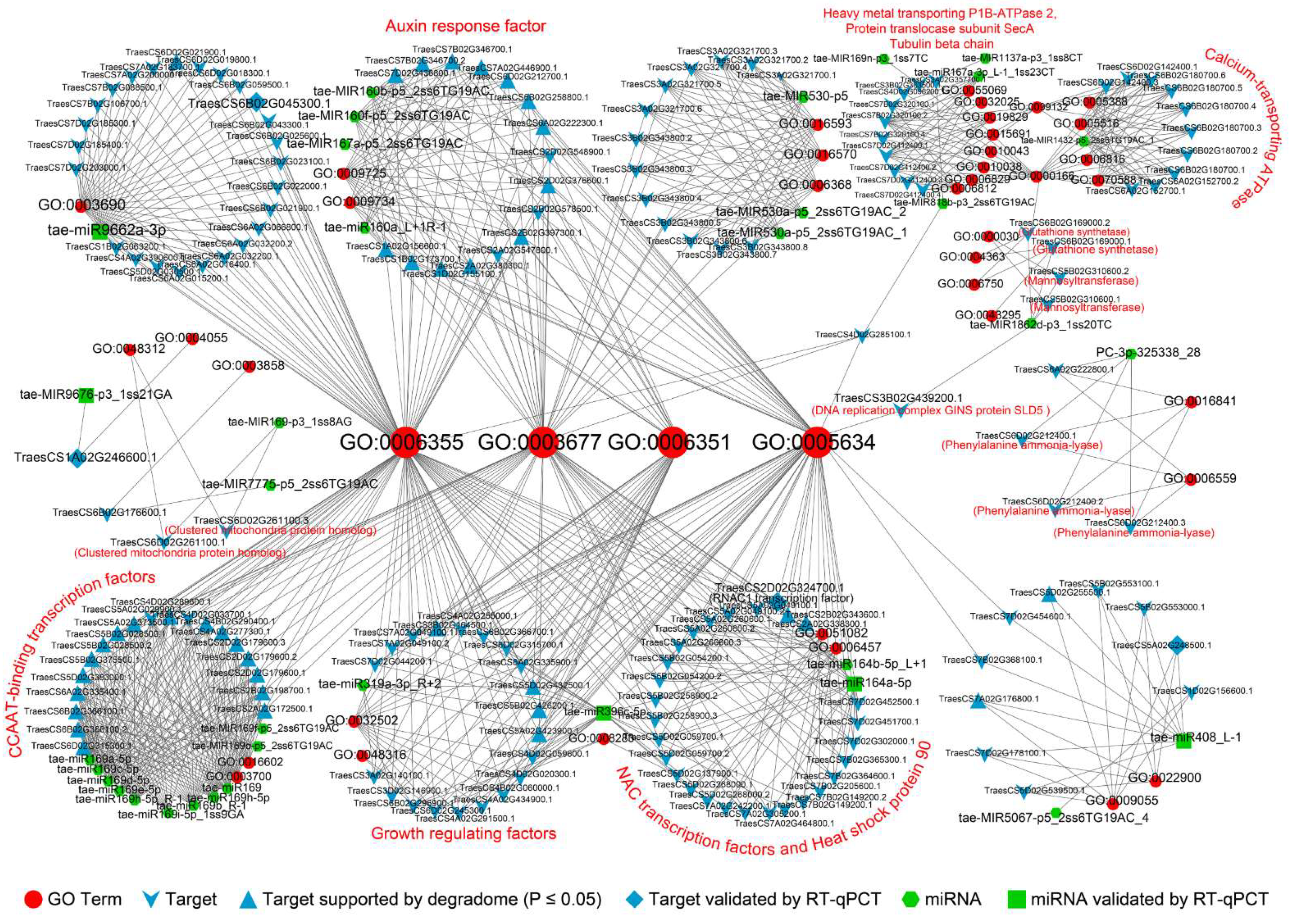
3.2. miRNA-Gene-GO Association Analysis of Drought Memory-Related miRNAs
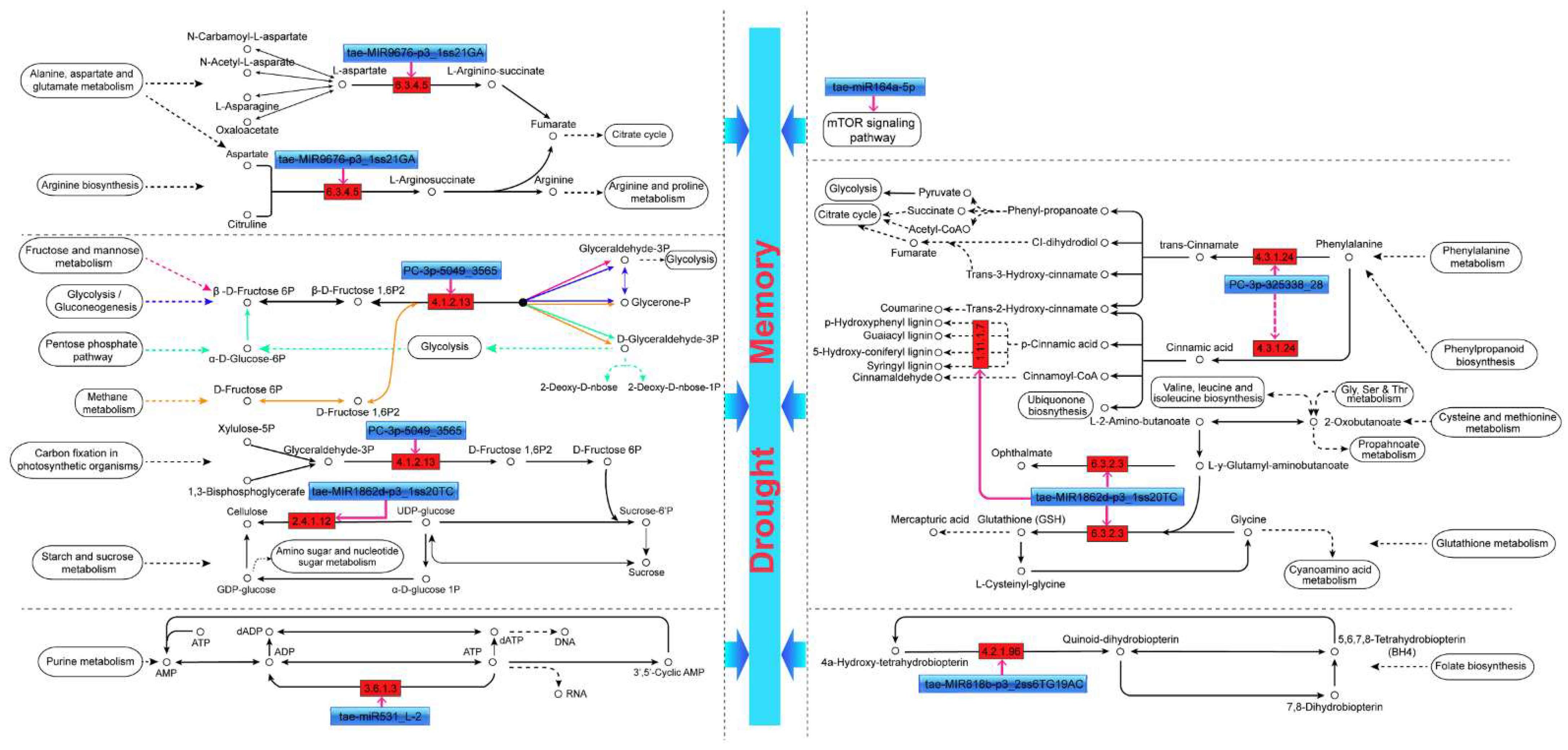
3.3. Proposed miRNA-Dependent Regulatory Pathways That Participate in Drought Memory
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Small RNA and Degradome Library Construction and Sequencing
4.3. Identification of Known and Potential Novel miRNAs
4.4. Analysis of Differentially Expressed miRNAs
4.5. Degradome Validation and Annotation of Target Genes of miRNAs
4.6. Validation of Differentially Expressed miRNAs and Their Target Genes
4.7. Overexpression of tae-miR531_L-2 in Arabidopsis thaliana
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fitton, N.; Alexander, P.; Arnell, N.; Bajzelj, B.; Calvin, K.; Doelman, J.; Gerber, J.S.; Havlik, P.; Hasegawa, T.; Herrero, M.; et al. The vulnerabilities of agricultural land and food production to future water scarcity. Glob. Environ. Chang.-Hum. Policy Dimens. 2019, 58, 101944. [Google Scholar] [CrossRef]
- Carraro, C.; Edenhofer, O.; Flachsland, C.; Kolstad, C.; Stavins, R.; Stowe, R. The IPCC at a crossroads: Opportunities for reform. Science 2015, 350, 34–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Borisjuk, N.; Kishchenko, O.; Eliby, S.; Schramm, C.; Anderson, P.; Jatayev, S.; Kurishbayev, A.; Shavrukov, Y. Genetic modification for wheat improvement: From transgenesis to genome editing. BioMed Res. Int. 2019, 2019, 6216304. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global synthesis of drought effects on maize and wheat production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef] [PubMed]
- Avramova, Z. Transcriptional ‘memory’ of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef]
- Fleta-Soriano, E.; Pinto-Marijuan, M.; Munne-Bosch, S. Evidence of drought stress memory in the facultative CAM, aptenia cordifolia: Possible role of phytohormones. PLoS ONE 2015, 10, e013539. [Google Scholar] [CrossRef] [Green Version]
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Munne-Bosch, S. Cross-stress tolerance and stress “memory” in plants: An integrated view. Environ. Exp. Bot. 2013, 94, 1–88. [Google Scholar] [CrossRef]
- Walter, J.; Nagy, L.; Hein, R.; Rascher, U.; Beierkuhnlein, C.; Willner, E.; Jentsch, A. Do plants remember drought? hints towards a drought-memory in grasses. Environ. Exp. Bot. 2011, 71, 34–40. [Google Scholar] [CrossRef]
- Li, X.; Topbjerg, H.B.; Jiang, D.; Liu, F. Drought priming at vegetative stage improves the antioxidant capacity and photosynthesis performance of wheat exposed to a short-term low temperature stress at jointing stage. Plant Soil 2015, 393, 307–318. [Google Scholar] [CrossRef]
- Hilker, M.; Schmulling, T. Stress priming, memory, and signalling in plants. Plant Cell Environ. 2019, 42, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Aditya; Banerjee; Aryadeep; Roychoudhury, Epigenetic regulation during salinity and drought stress in plants: Histone modifications and DNA methylation. Plant Gene 2017, 11, 199–204. [CrossRef]
- Roychoudhury, A.; Banerjee, A.; Lahiri, V. Metabolic and molecular-genetic regulation of proline signaling and its cross-talk with major effectors mediates abiotic stress tolerance in plants. Turk. J. Bot. 2015, 39, 887–910. [Google Scholar] [CrossRef]
- Luo, X.; Ou, Y.; Li, R.; He, Y. Maternal transmission of the epigenetic ‘memory of winter cold’ in Arabidopsis. Nat. Plants 2020, 6, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Feng, L.; Gu, X.; Deng, X.; Qiu, Q.; Li, Q.; Zhang, Y.; Wang, M.; Deng, Y.; Wang, E.; et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019, 29, 379–390. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Sanchez, R.; Kundariya, H.; Maher, T.; Dopp, I.; Schwegel, R.; Virdi, K.; Axtell, M.J.; Mackenzie, S.A. Segregation of an MSH1 RNAi transgene produces heritable non-genetic memory in association with methylome reprogramming. Nat. Commun. 2020, 11, 2214. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [Green Version]
- Virlouvet, L.; Ding, Y.; Fujii, H.; Avramova, Z.; Fromm, M. ABA signaling is necessary but not sufficient for RD29B transcriptional memory during successive dehydration stresses in Arabidopsis thaliana. Plant J. 2014, 79, 150–161. [Google Scholar] [CrossRef]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abid, M.; Hakeem, A.; Shao, Y.; Liu, Y.; Zahoor, R.; Fan, Y.; Jiang, S.; Ata-Ul-Karim, S.T.; Tian, Z.; Jiang, D.; et al. Seed osmopriming invokes stress memory against post-germinative drought stress in wheat (Triticum aestivum L.). Environ. Exp. Bot. 2018, 145, 12–20. [Google Scholar] [CrossRef]
- Ding, Y.; Virlouvet, L.; Liu, N.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Pagano, L.; Rossi, R.; Paesano, L.; Marmiroli, N.; Marmiroli, M. miRNA regulation and stress adaptation in plants. Environ. Exp. Bot. 2021, 184, 104369. [Google Scholar] [CrossRef]
- Urbanaviciute, I.; Bonfiglioli, L.; Pagnotta, M.A. One hundred candidate genes and their roles in drought and salt tolerance in wheat. Int. J. Mol. Sci. 2021, 22, 6378. [Google Scholar] [CrossRef]
- Shi, G.Q.; Fu, J.Y.; Rong, L.J.; Zhang, P.Y.; Guo, C.J.; Xiao, K. TaMIR1119, a miRNA family member of wheat (Triticum aestivum L.), is essential in the regulation of plant drought tolerance. J. Integr. Agric. 2018, 17, 2369–2378. [Google Scholar] [CrossRef] [Green Version]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Kantar, M.; Budak, H. Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct. Integr. Genom. 2015, 15, 587–598. [Google Scholar] [CrossRef]
- Galis, I.; Gaquerel, E.; Pandey, S.P.; Baldwin, I.T. Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell Environ. 2009, 32, 617–627. [Google Scholar] [CrossRef]
- Shah, S.M.S.; Ullah, F. A comprehensive overview of miRNA targeting drought stress resistance in plants. Braz. J. Biol. 2021, 83, e242708. [Google Scholar] [CrossRef] [PubMed]
- Perrino, E.V.; Wagensommer, R.P. Crop wild relatives (CWRs) threatened and endemic to italy: Urgent actions for protection and use. Biology 2022, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, L.; Milanovic, M.; Procopio, A.C.; Spampinato, G.; Maruca, G.; Perrino, E.V.; Mannino, G.C.; Fagoonee, S.; Luzza, F.; Musarella, C.M. Ancient wheats: Beneficial effects on insulin resistance. Minerva Med. 2021, 112, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Perrino, E.V.; Wagensommer, R.P.; Medagli, P. Aegilops (Poaceae) in Italy: Taxonomy, geographical distribution, ecology, vulnerability and conservation. Syst. Biodivers. 2014, 12, 331–349. [Google Scholar] [CrossRef]
- Vaucheret, H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE 2009, 4, e6442. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Baurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [Green Version]
- de Freitas Guedes, F.A.; Nobres, P.; Ferreira, D.C.R.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Hwang, E.W.; Shin, S.J.; Yu, B.K.; Byun, M.O.; Kwon, H.B. miR171 family members are involved in drought response in solanum tuberosum. J. Plant Biol. 2011, 54, 43–48. [Google Scholar] [CrossRef]
- Chen, L.; Sun, H.; Wang, F.; Yue, D.; Shen, X.; Sun, W.; Zhang, X.; Yang, X. Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14–GhMKK11–GhMPK31 pathway is involved in the drought response in cotton. Plant Mol. Biol. 2020, 103, 211–223. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2019, 16, 862–877. [Google Scholar] [CrossRef]
- Yang, W.; Fan, T.; Hu, X.Y.; Cheng, T.H.; Zhang, M.Y. Overexpressing osa-miR171c decreases salt stress tolerance in rice. J. Plant Biol. 2017, 60, 485–492. [Google Scholar] [CrossRef]
- Kamal, A.H.; Cho, K.; Kim, D.E.; Uozumi, N.; Chung, K.Y.; Lee, S.Y.; Choi, J.S.; Cho, S.W.; Shin, C.S.; Woo, S.H. Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol. Biol. Rep. 2012, 39, 9059–9074. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.W.; Roy, S.K.; Kamal, A.H.; Cho, K.; Cho, S.W.; Park, C.S.; Choi, J.S.; Komatsu, S.; Woo, S.H. Proteome analysis of roots of wheat seedlings under aluminum stress. Mol. Biol. Rep. 2014, 41, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Verdus, M.C.; Thellier, M.; Ripoll, C. Storage of environmental signals in flax. Their morphogenetic effect as enabled by a transient depletion of calcium. Plant J. 1997, 12, 1399–1410. [Google Scholar] [CrossRef]
- Wang, W.H.; Chen, J.; Liu, T.W.; Chen, J.; Han, A.D.; Simon, M.; Dong, X.J.; He, J.X.; Zheng, H.L. Regulation of the calcium-sensing receptor in both stomatal movement and photosynthetic electron transport is crucial for water use efficiency and drought tolerance in Arabidopsis. J. Exp. Bot. 2014, 65, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Labudda, M.; Azam, F.M.S. Glutathione-dependent responses of plants to drought: A review. Acta Soc. Bot. Pol. 2014, 83, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant. Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Mohanty, B.; Kitazumi, A.; Cheung, C.Y.M.; Lakshmanan, M.; de Los Reyes, B.G.; Jang, I.C.; Lee, D.Y. Identification of candidate network hubs involved in metabolic adjustments of rice under drought stress by integrating transcriptome data and genome-scale metabolic network. Plant Sci. 2016, 242, 224–239. [Google Scholar] [CrossRef]
- Sun, C.H.; Li, X.H.; Hu, Y.L.; Zhao, P.Y.; Xu, T.; Sun, J.; Gao, X.L. Proline, sugars, and antioxidant enzymes respond to drought stress in the leaves of strawberry plants. Hortic. Sci. Technol. 2015, 33, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Balibrea, M.E.; RusAlvarez, A.M.; Bolarin, M.C.; PerezAlfocea, F. Fast changes in soluble carbohydrates and proline contents in tomato seedlings in response to ionic and non-ionic iso-osmotic stresses. J. Plant Physiol. 1997, 151, 221–226. [Google Scholar] [CrossRef]
- Verslues, P.E.; Bray, E.A. Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 2006, 57, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, R. The water culture methods for growing plants without soil. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Feng, X.J.; Li, J.R.; Qi, S.L.; Lin, Q.F.; Jin, J.B.; Hua, X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8335–E8343. [Google Scholar] [CrossRef] [Green Version]
- Kuhlgert, S.; Austic, G.; Zegarac, R.; Osei-Bonsu, I.; Hoh, D.; Chilvers, M.I.; Roth, M.G.; Bi, K.; TerAvest, D.; Weebadde, P.; et al. MultispeQ Beta: A tool for large-scale plant phenotyping connected to the open PhotosynQ network. R. Soc. Open Sci. 2016, 3, 160592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Wu, P.H.; Beane, T.; Zamore, P.D.; Weng, Z.P. Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genom. 2018, 19, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Shahid, M.Q.; Wu, J.W.; Wang, L.; Liu, X.D.; Lu, Y.G. Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int. J. Mol. Sci. 2016, 17, 499. [Google Scholar] [CrossRef]
- Ambady, S.; Wu, Z.; Dominko, T. Identification of novel microRNAs in Xenopus laevis metaphase II arrested eggs. Genesis 2012, 50, 286–299. [Google Scholar] [CrossRef] [Green Version]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Li, H.P.; Peng, T.; Wang, Q.; Wu, Y.F.; Chang, J.F.; Zhang, M.B.; Tang, G.L.; Li, C.H. Development of incompletely fused carpels in maize ovary revealed by miRNA, target gene and phytohormone analysis. Front. Plant Sci. 2017, 8, 463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]

| Sample | UMI Labeled Non-Redundant Data | Known miRNAs | Conserved miRNAs | Novel miRNAs | ||||
|---|---|---|---|---|---|---|---|---|
| gp1a | gp1b | gp2a | gp2b | gp3 | total | gp4 | ||
| CG/DD_0 h | 8244816 | 461 | 210 | 330 | 42 | 71 | 653 | 1431 |
| CG_1 h | 9284052 | 414 | 217 | 344 | 38 | 65 | 664 | 1306 |
| CG_6 h | 8631839 | 495 | 211 | 282 | 39 | 68 | 600 | 1470 |
| CG_12 h | 11244731 | 437 | 211 | 356 | 42 | 68 | 677 | 1294 |
| DD_1 h | 8829616 | 372 | 212 | 325 | 38 | 54 | 629 | 1292 |
| DD_6 h | 11247192 | 469 | 216 | 355 | 41 | 64 | 676 | 1430 |
| DD_12 h | 18968451 | 407 | 216 | 374 | 39 | 72 | 701 | 1419 |
| DM_0 h | 10502181 | 490 | 220 | 348 | 38 | 64 | 670 | 1754 |
| DM_1 h | 7799044 | 348 | 214 | 329 | 36 | 63 | 642 | 1226 |
| DM_6 h | 12679727 | 332 | 202 | 306 | 41 | 56 | 605 | 1333 |
| DM_12 h | 13278172 | 401 | 211 | 332 | 36 | 59 | 638 | 1159 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, H.; Zhang, H.; Su, N.; Sun, X.; Zhao, Q.; Weining, S.; Nie, X.; Yue, W. Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2022, 23, 5917. https://doi.org/10.3390/ijms23115917
Yue H, Zhang H, Su N, Sun X, Zhao Q, Weining S, Nie X, Yue W. Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences. 2022; 23(11):5917. https://doi.org/10.3390/ijms23115917
Chicago/Turabian StyleYue, Hong, Haobin Zhang, Ning Su, Xuming Sun, Qi Zhao, Song Weining, Xiaojun Nie, and Wenjie Yue. 2022. "Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.)" International Journal of Molecular Sciences 23, no. 11: 5917. https://doi.org/10.3390/ijms23115917
APA StyleYue, H., Zhang, H., Su, N., Sun, X., Zhao, Q., Weining, S., Nie, X., & Yue, W. (2022). Integrate Small RNA and Degradome Sequencing to Reveal Drought Memory Response in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences, 23(11), 5917. https://doi.org/10.3390/ijms23115917








