Effects of Bile Acid Modulation by Dietary Fat, Cholecystectomy, and Bile Acid Sequestrant on Energy, Glucose, and Lipid Metabolism and Gut Microbiota in Mice
Abstract
:1. Introduction
2. Results
2.1. Energy Metabolism
2.2. Glucose and Lipid Metabolism
2.3. Hepatic Metabolism
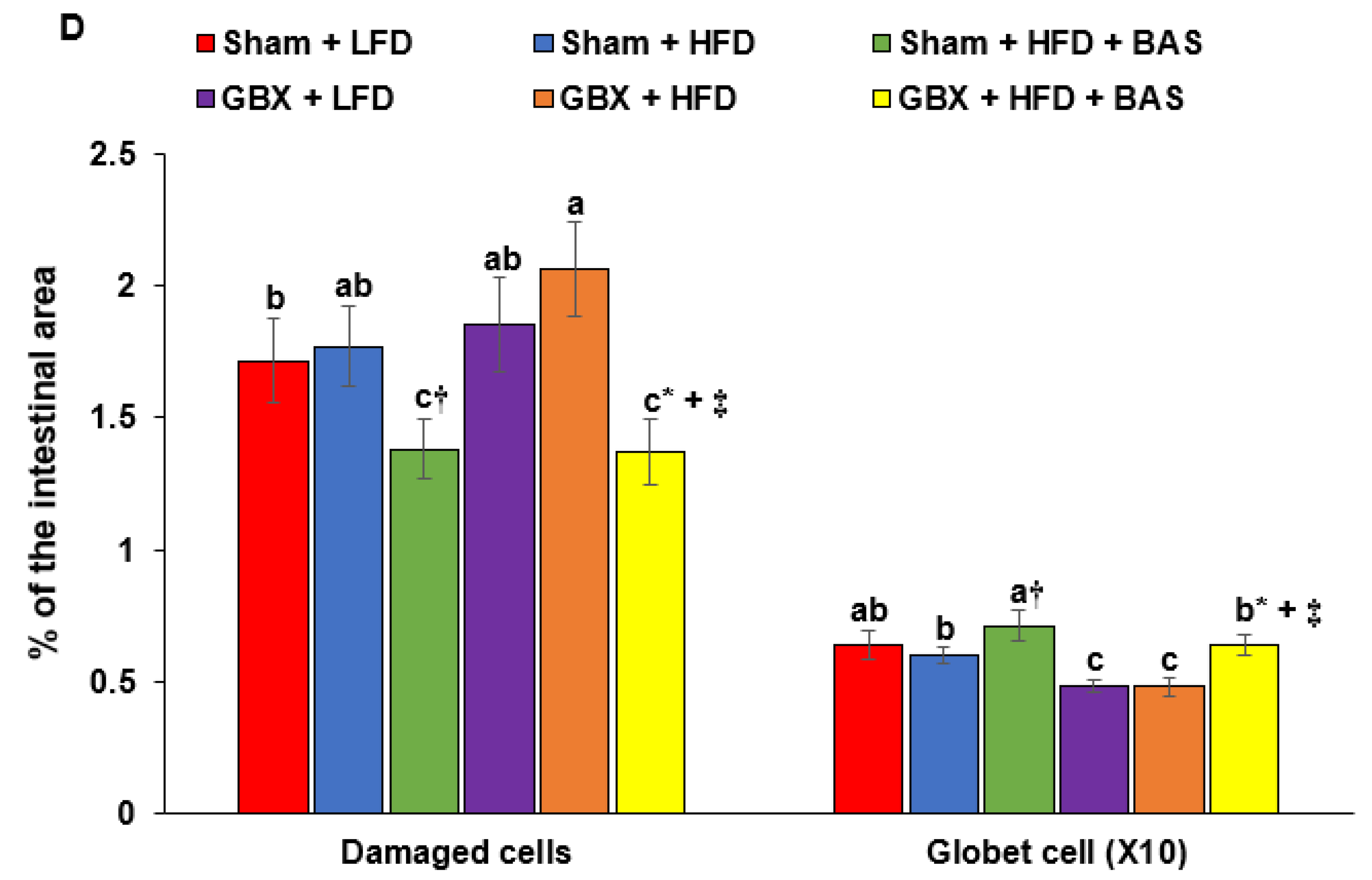
2.4. Histology of the Large Intestines
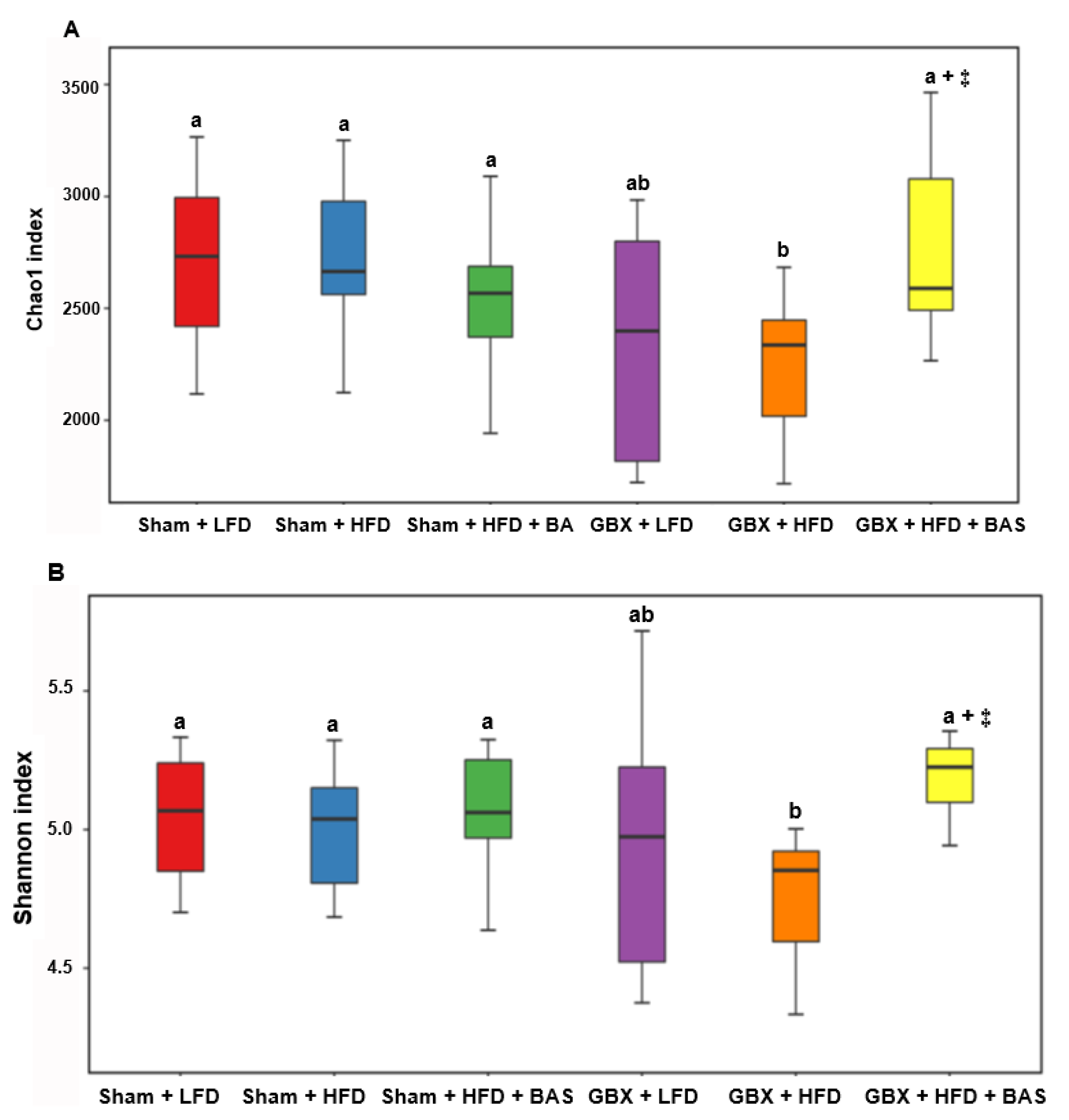
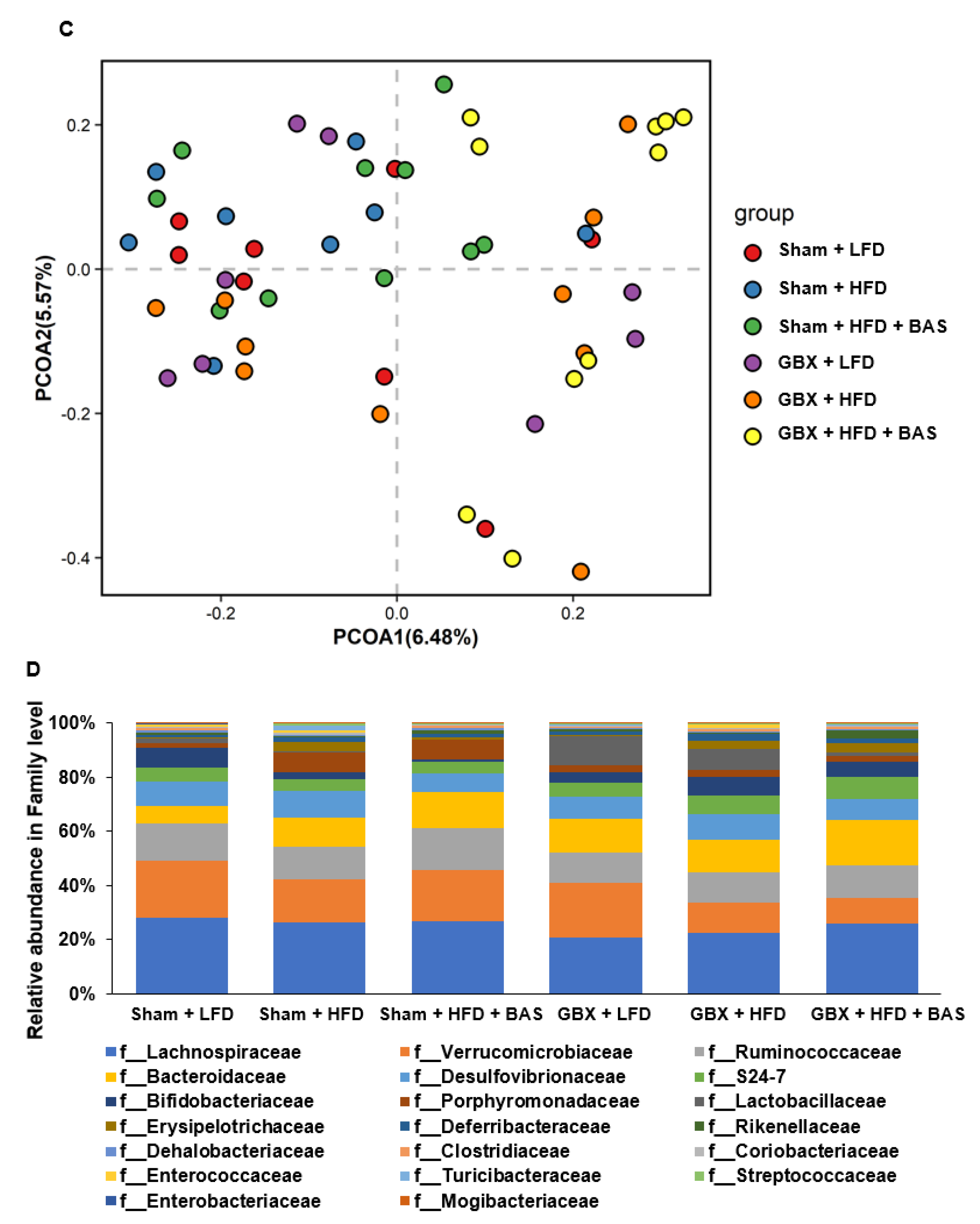
2.5. Fecal Microbiota Community
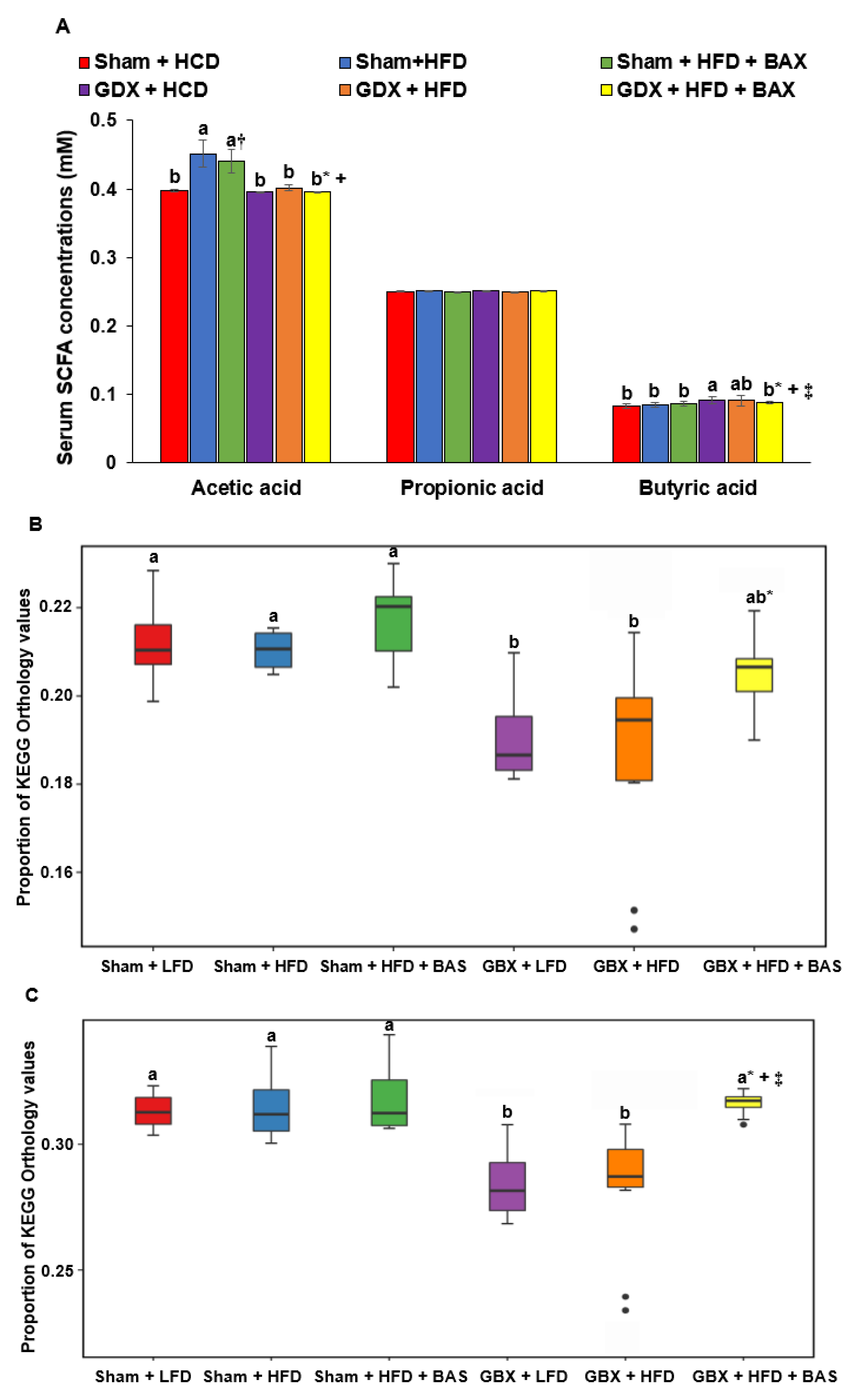
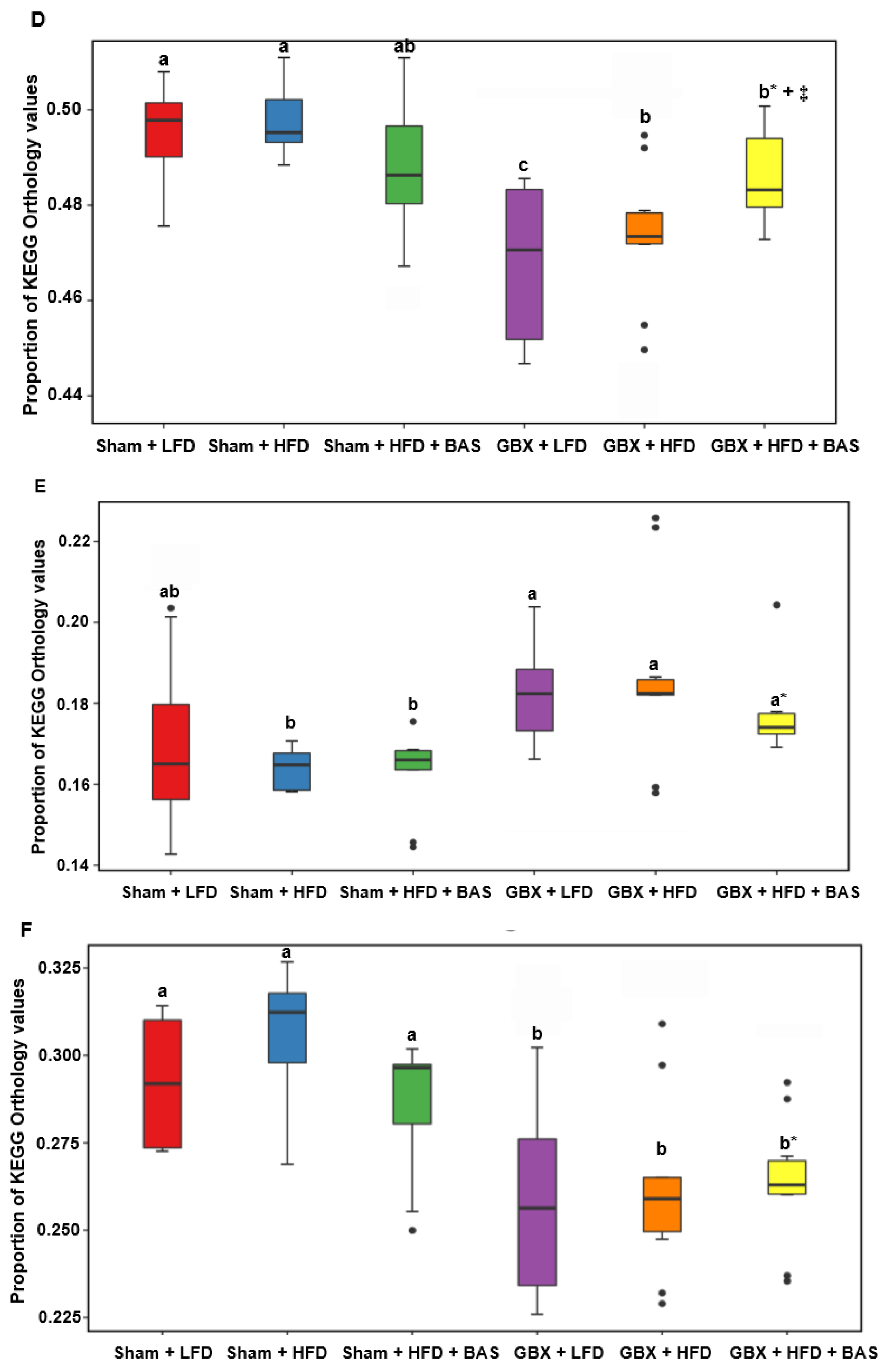
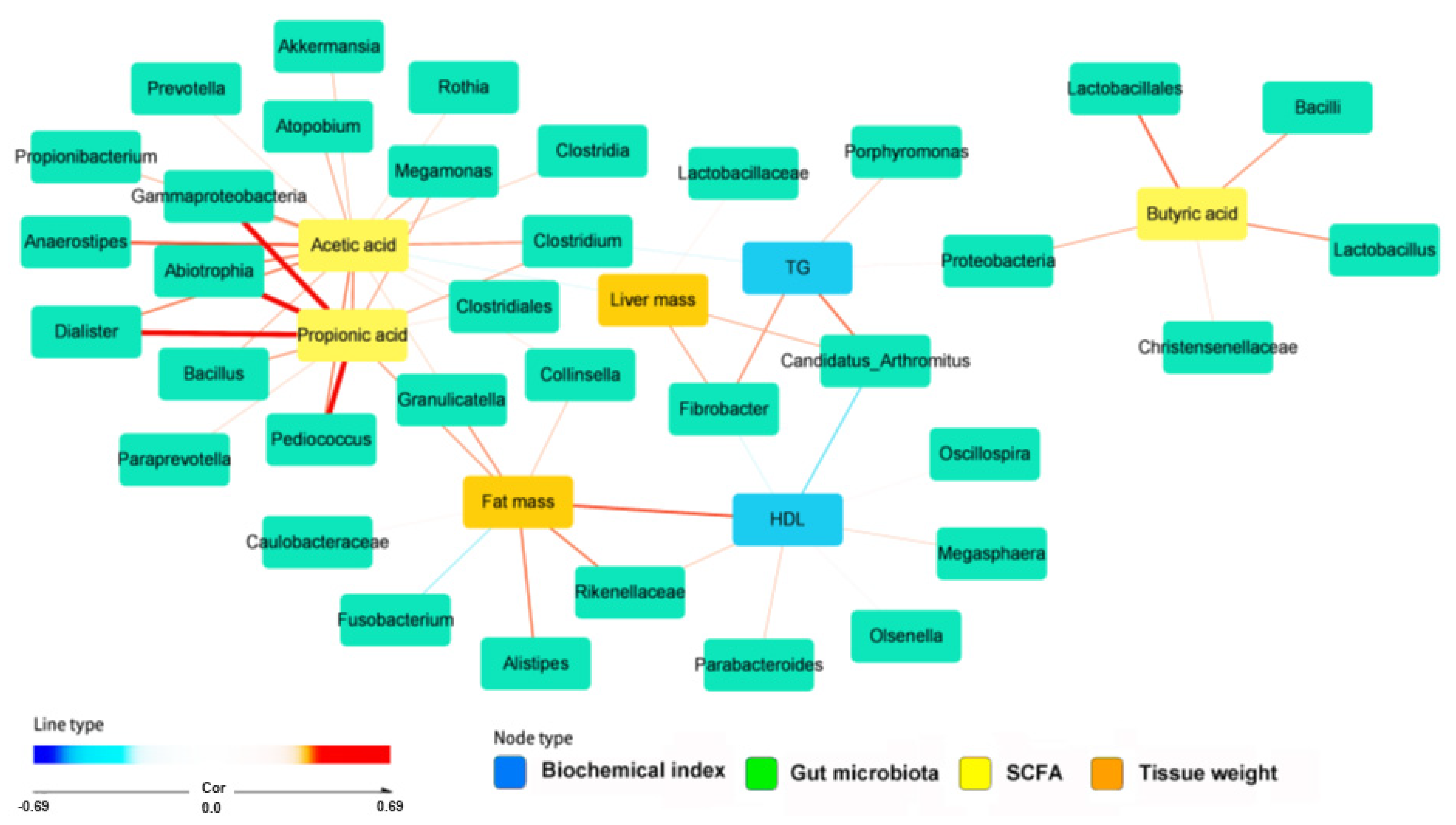
2.6. SCFA and Metagenome Functions of Fecal Bacteria
3. Discussion
4. Materials and Methods
4.1. Animal Care
4.2. Diet Preparation
4.3. GBX (Cholecystectomy)
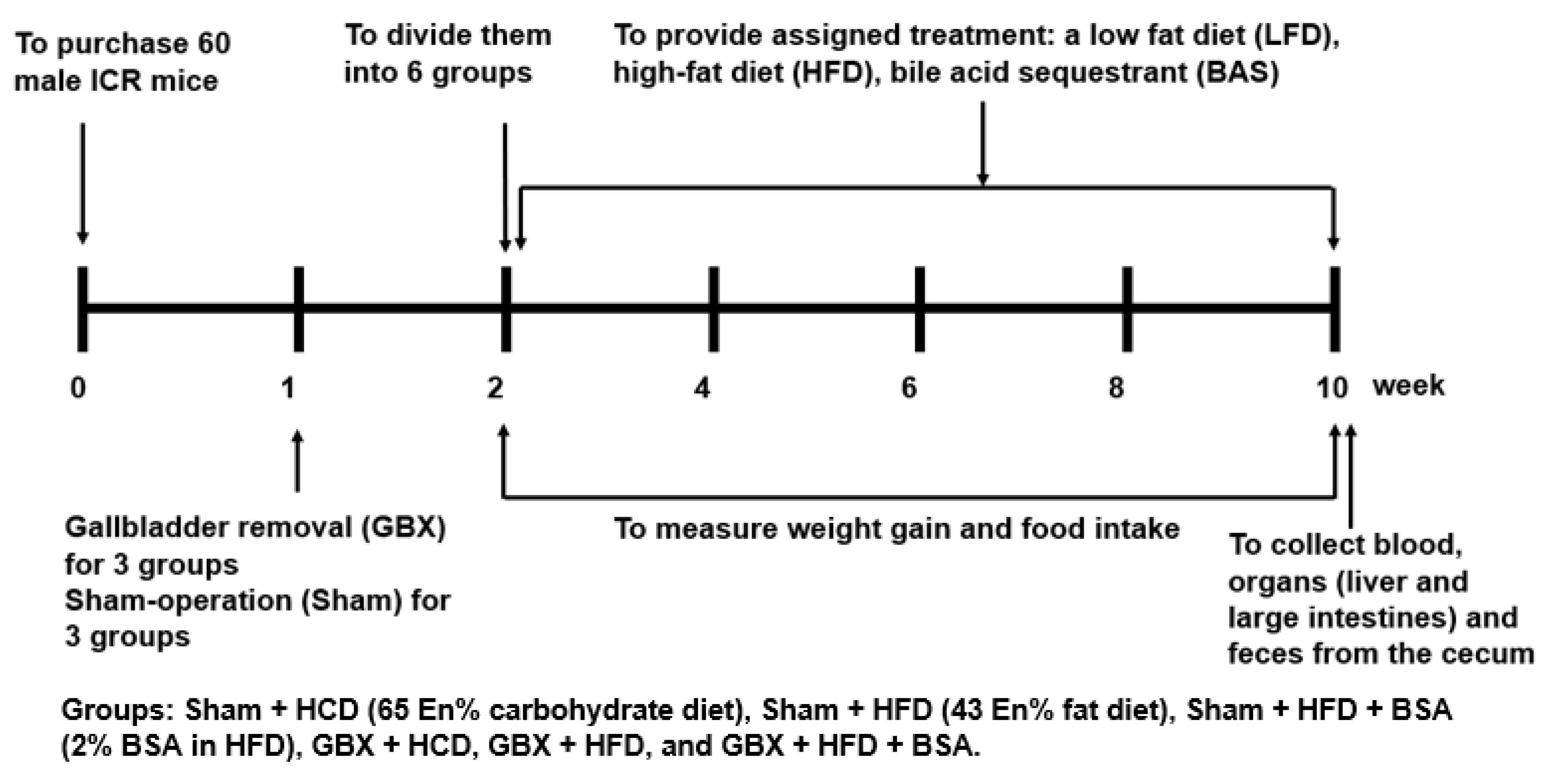
4.4. Experimental Design
4.5. Quantitative Real-Time PCR
4.6. Histology of the Large Intestines
4.7. Serum SCFA Concentrations and Gut Microbiome by Next-Generation Sequencing (NGS)
4.8. Determination of Metabolic Functions of the Gut Microbiome by PICRUSt2 Pipeline Analysis
4.9. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alemao, C.A.; Budden, K.F.; Gomez, H.M.; Rehman, S.F.; Marshall, J.E.; Shukla, S.D.; Donovan, C.; Forster, S.C.; Yang, I.A.; Keely, S.; et al. Impact of diet and the bacterial microbiome on the mucous barrier and immune disorders. Allergy 2021, 76, 714–734. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Unno, T.; Kang, S.; Park, S. A Korean-Style Balanced Diet Has a Potential connection with Ruminococcaceae enterotype and reduction of metabolic syndrome incidence in Korean adults. Nutrients 2021, 13, 495. [Google Scholar] [CrossRef] [PubMed]
- Daily, J.W.; Park, S. Sarcopenia is a cause and consequence of metabolic dysregulation in aging humans: Effects of gut dysbiosis, glucose dysregulation, diet and lifestyle. Cells 2022, 11, 338. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Joyce, S.A.; Shanahan, F.; Hill, C.; Gahan, C.G. Bacterial bile salt hydrolase in host metabolism: Potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 2014, 5, 669–674. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Ploton, M.; Mazuy, C.; Gheeraert, C.; Dubois, V.; Berthier, A.; Dubois-Chevalier, J.; Maréchal, X.; Bantubungi, K.; Diemer, H.; Cianférani, S.; et al. The nuclear bile acid receptor FXR is a PKA- and FOXA2-sensitive activator of fasting hepatic gluconeogenesis. J. Hepatol. 2018, 69, 1099–1109. [Google Scholar] [CrossRef] [Green Version]
- Cortés, V.A.; Barrera, F.; Nervi, F. Pathophysiological connections between gallstone disease, insulin resistance, and obesity. Obes. Rev. 2020, 21, e12983. [Google Scholar] [CrossRef]
- Sjöberg, B.G.; Straniero, S.; Angelin, B.; Rudling, M. Cholestyramine treatment of healthy humans rapidly induces transient hypertriglyceridemia when treatment is initiated. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E167–E174. [Google Scholar] [CrossRef]
- Ramírez-Pérez, O.; Cruz-Ramón, V.; Chinchilla-López, P.; Méndez-Sánchez, N. The role of the gut microbiota in bile acid metabolism. Ann. Hepatol. 2017, 16, S21–S26. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Druart, C.; Alligier, M.; Salazar, N.; Neyrinck, A.M.; Delzenne, N.M. Modulation of the gut microbiota by nutrients with prebiotic and probiotic properties. Adv. Nutr. 2014, 5, 624S–633S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wu, S.; Tian, Y. Cholecystectomy as a risk factor of metabolic syndrome: From epidemiologic clues to biochemical mechanisms. Lab. Investig. 2018, 98, 7–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.Y.; Portincasa, P.; Liu, M.; Tso, P.; Wang, D.Q.H. Mouse models of gallstone disease. Curr. Opin. Gastroenterol. 2018, 34, 59–70. [Google Scholar] [CrossRef]
- Zheng, X.; Huang, F.; Zhao, A.; Lei, S.; Zhang, Y.; Xie, G.; Chen, T.; Qu, C.; Rajani, C.; Dong, B.; et al. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017, 15, 120. [Google Scholar] [CrossRef] [Green Version]
- Housset, C.; Chrétien, Y.; Debray, D.; Chignard, N. Functions of the gallbladder. Compr. Physiol. 2016, 6, 1549–1577. [Google Scholar]
- Xie, C.; Huang, W.; Young, R.L.; Jones, K.L.; Horowitz, M.; Rayner, C.K.; Wu, T. Role of bile acids in the regulation of food intake, and their dysregulation in metabolic disease. Nutrients 2021, 13, 1104. [Google Scholar] [CrossRef]
- Rehfeld, J.F. Cholecystokinin—From local gut hormone to ubiquitous messenger. Front. Endocrinol. 2017, 8, 47. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G554–G573. [Google Scholar] [CrossRef]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A High-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Jiang, F.; Cheng, R.; Luo, Y.; Wang, J.; Luo, Z.; Li, M.; Shen, X.; He, F. A high-fat diet and high-fat and high-cholesterol diet may affect glucose and lipid metabolism differentially through gut microbiota in mice. Exp. Anim. 2021, 70, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Upadhyay, V.; Turnbaugh, J.A.; Ly, K.; Turnbaugh, P.J. Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe 2019, 26, 265–272.e4. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; An, Y.; Tang, H.; Wang, Y. Alterations of bile acids and gut microbiota in obesity induced by high fat diet in rat model. J. Agric. Food Chem. 2019, 67, 3624–3632. [Google Scholar] [CrossRef]
- Garruti, G.; Wang, D.Q.H.; Di Ciaula, A.; Portincasa, P. Cholecystectomy: A way forward and back to metabolic syndrome? Lab. Investig. 2018, 98, 4–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, W.J.; Kim, H.-N.; Park, E.; Ryu, S.; Chang, Y.; Shin, H.; Kim, H.-L.; Yi, S.Y. The impact of cholecystectomy on the gut microbiota: A case-control study. J. Clin. Med. 2019, 8, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullak-Ublick, G.A.; Paumgartner, G.; Berr, F. Long-term effects of cholecystectomy on bile acid metabolism. Hepatology 1995, 21, 41–45. [Google Scholar] [CrossRef]
- Zhang, B.; Kuipers, F.; de Boer, J.F.; Kuivenhoven, J.A. Modulation of bile acid metabolism to improve plasma lipid and lipoprotein profiles. J. Clin. Med. 2021, 11, 4. [Google Scholar] [CrossRef]
- Li, B.; Zhang, J.; Chen, Y.; Wang, Q.; Yan, L.; Wang, R.; Wei, Y.; You, Z.; Li, Y.; Miao, Q.; et al. Alterations in microbiota and their metabolites are associated with beneficial effects of bile acid sequestrant on icteric primary biliary Cholangitis. Gut Microbes 2021, 13, 1946366. [Google Scholar] [CrossRef]
- Frontoni, S.; Choi, S.B.; Banduch, D.; Rossetti, L. In vivo insulin resistance induced by amylin primarily through inhibition of insulin-stimulated glycogen synthesis in skeletal muscle. Diabetes 1991, 40, 568–573. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Kim, Y.S.; Ryu, S.Y.; Cha, M.R.; Yon, G.H.; Yang, H.J.; Kim, M.J.; Kang, S.; Park, S. Capsiate improves glucose metabolism by improving insulin sensitivity better than capsaicin in diabetic rats. J. Nutr. Biochem. 2013, 24, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Ryuk, J.A.; Ko, B.S.; Moon, N.R.; Park, S. Protection against neurological symptoms by consuming corn silk water extract in artery-occluded gerbils with reducing oxidative stress, inflammation, and post-stroke hyperglycemia through the gut-brain axis. Antioxidants 2022, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Peinnequin, A.; Mouret, C.; Birot, O.; Alonso, A.; Mathieu, J.; Clarençon, D.; Agay, D.; Chancerelle, Y.; Multon, E. Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green. BMC Immunol. 2004, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Hong, S.M.; Lee, J.E.; Sung, S.R. Exercise improves glucose homeostasis that has been impaired by a high-fat diet by potentiating pancreatic beta-cell function and mass through IRS2 in diabetic rats. J. Appl. Physiol. 2007, 103, 1764–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Yuan, H.; Zhang, T.; Wu, X.; Huang, S.K.; Cho, S.M. Long-term silk peptide intake promotes skeletal muscle mass, reduces inflammation, and modulates gut microbiota in middle-aged female rats. Biomed. Pharmacother. 2021, 137, 111415. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, D.S.; Kang, E.S.; Kim, D.B.; Kang, S. Low dose brain estrogen prevents menopausal syndrome while maintaining the diversity of the gut microbiomes in estrogen-deficient rats. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E99–E109. [Google Scholar] [CrossRef]
- Wu, X.; Kim, M.J.; Yang, H.J.; Park, S. Chitosan alleviated menopausal symptoms and modulated the gut microbiota in estrogen-deficient rats. Eur. J. Nutr. 2020, 60, 1907–1919. [Google Scholar] [CrossRef]
- Yang, H.J.; Zhang, T.; Wu, X.G.; Kim, M.J.; Kim, Y.H.; Yang, E.S.; Yoon, Y.S.; Park, S. Aqueous Blackcurrant Extract Improves Insulin Sensitivity and Secretion and Modulates the Gut Microbiome in Non-Obese Type 2 Diabetic Rats. Antioxidants 2021, 10, 756. [Google Scholar] [CrossRef]
- Matchado, M.S.; Lauber, M.; Reitmeier, S.; Kacprowski, T.; Baumbach, J.; Haller, D.; List, M. Network analysis methods for studying microbial communities: A mini review. Comput. Struct. Biotechnol. J. 2021, 19, 2687–2698. [Google Scholar] [CrossRef]


| Sham + LFD (n = 10) | Sham + HFD (n = 10) | Sham + HFD + BAS (n = 10) | GBX + LFD (n = 10) | GBX + HFD (n = 10) | GBX + HFD + BAS (n = 10) | |
|---|---|---|---|---|---|---|
| Final body weight (g) | 36.0 ± 1.86 b | 37.6 ± 2.8 ab | 39.0 ± 2.8 a† | 36.4 ± 1.52 b | 39.6 ± 3.48 a | 39.8 ± 3.64 a*+‡ |
| Body weight gain (g) | 7.1 ± 0.61 c | 9.0 ± 1.1 b | 9.1 ± 0.44 b† | 8.4 ± 0.60 b | 11.8 ± 0.55 a | 12.4 ± 0.80 a*+‡ |
| Food intake (g/day) | 6.09 ± 0.53 a | 4.59 ± 0.59 b | 4.71 ± 0.51 b† | 5.75 ± 0.51 a | 4.89 ± 0.56 b | 5.03 ± 0.66 b*‡ |
| Caloric intake (kcal/day) | 26.7 ± 2.76 | 23.3 ± 3.00 | 23.9 ± 2.59 | 25.2 ± 2.50 | 24.8 ± 2.84 | 25.6 ± 3.35 |
| Food efficiency (g/day) | 1.17 ± 0.44 c | 1.84 ± 0.54 b | 1.85 ± 0.58 b† | 1.33 ± 0.51 c | 1.98 ± 0.69 ab | 2.49 ± 0.64 a*+‡ |
| Visceral fat mass (g) | 1.42 ± 0.18 c | 1.74 ± 0.20 b | 2.26 ± 0.22 a† | 1.67 ± 0.21 b | 2.43 ± 0.58 a | 2.31 ± 0.30 a*+‡ |
| Liver mass (g) | 1.89 ± 0.07 b | 1.63 ± 0.06 c | 1.66 ± 0.07 c† | 2.06 ± 0.17 a | 1.91 ± 0.20 ab | 1.91 ± 0.14 ab*+ |
| Sham + LFD (n = 10) | Sham + HFD (n = 10) | Sham + HFD + BAS (n = 10) | GBX + LFD (n = 10) | GBX + HFD (n = 10) | GBX + HFD + BAS (n = 10) | |
|---|---|---|---|---|---|---|
| Fasting serum glucose (mg/dL) | 93.0 ± 8.8 | 85.9 ± 8.54 | 86.3 ± 6.32 | 82.3 ± 8.78 | 87.9 ± 6.83 | 92.3 ± 8.93 |
| Post-prandial serum glucose (mg/dL) | 168 ± 13.4 b | 157 ± 10.6 b | 161 ± 7.89 b | 160 ± 9.1 b | 168 ± 11.1 b | 182 ± 10.9 a*+‡ |
| Fasting serum insulin (ng/mL) | 1.05 ± 0.21 d | 1.54 ± 0.27 b | 1.51 ± 0.24 b† | 1.33 ± 0.23 c | 1.81 ± 0.27 a | 1.83 ± 0.25 a*+‡ |
| HOMA-IR | 5.21 ± 0.48 c | 7.06 ± 0.85 b | 6.95 ± 1.04 b† | 5.84 ± 0.88 c | 8.66 ± 1.44 a | 9.01 ± 1.30 a*+‡ |
| Total cholesterol (mg/dL) | 195 ± 8.37 c | 224 ± 10.9 a | 206 ± 6.95 b | 162 ± 9.83 d | 189 ± 10.6 c | 193 ± 10.1 c*+‡ |
| HDL (mg/dL) | 52.5 ± 2.98 a | 51.3 ± 2.35 a | 53.6 ± 5.05 a | 40.7 ± 5.34 b | 43.7 ± 4.10 b | 41.7 ± 5.12 b*+ |
| LDL (mg/dL) | 115 ± 7.35 c | 150 ± 8.68 a | 110 ± 6.51 c† | 95.4 ± 5.47 d | 110 ± 8.47 c | 128 ± 10.5 b*+‡ |
| Triglyceride (mg/dL) | 138 ± 11.8 b | 118 ± 9.75 d | 111 ± 8.81 d† | 135 ± 11.9 b | 176 ± 15.9 a | 128 ± 9.47 c*+‡ |
| Sham + LFD (n = 10) | Sham + HFD (n = 10) | Sham + HFD + BAS (n = 10) | GBX + LFD (n = 10) | GBX + HFD (n = 10) | GBX + HFD + BAS (n = 10) | |
|---|---|---|---|---|---|---|
| Hepatic glycogen (mg/g tissue) | 88.8 ± 10.4 a | 69.0 ± 6.92 b | 71.3 ± 10.4 b† | 69.3 ± 4.81 b | 61.2 ± 6.69 c | 59.5 ± 6.92 c*+‡ |
| Hepatic triglyceride (mg/g tissue) | 195 ± 14.1 c | 205 ± 17.3 c | 265 ± 18.3 a† | 186 ± 18.9 c | 226 ± 20.2 b | 245 ± 21.7 ab*+‡ |
| Hepatic TNF-α mRNA (AU) | 0.88 ± 0.11 d | 1 ± 0 c | 0.97 ± 1.06 cd† | 1.05 ± 0.17 c | 1.24 ± 0.19 b | 1.49 ± 0.22 a*+‡ |
| Hepatic SREBP1c mRNA (AU) | 1.04 ± 0.16 c | 1 ± 0 c | 1.34 ± 0.22 b† | 1.27 ± 0.17 b | 1.37 ± 0.19 b | 1.78 ± 0.27 a*+‡ |
| Hepatic PPAR-γ mRNA (AU) | 0.67 ± 0.16 d | 1 ± 0 c | 1.03 ± 0.15 c† | 0.74 ± 0.26 d | 1.43 ± 0.32 b | 1.89 ± 0.33 a*+‡ |
| Hepatic lipid peroxides (nmol/g tissue) | 21.5 ± 1.97 d | 28.6 ± 2.35 c | 29.1 ± 3.07 c† | 33.8 ± 3.42 b | 38.9 ± 4.05 a | 39.6 ± 3.87 a*+‡ |
| Serum ALT (U/L) | 29.8 ± 3.14 c | 29.7 ± 3.23 c | 28.5 ± 2.94 c | 34.2 ± 3.18 b | 39.4 ± 3.51 a | 38.8 ± 3.12 a*+‡ |
| Serum AST (U/L) | 34.2 ± 3.03 a | 31.4 ± 0.92 b | 30.5 ± 3.04 b | 33.2 ± 3.23 ab | 32.9 ± 1.89 ab | 33.1 ± 3.20 ab* |
| Sham + LFD (n = 10) | Sham + HFD (n = 10) | Sham + HFD + BAS (n = 10) | GBX + HCD (n = 10) | GBX + HFD (n = 10) | GBX + HFD + BAS (n = 10) | |
|---|---|---|---|---|---|---|
| Akkermansia | 21.02 ± 2.59 a | 14.68 ± 2.12 ab | 18.16 ± 4.29 a† | 19.52 ± 3.26 a | 10.85 ± 3.03 b | 8.99 ± 1.91 b*+‡ |
| Lachnospiraceae_unclassified | 24.58 ± 2.87 a | 20.69 ± 2.93 ab | 21.00 ± 1.66 ab | 17.34 ± 1.48 b | 17.9 ± 2.13 b | 17.46 ± 1.88 b* |
| Bacteroides | 6.18 ± 1.64 c | 9.50 ± 1.58 b | 12.73 ± 2.15 ab† | 12.55 ± 2.90 ab | 11.4 ± 3.14 ab | 15.34 ± 2.30 a*+‡ |
| Lactobacillus | 1.48 ± 0.57 b | 0.58 ± 0.13 bc | 0.12 ± 0.06 c† | 10.29 ± 2.26 a | 7.47 ± 3.56 a | 1.04 ± 0.60 b*+‡ |
| Oscillospira | 7.87 ± 1.22 b | 7.58 ± 0.62 b | 11.25 ± 0.68 a† | 8.14 ± 2.69 ab | 6.89 ± 1.37 b | 7.77 ± 0.83 b+ |
| Desulfovibrio | 9.16 ± 1.75 | 9.03 ± 1.26 | 6.50 ± 1.07 | 7.72 ± 1.21 | 9.29 ± 1.34 | 7.07 ± 0.70 |
| Bifidobacterium | 7.25 ± 4.03 a | 2.50 ± 0.93 b | 1.80 ± 0.32 b† | 3.75 ± 2.06 b | 6.57 ± 2.63 a | 5.15 ± 2.88 ab*+‡ |
| Parabacteroides | 6.74 ± 4.01 a | 2.69 ± 0.40 b | 6.96 ± 3.48 a† | 2.53 ± 0.33 b | 2.68 ± 0.85 b | 2.28 ± 0.44 b* |
| Ruminococcus | 1.36 ± 0.20 | 0.85 ± 0.13 | 0.96 ± 0.07 | 1.55 ± 0.49 | 1.71 ± 0.36 | 1.034 ± 0.17 |
| Clostridium | 1.88 ± 0.32 b | 2.41 ± 0.25 ab | 3.42 ± 0.59 a† | 1.13 ± 0.09 b | 1.78 ± 0.43 b | 4.26 ± 0.71 a+ |
| Anaerotruncus | 2.86 ± 0.76 a | 1.26 ± 0.26 ab | 0.88 ± 0.19 b† | 1.04 ± 0.20 b | 0.96 ± 0.21 b | 1.32 ± 0.26 ab* |
| Alistipes | 0.04 ± 0.01 b | 0.07 ± 0.02 b | 0.11 ± 0.03 ab | 0.13 ± 0.02 b | 0.07 ± 0.02 b | 0.26 ± 0.07 a*+‡ |
| Dorea | 0.06 ± 0.03 b | 0.12 ± 0.03 ab | 0.05 ± 0.01 b | 0.11 ± 0.03 ab | 0.05 ± 0.01 b | 0.50 ± 0.13 a*‡ |
| Blautia | 0.001 ± 0.001 b | 0.06 ± 0.03 a | 0.04 ± 0.03 a† | 0 ± 0 b | 0 ± 0 b | 0.43 ± 0.11 a+‡ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Zhang, T.; Yue, Y.; Wu, X. Effects of Bile Acid Modulation by Dietary Fat, Cholecystectomy, and Bile Acid Sequestrant on Energy, Glucose, and Lipid Metabolism and Gut Microbiota in Mice. Int. J. Mol. Sci. 2022, 23, 5935. https://doi.org/10.3390/ijms23115935
Park S, Zhang T, Yue Y, Wu X. Effects of Bile Acid Modulation by Dietary Fat, Cholecystectomy, and Bile Acid Sequestrant on Energy, Glucose, and Lipid Metabolism and Gut Microbiota in Mice. International Journal of Molecular Sciences. 2022; 23(11):5935. https://doi.org/10.3390/ijms23115935
Chicago/Turabian StylePark, Sunmin, Ting Zhang, Yu Yue, and Xuangao Wu. 2022. "Effects of Bile Acid Modulation by Dietary Fat, Cholecystectomy, and Bile Acid Sequestrant on Energy, Glucose, and Lipid Metabolism and Gut Microbiota in Mice" International Journal of Molecular Sciences 23, no. 11: 5935. https://doi.org/10.3390/ijms23115935
APA StylePark, S., Zhang, T., Yue, Y., & Wu, X. (2022). Effects of Bile Acid Modulation by Dietary Fat, Cholecystectomy, and Bile Acid Sequestrant on Energy, Glucose, and Lipid Metabolism and Gut Microbiota in Mice. International Journal of Molecular Sciences, 23(11), 5935. https://doi.org/10.3390/ijms23115935








