Effect of Separate and Combined Toxicity of Bisphenol A and Zinc on the Soil Microbiome
Abstract
:1. Introduction
2. Results
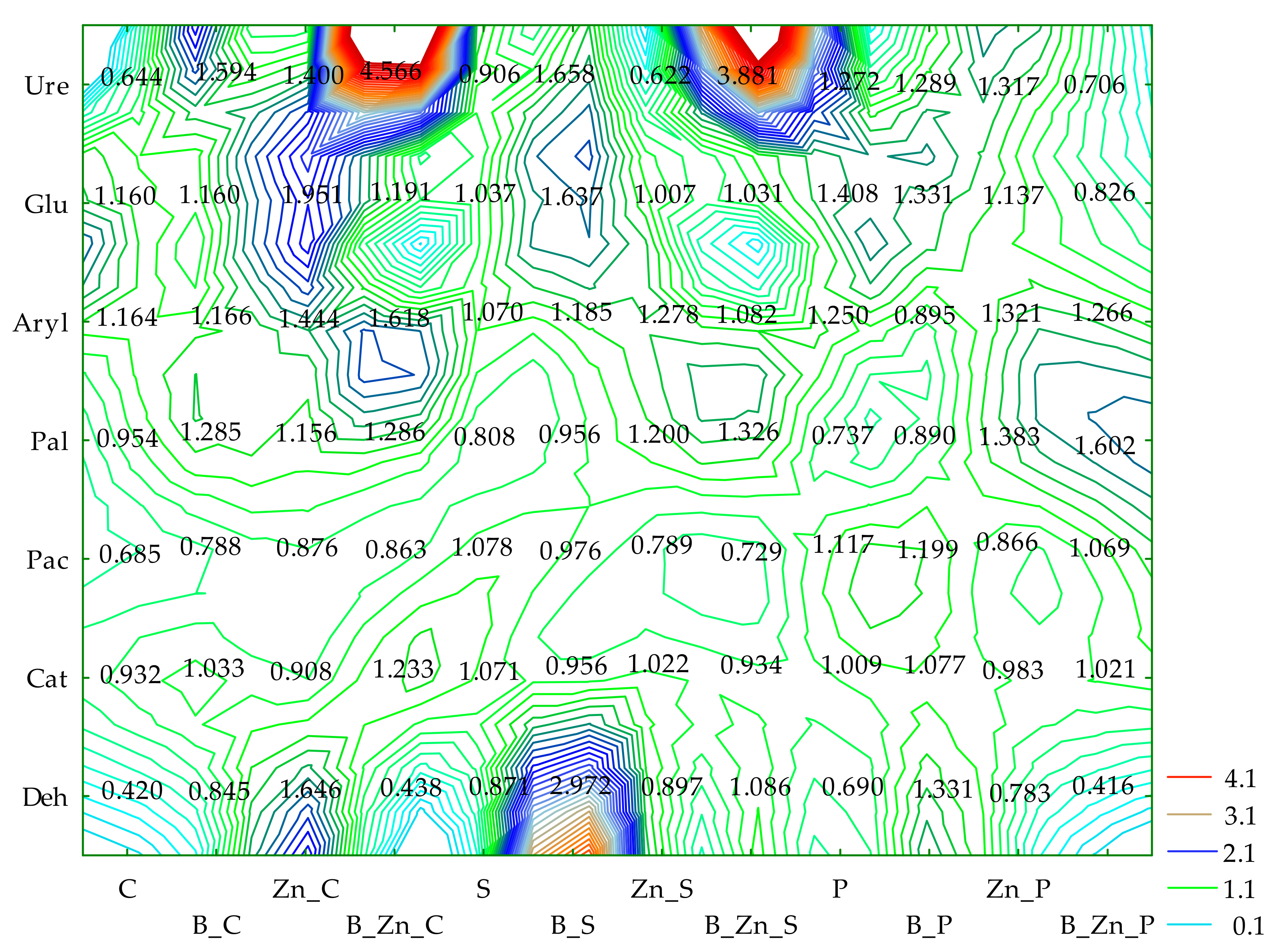
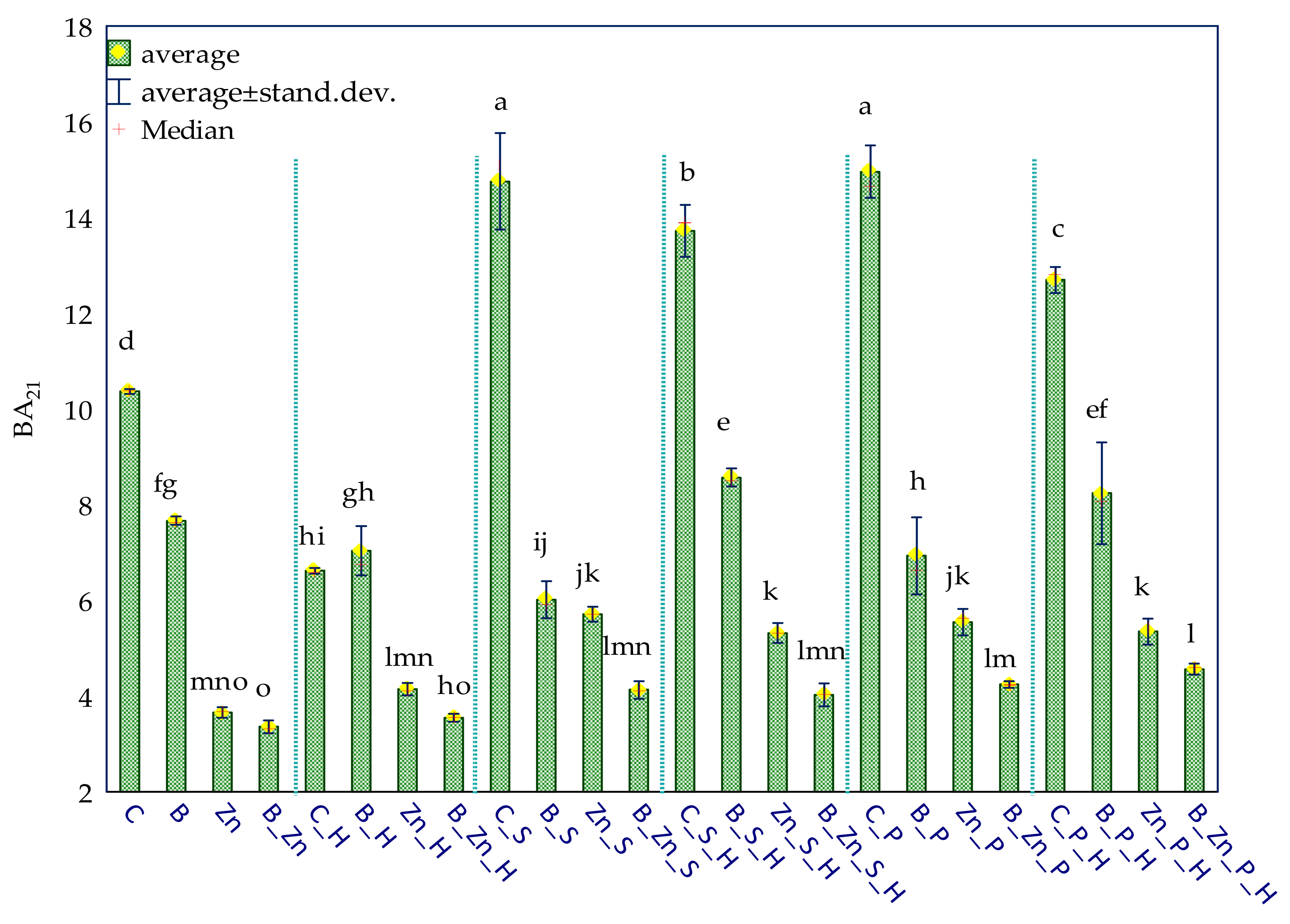
2.1. Enzyme Activity
2.2. Response of Microorganisms to Soil Contamination with Bisphenol A and Zinc
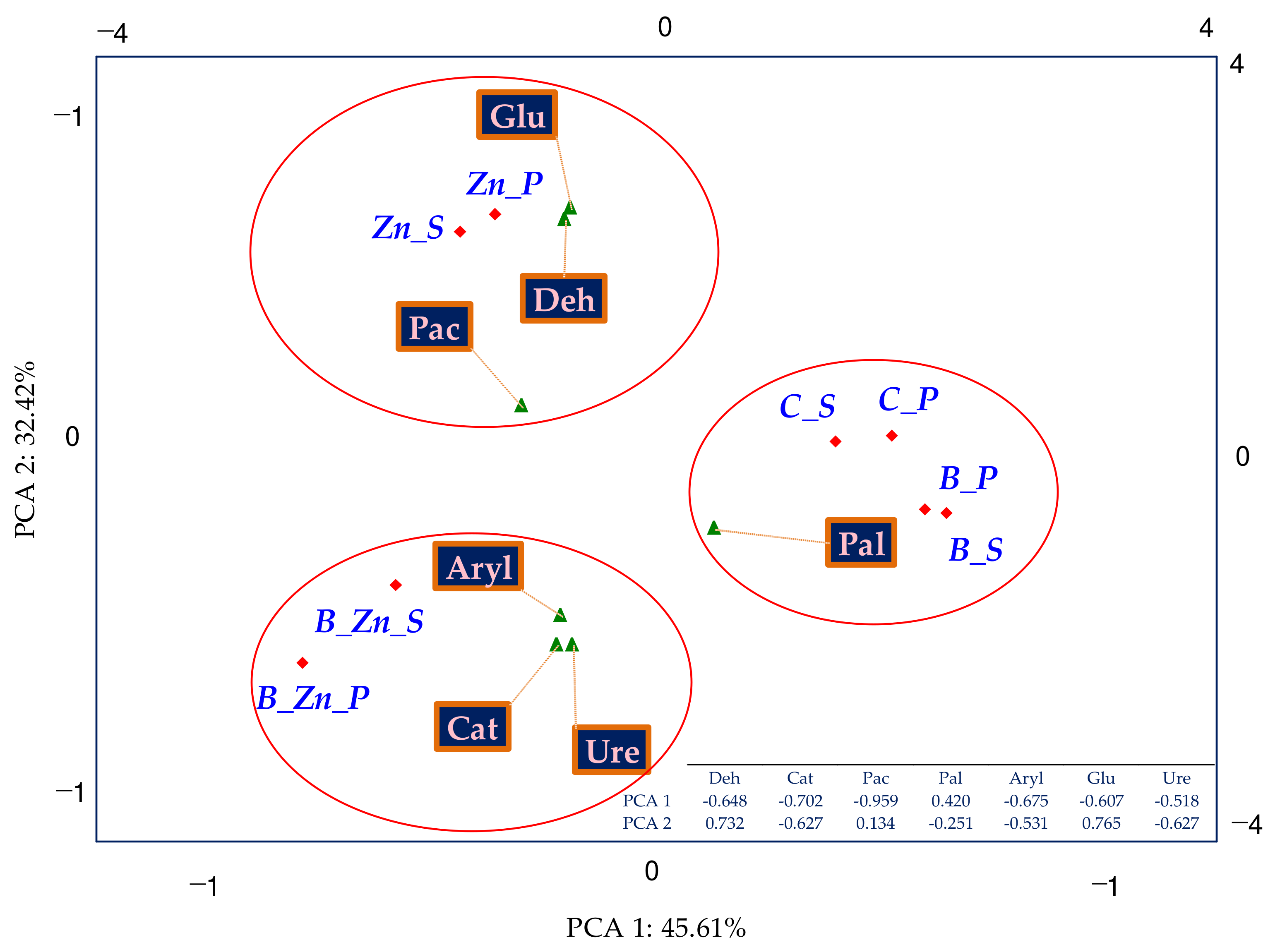
2.2.1. Culturable Microorganisms
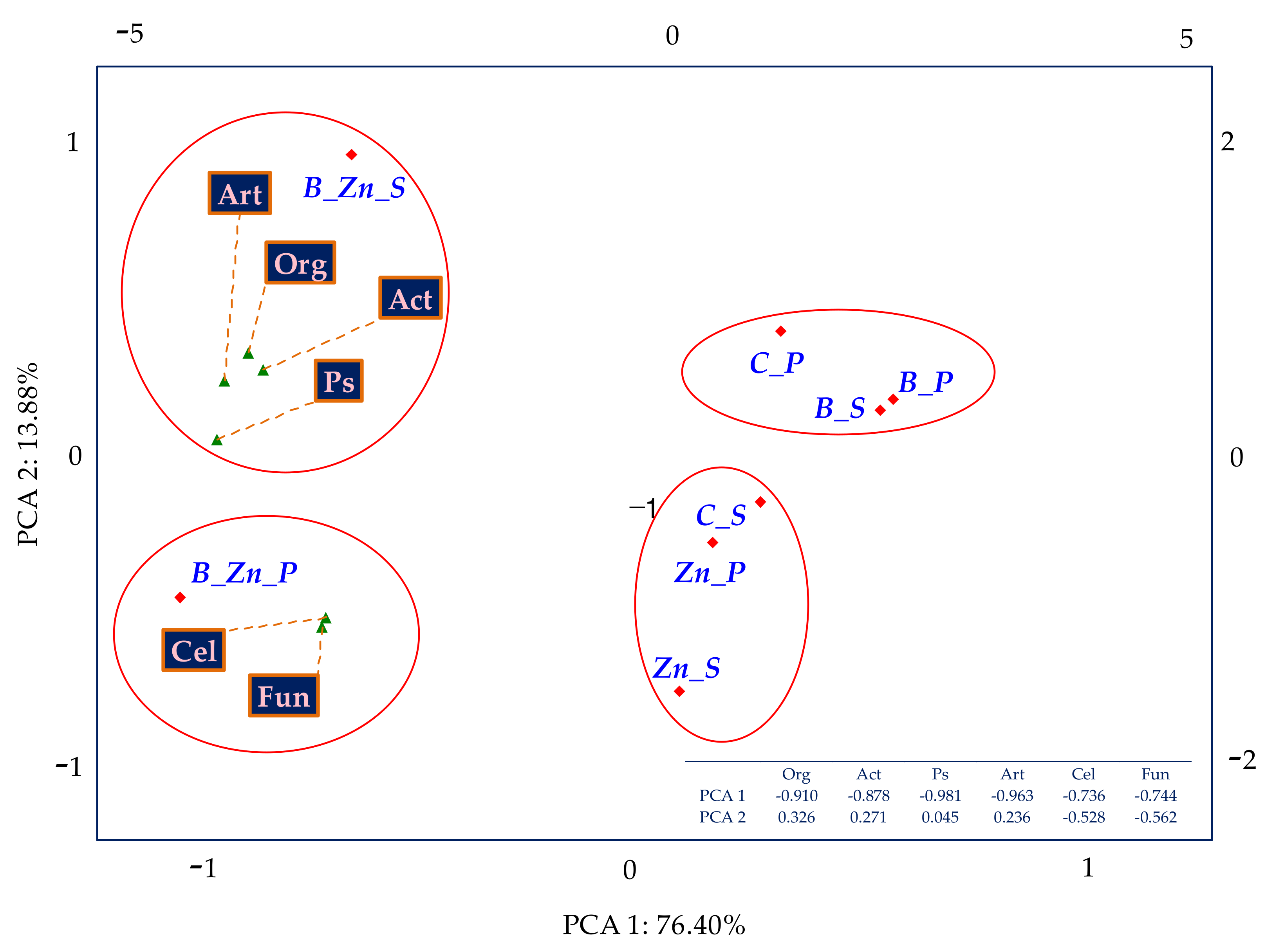
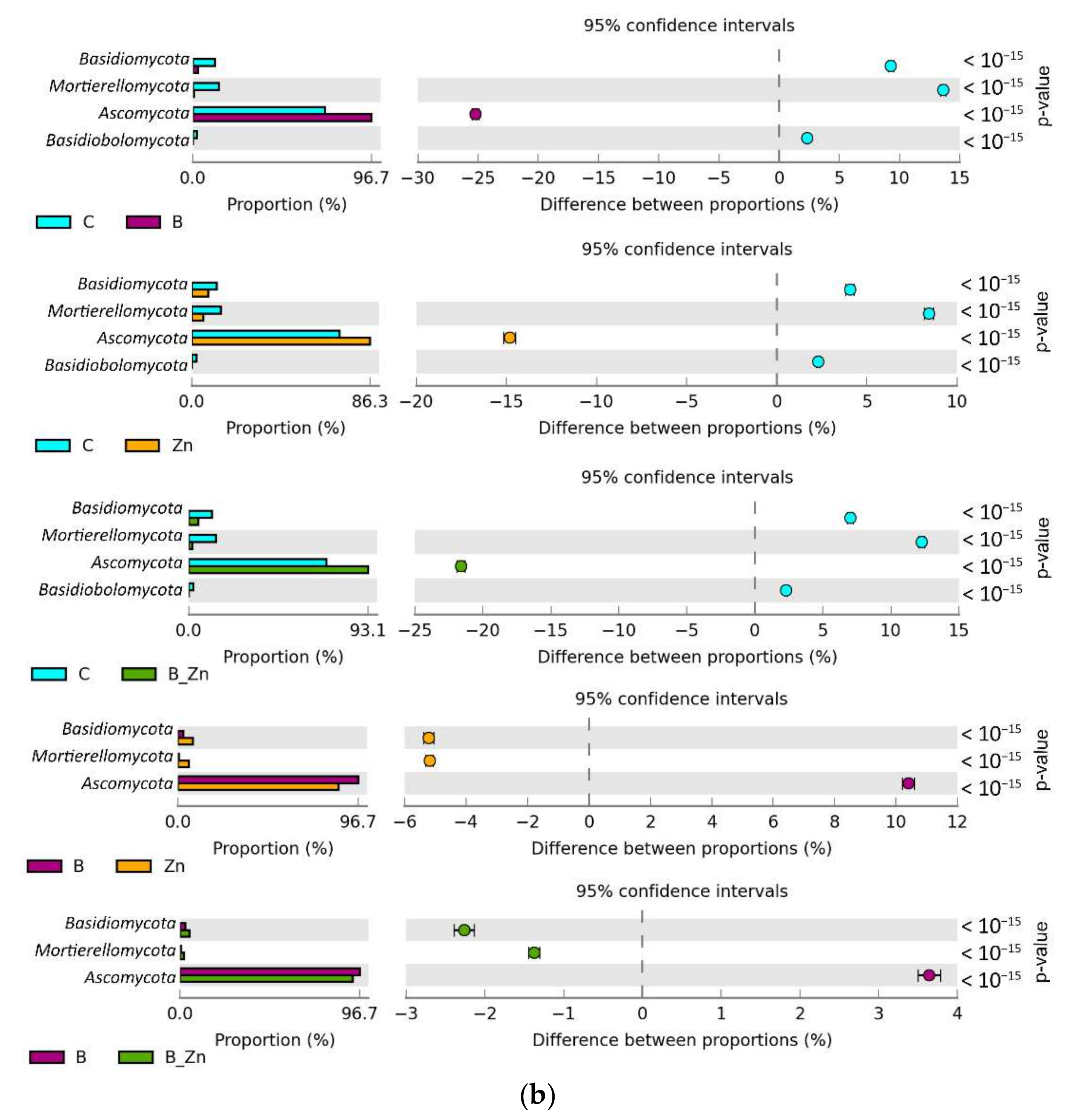
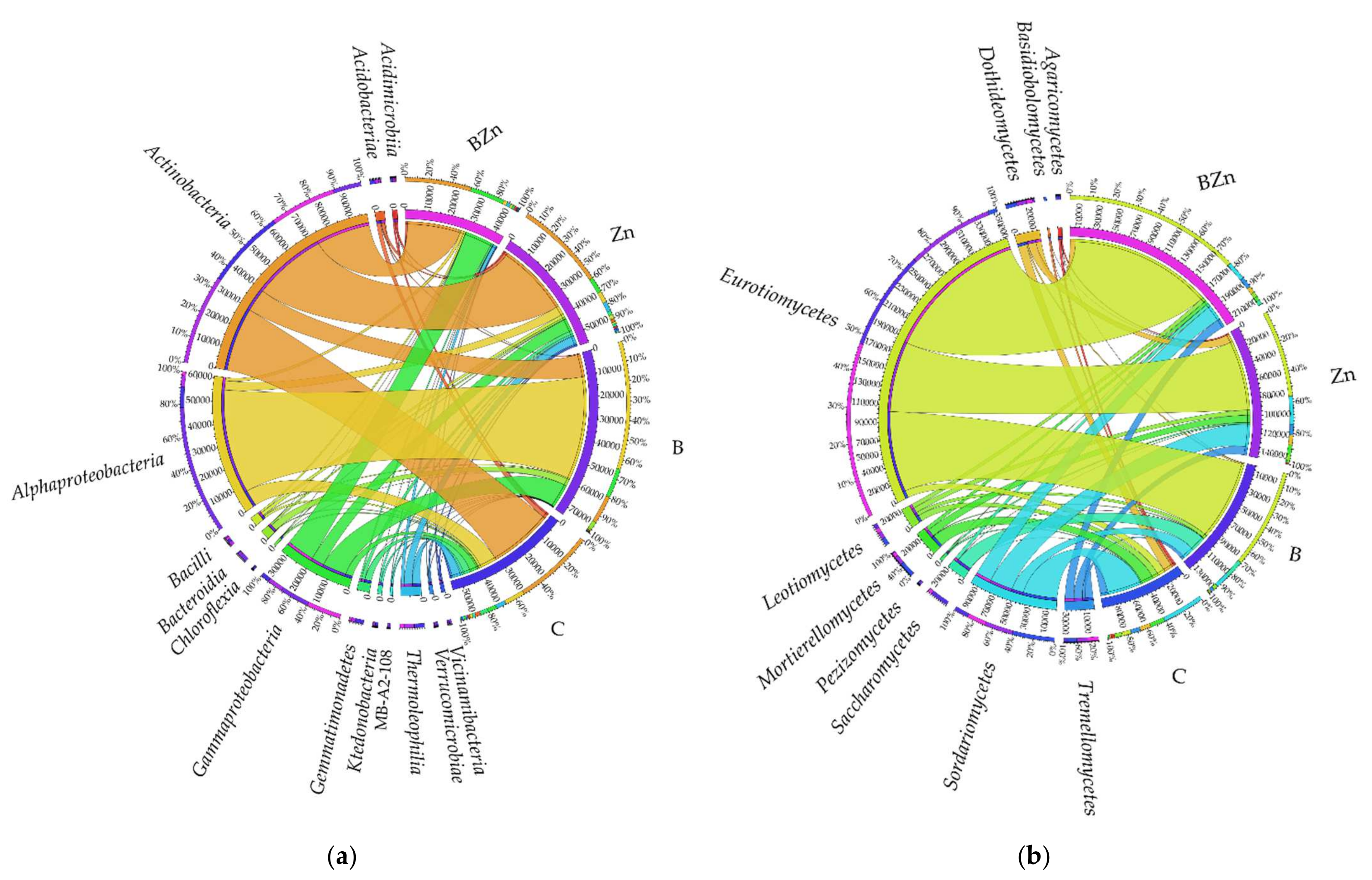
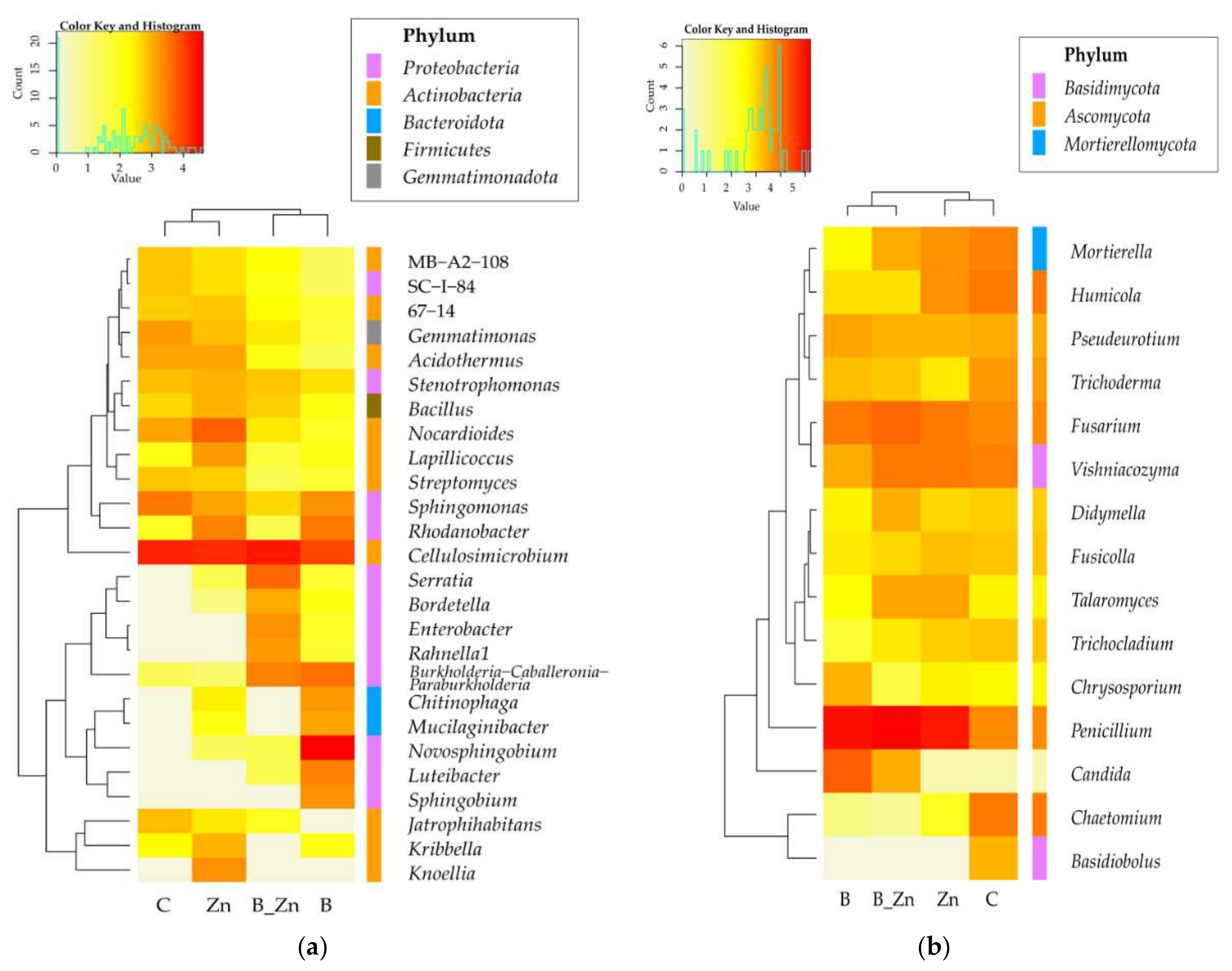
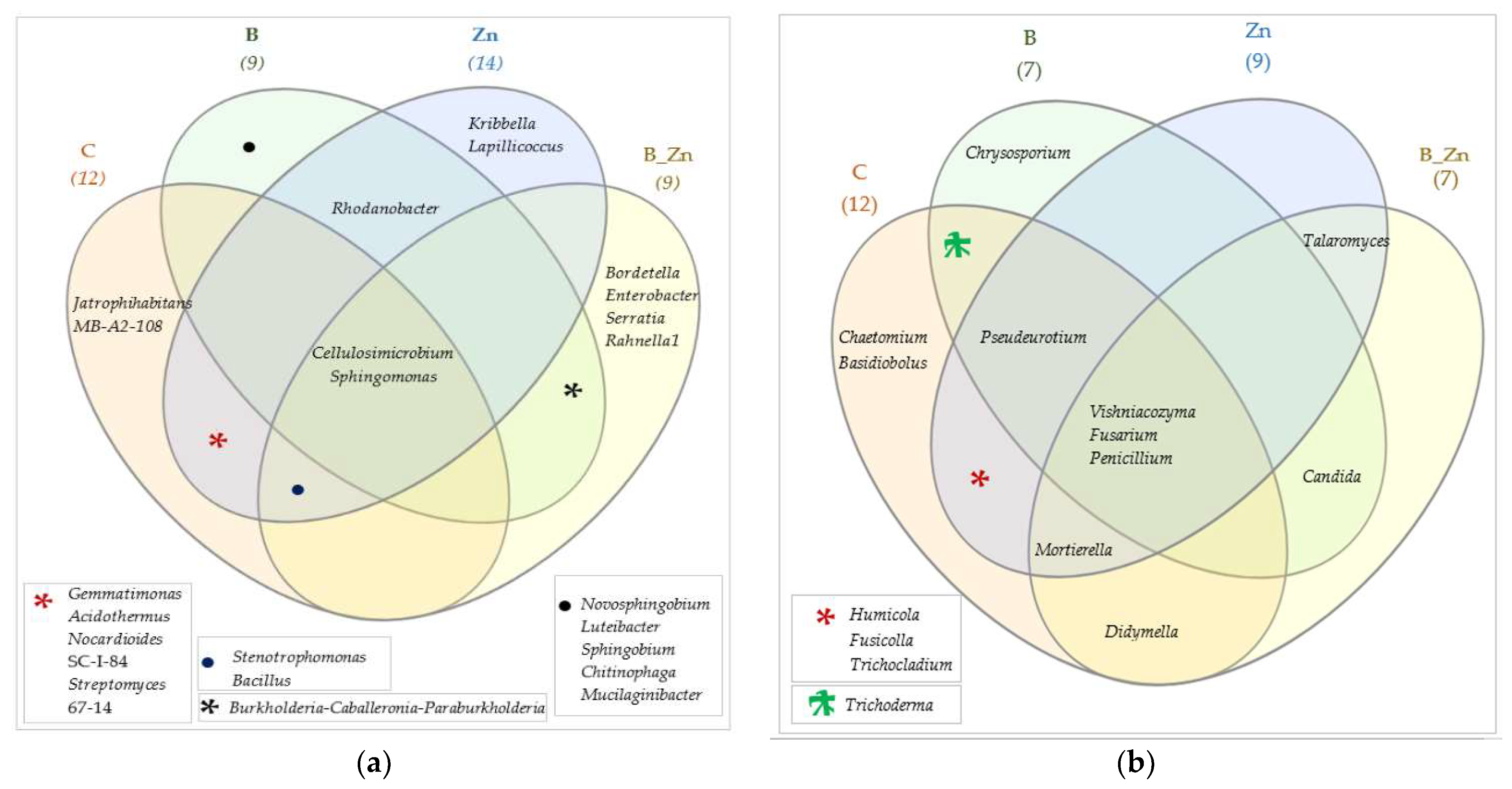
2.2.2. Microorganisms Identified by the NGS Method
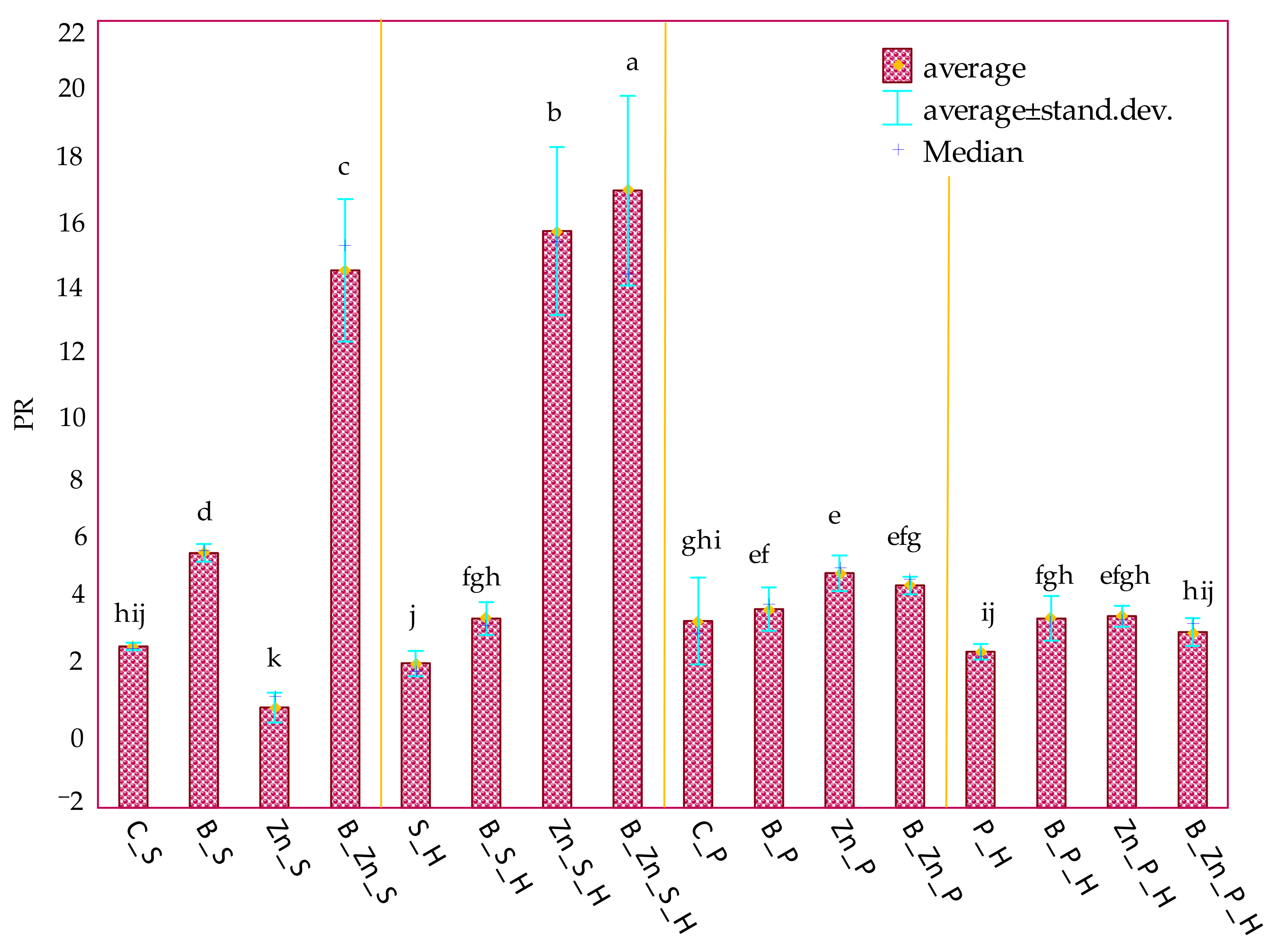
2.3. Response of Plants to Soil Contamination with Bisphenol A and Zinc
3. Discussion
3.1. Soil Enzymes
3.2. Number and Variety of Bacteria and Fungi
3.3. The Response of Sorghum and Millet to B, B + Zn2+ and Zn2+ Pressure
4. Materials and Methods
4.1. Characteristics of the Soil
4.2. Design of the Experiment
4.3. Plants
4.4. Physicochemical Analyses
4.5. Biochemical Analyzes
4.6. Microbiological Analyses
4.6.1. Determination of Counts of Soil Microorganisms
4.6.2. DNA Isolation
4.6.3. Metagenomic Analysis of Bacterial and Fungal Taxa
4.6.4. Statistical Analysis of Data and Methodology of Calculations
- IFx—The coefficient of the impact of soil contaminated with bisphenol A (B), Zn2+, B + Zn2+,
- IFH—coefficient of soil biostimulation with humic acid,
- IFP—plant influence coefficient,
- IFx_H_P < 1—inhibition of the activity of individual enzymes and the number of microorganisms; >1—stimulation of the activity of individual enzymes and the number of microorganisms,
- Ax—activity of individual enzymes and groups of microorganisms in soil contaminated with bisphenol A (B); Zn2+; B + Zn2+,
- AC—activity of individual enzymes and the number of microorganisms in the control soil (uncontaminated soil).
- TI—tolerance index of plants (aerial part and roots for soil contaminated with bisphenol A (B), Zn2+, B + Zn2+ (TI < 100—inhibitory effect of xenobiotics; TI > 100—stimulating effect of xenobiotics),
- Yp—yield of aerial parts and roots of plants in soil contaminated with xenobiotics,
- YC—yield of aerial parts and roots of plants in the control soil, uncontaminated with xenobiotics.
- Deh—dehydrogenase, Cat—catalase, Pal—alkaline phosphatase, Pac—acid phosphatase, Ure—urease, Glu—β-glucosidase, and Aryl—arylsulfatase.
- N1, N2, N3… N10—sum of ratios of the colony numbers identified on each day (1, 2, 3, … 10) and the sum of all the colonies identified during the entire experiment
- pi—the number of microbial colonies on a given day divided by the number of all colonies.
- PR—ratio of the mass of the aerial parts to the mass of the roots of plants, P—dry matter yield of aerial parts, and R—dry matter yield of roots.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Corrales, J.; Kristofco, L.A.; Steele, W.B.; Yates, B.S.; Breed, C.S.; Williams, E.S.; Brooks, B.W. Global assessment of bisphenol A in the environment: Review and analysis of its occurrence and bioaccumulation. Dose-Response 2015, 13, 1559325815598308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, S.A. The politics of plastics: The making and unmaking of Bisphenol a “safety”. Am. J. Public Health 2009, 99, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Environment and Climate Change Canada Environmental Monitoring and Surveillance in Support of the Chemicals Management Plan. 2020. Available online: https://www.canada.ca/en/environment-climate-change/services/evaluating-existing-substances/environmental-monitoring-surveillance-support-chemicals-management-plan-bisphenol-a-canadian-environment.html (accessed on 8 April 2022).
- Research and Markets. ComGlobal Bisphenol A Market Report 2018: Analysis 2013–2017 & Forecasts 2018–2023 [WWW Document]. 2018. Available online: https://www.prnewswire.com/news-releases/global-bisphenol-a-market-report-2018-analysis-2013-2017--forecasts-2018-2023-300757673.html (accessed on 8 April 2022).
- Li, J.; Wang, G. Airborne particulate endocrine disrupting compounds in China: Compositions, size distributions and seasonal variations of phthalate esters and bisphenol A. Atmos. Res. 2015, 154, 138–145. [Google Scholar] [CrossRef]
- Hahladakis, J.N.; Velis, C.A.; Weber, R.; Iacovidou, E.; Purnell, P. An overview of chemical additives present in plastics: Migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard. Mater. 2018, 344, 179–199. [Google Scholar] [CrossRef] [PubMed]
- Geens, T.; Aerts, D.; Berthot, C.; Bourguignon, J.P.; Goeyens, L.; Lecomte, P.; Maghuin, R.G.; Pironnet, A.M.; Pussemier, L.; Scippo, M.L.; et al. A review of dietary and non-dietary exposure to bisphenol-A. Food Chem. Toxicol. 2012, 50, 3725–3740. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.A.; Kjeldal, H.; Gough, H.L.; Nielsen, J.L. Identification of putative genes involved in bisphenol a degradation using differential protein abundance analysis of Sphingobium sp. BiD32. Environ. Sci. Technol. 2015, 49, 12232–12241. [Google Scholar] [CrossRef]
- ECHA. European Chemicals Agency. 2020. Available online: https://echa.europa.eu/substanceinformation/-/substanceinfo/100.001.133 (accessed on 20 February 2022).
- Arp, H.P.H.; Morin, N.A.O.; Hale, S.E.; Okkenhaug, G.; Breivik, K.; Sparrevik, M. The mass flow and proposed management of bisphenol A in selected Norwegian waste streams. Waste Manag. 2017, 60, 775–785. [Google Scholar] [CrossRef]
- Teeguarden, J.G.; Twaddle, N.C.; Churchwell, M.I.; Yang, X.; Fisher, J.W.; Seryak, L.M.; Doerge, D.R. 24-hour human urine and serum profiles of bisphenol A following ingestion in soup: Individual pharmacokinetic data and emographics. Data Brief 2015, 4, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Stein, T.P.; Schluter, M.D.; Steer, R.A.; Guo, L.; Ming, X. Bisphenol A exposure in children with autism spectrum disorders. Autism Res. 2015, 8, 272–283. [Google Scholar] [CrossRef] [Green Version]
- Kahn, L.G.; Philippat, C.; Nakayama, S.F.; Slama, R.; Trasande, L. Endocrine disrupting chemicals: Implications for human health. Lancet Diabetes Endocrinol. 2020, 8, 703–718. [Google Scholar] [CrossRef]
- Staples, C.; van der Hoeven, N.; Clark, K.; Mihaich, E.; Woelz, J.; Hentges, S. Distributions of concentrations of bisphenol A in North American and European surface waters and sediments determined from 19 years of monitoring data. Chemosphere 2018, 201, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Langdon, K.A.; Warne, M.S.J.; Smernik, R.J.; Shareef, A.; Kookana, R.S. Field dissipation of 4-nonylphenol, 4-t-octylphenol, triclosan and bisphenol A following land application of biosolids. Chemosphere 2012, 86, 1050–1105. [Google Scholar] [CrossRef] [PubMed]
- Diagboya, P.N.; Olu-Owolabi, B.I.; Adebowale, K.O. Distribution and interactions of pentachlorophenol in soils: The roles of soil iron oxides and organic matter. J. Contam. Hydrol. 2016, 191, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehr, M.R.; Shakeri, A.; Amjadian, K.; Poshtegal, M.K.; Sharifi, R. Bioavailability, distribution and health risk assessment of arsenic and heavy metals (HMs) in agricultural soils of Kermanshah Province, west of Iran. J. Environ. Health Sci. Eng. 2021, 19, 107–120. [Google Scholar] [CrossRef]
- Milieu Ltd., RPA, WRc, Environmental, Economic and Social Impacts of the Use of Sewage Sludge on Land—Consultation Report on Options and Impacts. 2010. pp. 1–32. Available online: https://ec.europa.eu/environment/archives/waste/sludge/pdf/part_i_report.pdf (accessed on 8 April 2022).
- Liu, L.; Li, W.; Song, W.; Guo, M. Remediation techniques for heavy metalcontaminated soils: Principles and applicability. Sci. Total Environ. 2018, 633, 206–219. [Google Scholar] [CrossRef]
- Uchimiya, M.; Bannon, D.; Nakanishi, H.; McBride, M.B.; Williams, M.A.; Yoshihara, T. Chemical speciation, plant uptake, and toxicity of heavy metals in agricultural soils. J. Agric. Food Chem. 2020, 68, 12856–12869. [Google Scholar] [CrossRef]
- Ahmed, T.; Noman, M.; Ijaz, M.; Ali, S.; Rizwan, M.; Ijaz, U.; Hameed, A.; Ahmad, U.; Wang, Y.; Sun, G.; et al. Current trends and future prospective in nanoremediation of heavy metals contaminated soils: A way forward towards sustainable agriculture. Ecotoxicol. Environ. Saf. 2021, 227, 112888. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.Z.; Zhang, I. Status and associated human health risk of zinc accumulation in agricultural soils across China. Process Saf. Environ. Prot. 2021, 146, 867–876. [Google Scholar] [CrossRef]
- Natasha, N.; Shahid, M.; Bibi, I.; Iqbal, J.; Sana, K.; Murtaza, B.; Bakhat, H.F.; Farooq, A.B.U.; Hafiz, M.A.; Mohkum, H.N.; et al. Zinc in soil-plant-human system: A data-analysis review. Sci. Total Environ. 2022, 808, 152024. [Google Scholar] [CrossRef]
- Nriagu, J. Zinc Toxicity in Humans. Reference Module in Earth Systems and Environmental Sciences. In Encyclopedia of Environmental Health, 2nd ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 500–508. [Google Scholar] [CrossRef]
- Menz, J.; Müller, J.; Olsson, O.; Kümmerer, K. Bioavailability of antibiotics at soil–water interfaces: A comparison of measured activities and equilibrium partitioning estimates. Environ. Sci. Technol. 2018, 52, 6555–6564. [Google Scholar] [CrossRef]
- Thomas, J.C.; Oladeinde, A.; Kieran, T.J.; Finger, J.W.; Bayona-Vasquez, N.J.; Cartee, J.C.; Beasley, J.C.; Seaman, J.C.; McArthur, J.V.; Rhodes, O.E.; et al. Cooccurrence of antibiotic, biocide, and heavy metal resistance genes in bacteria from metal and radionuclide contaminated soils at the Savannah River Site. Microb. Biotechnol. 2020, 13, 1179–1200. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, B.; Yu, G. Antibiotic resistance genes in China: Occurrence, risk, and correlation among different parameters. Environ. Sci. Pollut. Res. 2018, 25, 21467–21482. [Google Scholar] [CrossRef] [PubMed]
- Wyszkowska, J.; Boros-Lajszner, E.; Borowik, A.; Kucharski, J.; Baćmaga, M.; Tomkiel, M. Changes in the microbiological and biochemical properties of soil contaminated with zinc. J. Elementol. 2017, 22, 437–451. [Google Scholar] [CrossRef]
- Staples, C.A.; Dom, P.B.; Klecka, G.M.; Sandra, T.O.; Harris, L.R. A review of the environmental fate, effects, and exposures of bisphenol A. Chemosphere 1998, 36, 2149–2173. [Google Scholar] [CrossRef]
- Cousins, I.T.; Staples, C.A.; Klecka, G.M.; Mackay, D. A multimedia assessment of the environmental fate of bisphenol A. Hum. Ecol. Risk Assess. Int. J. 2002, 8, 1107–1135. [Google Scholar] [CrossRef]
- Rijkswaterstaat Institute. Chemical study on Bisphenol A [WWW Document]. 2001. Available online: https://edepot.wur.nl/174301 (accessed on 8 April 2022).
- Zhao, J.; Zhou, D.; Zhang, C.; Li, F.; Chu, G.; Wu, M.; Pan, B.; Steinberg, C.E.W. The contrasting role of minerals in biochars in bisphenol A and sulfamethoxazole sorption. Chemosphere 2021, 264, 128490. [Google Scholar] [CrossRef]
- Setlhare, B.; Kumar, A.; Mokoena, M.P.; Pillay, B.; Olaniran, A.O. Phenol hydroxylase from Pseudomonas sp. KZNSA: Purification, characterization and prediction of three-dimensional structure. Int. J. Biol. Macromol. 2020, 146, 1000–1008. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Xu, L.; Zhu, K.; Feng, Y.; Pan, J.; Si, M.; Zhang, L.; Shen, X. Transcriptional control of the phenol hydroxylase gene phe of Corynebacterium glutamicum by the AraC-type regulator PheR. Microbiol. Res. 2018, 209, 14–20. [Google Scholar] [CrossRef]
- Zaborowska, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Bisphenol A—A dangerous pollutant distorting the biological properties of soil. Int. J. Mol. Sci. 2021, 22, 12753. [Google Scholar] [CrossRef]
- Wang, S.; Liu, F.; Wu, W.; Hu, Y.; Liao, R.; Chen, G.; Wang, J.; Li, J. Migration and health risks of nonylphenol and bisphenol A in soil-winter wheat systems with long-term reclaimed water irrigation. Ecotoxicol. Environ. Saf. 2018, 158, 28–36. [Google Scholar] [CrossRef]
- Li, S.W.; Leng, Y.; Feng, L.; Zeng, X.Y. Involvement of abscisic acid in regulating antioxidative defense systems and IAA-oxidase activity and improving adventitious rooting in mung bean [Vigna radiata L. Wilczek] seedlings under cadmium stress. Environ. Sci. Pollut. Res. 2014, 21, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Zhou, Q.; Huang, X. Reactive oxygen species initiate a protective response in plant roots to stress induced by environmental bisphenol A. Ecotoxicol. Environ. Saf. 2018, 154, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Moeskops, B.; Buchan, D.; Sleutel, S.; Herawaty, L.; Husen, E.; Saraswati, R.; Setyorini, D.; De Neve, S. Soil Microbial Communities and Activities Under Intensive Organic and Conventional Vegetable Farming In West Java, Indonesia. Appl. Soil Ecol. 2010, 45, 112120. [Google Scholar] [CrossRef]
- Frankenberger, W.; Johanson, J. Effect of pH On Enzyme Stability in Soils. Soil Biol. Biochem. 1982, 14, 433437. [Google Scholar] [CrossRef]
- Siczek, A.; Frąc, M.; Gryta, A.; Kalembasa, S.; Kalembasa, D. Variation in soil microbial population and enzyme activities under faba bean as affected by pentachlorophenol. Appl. Soil Ecol. 2020, 150, 103466. [Google Scholar] [CrossRef]
- Zaborowska, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S. Int. J. Mol. Sci. 2020, 21, 3529. [Google Scholar] [CrossRef]
- Borisov, V.B.; Verkhovsky, M.I. Oxygen as acceptor. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef]
- Perotti, E.B.R. Impact of hydroquinone used as a redox effector model on potential denitrification, microbial activity and redox condition of a cultivable soil. Rev. Argent. Microbiol. 2015, 47, 212–218. [Google Scholar] [CrossRef] [Green Version]
- Mazzei, L.; Cianci, M.; Musiani., F.; Ciurli, S. Inactivation of urease by 1,4-benzoquinone: Chemistry at the protein surface. Dalton Trans. 2016, 45, 5455–5459. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.M.; Cao, W.Q.; Yu, B.G.; Lang, M.; Chen, X.P.; Zou, C.Q. The responses of soil enzyme activities, microbial biomass and microbial community structure to nine years of varied zinc application rates. Sci. Total Environ. 2020, 737, 140245. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J.; Baćmaga, M.; Boros-Lajszner, E.; Tomkiel, M. Sensitivity of soil enzymes to excessive zinc concentrations. J. Elementol. 2014, 19, 637–648. [Google Scholar] [CrossRef]
- Kucharski, J.; Wieczorek, K.; Wyszkowska, J. Changes in the enzymatic activity in sandy loam soil exposed to zinc pressure. J. Elementol. 2011, 16, 577–589. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Boros-Lajszner, E.; Borowik, A.; Baćmaga, M.; Kucharski, J.; Tomkiel, M. Implication of zinc excess on soil health. J. Environ. Sci. Health B 2016, 51, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Strachel, R.; Wyszkowska, J.; Baćmaga, M. Bioaugmentation of Soil Contaminated with Zinc. Water Air Soil Pollut. 2020, 231, 443. [Google Scholar] [CrossRef]
- Iyer, S.; Holloway, D.E.; Acharya, K.R. Crystal structures of murine angiogenins-2 and-3-probing ‘‘structure–function’’ relationships amongst angiogenin. FEBS J. 2013, 280, 302–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spivack, J.; Leib, T.K.; Lobos, J.H. Novel pathway for bacterial metabolism of bisphenol A. Rearrangements and stilbene cleavage in bisphenol A metabolism. J. Biol. Chem. 1994, 269, 7323–7329. [Google Scholar] [CrossRef]
- Kolvenbach, B.A.; Helbling, D.E.; Kohler, H.E.; Corvini, P.F. Science Direct Emerging chemicals and the evolution of biodegradation capacities and pathways in bacteria. Curr. Opin. Biotechnol. 2014, 27, 8–14. [Google Scholar] [CrossRef]
- Xiong, J.; Koopal, L.K.; Tan, W.F.; Fang, L.C.; Wang, M.X.; Zhao, W.; Liu, F.; Zhang, J.; Weng, L.P. Lead binding to soil fulvic and humic acids: Nica-donnan modeling and xafs spectroscopy. Environ. Sci. Technol. 2013, 47, 11634–11642. [Google Scholar] [CrossRef]
- Gondar, D.; Lopez, R.; Fiol, S.; Antelo, J.M.; Arce, F. Cadmium, lead, and copper binding to humic acid and fulvic acid extracted from an ombrotrophic peat bog. Geoderma 2006, 135, 196–203. [Google Scholar] [CrossRef]
- Garcia-Mina, J.M. Stability, solubility and maximum metal binding capacity in metal–humic complexesinvolving humic substances extracted from peat and organic compost. Org. Geochem. 2006, 37, 1960–1972. [Google Scholar] [CrossRef]
- Irving, H.; Williams, R.J.P. Order of stability of metal complexes. Nature 1948, 161, 436–437. [Google Scholar] [CrossRef]
- Lin, K.; Liu, W.; Gan, J. Oxidative removal of bisphenol A by manganese dioxide: Efficacy, products, and pathways. Environ. Sci. Technol. 2009, 43, 3860–3864. [Google Scholar] [CrossRef] [PubMed]
- Zwetsloot, M.J.; Ucros, J.M.; Wickings, K.; Wilhelm, R.C.; Sparks, J.; Buckley, D.H.; Bauerle, T.L. Prevalent root-derived phenolics drive shifts in microbial community composition and prime decomposition in forest soil. Soil Biol. Biochem. 2020, 145, 107797. [Google Scholar] [CrossRef]
- Fischer, J.; Kappelmeyer, U.; Kastner, M.; Schauer, F.; Heipieper, H.J. The degradation of bisphenol A by the newly isolated bacterium Cupriavidus basilensis JF1 can be enhanced by biostimulation with phenol. Int. Biodeterior. Biodegrad. 2010, 64, 324–330. [Google Scholar] [CrossRef]
- Gulve, R.M.; Deshmukh, A.M. Antimicrobial activity of the marine Actinomycetes. Int. Multidiscip. Res. J. 2012, 2, 16–22. [Google Scholar]
- Ogata, Y.; Toyama, T.; Yu, N.; Wang, X.; Sei, K.; Ike, M. Occurrence of 4-tert-butylphenol (4-t-BP) biodegradation in an aquatic sample caused by the presence of Spirodela polyrrhiza and isolation of a 4-t-BP-utilizing bacterium. Biodegradation 2013, 24, 191–202. [Google Scholar] [CrossRef]
- Harwood, C.S.; Burchhardt, G.; Herrmann, H.; Fuchs, G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 1999, 22, 439–458. [Google Scholar] [CrossRef]
- Rudolphi, A.; Tschech, A.; Fuchs, G. Anaerobic degradation of cresols by denitrifying bacteria. Arch. Microbiol. 1991, 155, 238–248. [Google Scholar] [CrossRef]
- Zühlke, M.K.; Schlüter, R.; Henning, A.K.; Lipka, M.; Mikolasch, A.; Schumann, P.; Giersberg, M.; Kunze, G. A novel mechanism of conjugate formation of bisphenol A and its analogues by Bacillus amyloliquefaciens: Detoxification and reduction of estrogenicity of bisphenols. Int. Biodeterior. Biodegrad. 2016, 109, 165–173. [Google Scholar] [CrossRef]
- Sarathchandra, S.U.; Burch, G.; Cox, N.R. Growth patterns of bacterial communities in the rhizoplane and rhizosphere of with clover (Trifolium repens L.) and perennial ryegrass (Lolium perenne L.) in long—term pasture. Appl. Soil Ecol. 1997, 6, 293–299. [Google Scholar] [CrossRef]
- Cajthaml, T.; Křesinová, Z.; Svobodová, K.; Möder, M. Biodegradation of endocrine-disrupting compounds and suppression of estrogenic activity by ligninolytic fungi. Chemosphere 2009, 75, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Hugentobler, K.G.; Müller, M. Towards semisynthetic natural compounds with a biaryl axis: Oxidative phenol coupling in Aspergillus niger. Bioorg. Med. Chem. 2018, 26, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, K.; Chandirika, J.U.; Vinothkanna, A.; Yin, H.; Liu, X.; Meng, D. Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment—A review. Ecotoxicol. Environ. Saf. 2021, 226, 112863. [Google Scholar] [CrossRef] [PubMed]
- Bourles, A.; Amir, H.; Gensous, S.; Cussonneau, F.; Medevielle, V.; Juillot, F.; Bazire, A.; Meyer, M.; Burtet-Sarramegna, V.; Cavaloc, Y. Investigating some mechanisms underlying stress metal adaptations of two Burkholderia sensu lato species isolated from New Caledonian ultramafic soils. Eur. J. Soil Biol. 2020, 97, 103166. [Google Scholar] [CrossRef]
- Fashola, M.O.; Ngole-Jeme, V.M.; Babalola, O.O. Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int. J. Environ. Res. Public Health 2016, 13, 1047. [Google Scholar] [CrossRef] [Green Version]
- Sobol, Z.; Schiestl, R.H. Intracellular and extracellular factors influencing Cr(VI) and Cr(III) genotoxicity. Environ. Mol. Mutagenesis 2012, 53, 94–100. [Google Scholar] [CrossRef]
- Zhu, X.; Wu, X.; Yao, J.; Wang, F.; Liu, W.; Luo, Y.; Jiang, X. Toxic effects of binary toxicants of cresol frother and Cu (II) on soil microorganisms. Int. Biodeterior. Biodegrad. 2018, 128, 155–163. [Google Scholar] [CrossRef]
- Moridani, M.Y.; Siraki, A.; Chevaldina, T.; Scobie, H.; Obrie, N.P.J. Quantitative structure toxicity relationship for catechols in isolated rat hepatocytes. Chem. Biol. Interact. 2004, 147, 297–307. [Google Scholar] [CrossRef]
- Heider, J.; Fuchs, G. Microbial Anaerobic Aromatic Metabolism. Anaerobe 1997, 3, 1–22. [Google Scholar] [CrossRef]
- Karthik, C.; Oves, M.; Thangabalu, R.; Sharma, R.; Santhosh, S.B.; Indra Arulselvi, P. Cellulosimicrobium funkei-like enhances the growth of Phaseolus vulgaris by modulating oxidative damage under Chromium(VI) toxicity. J. Adv. Res. 2016, 7, 839–850. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salam, M.; Varma, A. Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electron. J. Biotechnol. 2019, 41, 72–80. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Xue, S.; Wang, G. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, Y.; Huang, H.; Mou, L.; Ru, J.; Zhao, J.; Xiao, S. Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci. Total Environ. 2018, 637, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yin, K.; Chen, L. Bacteria-mediated bisphenol A degradation. Appl. Microbiol. Biotechnol. 2013, 97, 5681–5689. [Google Scholar] [CrossRef]
- Fonseca, F.; Bromley, E.H.C.; Saavedra, M.J.; Correia, A.; Spencer, J. Crystal Structure of Serratia fonticola Sfh-I: Activation of the Nucleophile in Mono-Zinc Metallo-β-Lactamases. J. Mol. Biol. 2011, 411, 951–959. [Google Scholar] [CrossRef]
- Al-Zaban, M.I.; Al-Harbi, M.A.; Mahmoud, M.A. Hydrocarbon biodegradation and transcriptome responses of cellulase, peroxidase, and laccase encoding genes inhabiting rhizospheric fungal isolates. Saudi J. Biol. Sci. 2021, 28, 2083–2090. [Google Scholar] [CrossRef]
- Sychta, K.; Słomka, A.; Kuta, E. Insights into plant programmed cell death induced by heavy metals—Discovering a terra incognita. Cells 2021, 10, 65. [Google Scholar] [CrossRef]
- Stanton, C.; Sanders, D.; Kramer, U.; Podar, D. Zinc in plants: Integrating homeostasis and biofortification. Mol. Plant 2022, 15, 65–85. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Sesin, V.; Kisiala, A.; Emery, R.N. Phytohormonal roles in plant responses to heavy metal stress: Implications for using macrophytes in phytoremediation of aquatic ecosystems. Environ. Toxicol. Chem. 2021, 40, 7–22. [Google Scholar] [CrossRef]
- Noureddin, I.M.; Furumoto, T.; Ishida, Y.; Fukui, H. Absorption and metabolism of bisphenol A, a possible endocrine disruptor, in the aquatic edible plant, water convolvulus (Ipomoea aquatica). Biosci. Biotechnol. Biochem. 2004, 68, 1398–1402. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Shimoda, K.; Miyahara, K.; Hamada, H.; Hamada, H. Regioselective hydroxylation, reduction, and glycosylation of diphenyl compounds by cultured plant cells of Eucalyptus perriniana. Plant Biotechnol. 2006, 23, 291–296. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Kwak, J.I.; An, Y.J. Effects of bisphenol A in soil on growth, photosynthesis activity. and genistein levels in crop plants (Vigna radiata). Chemosphere 2018, 209, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Liang, Y.; Li, Y.N.; Che, X.K.; Zhao, S.J.; Zhang, Z.S.; Gao, H.Y. Mechanisms by which bisphenol A affect the photosynthetic apparatus in cucumber (Cucumis sativus L.) leaves. Sci. Rep. 2018, 8, 4253. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, Y.; Huang, H.; Mou, L.; Ru, J.; Zhao, J.; Xiao, S. Recent advances in exploring the heavy metal(loid) resistant microbiome. Comput. Struct. Biotechnol. J. 2021, 19, 94–109. [Google Scholar]
- Broadley, M.R.; White, P.J.; Hammond, J.P.; Zelko, I.; Lux, A. Zinc in plants. New Phytol. 2007, 173, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Begum, M.C.; Islam, M.; Sarkar, M.R.; Azad, M.A.S.; Huda, A.N.; Kabir, A.H. Auxin signaling is closely associated with Zn efficiency in rice (Oryza sativa L.). J. Plant Interact. 2016, 11, 124–129. [Google Scholar] [CrossRef]
- Marschner, H. Diagnosis of deficiency and toxicity of mineral nutrients. In Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995; pp. 461–479. [Google Scholar]
- Leskova, A.; Giehl, R.F.H.; Hartmann, A.; Fargasova, A.; von Wiren, N. Heavy metals induce iron deficiency responses at different hierarchic and regulatory levels. Plant Physiol. 2017, 174, 1648–1668. [Google Scholar] [CrossRef] [Green Version]
- Tsednee, M.; Yang, S.C.; Lee, D.C.; Yeh, K.C. Root-secreted nicotianamine from Arabidopsis halleri facilitates zinc hypertolerance by regulating zinc bioavailability. Plant Physiol. 2014, 166, 839–852. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Ram, H.; Kumar, B. Mechanism of zinc absorption in plants: Uptake, transport, translocation and accumulation. Rev. Environ. Sci. Biotechnol. 2016, 15, 89–109. [Google Scholar] [CrossRef]
- Zhou, G.; Delhaize, E.; Zhou, M.; Ryan, P.R. The barley MATE gene, HvAACT1, increases citrate efflux and Al(3+) tolerance when expressed in wheat and barley. Ann. Bot. 2013, 112, 603–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianco-Gomes, A.C.; Nogueira, L.D.S.; Bono-Lopes, N.V.M.; Gouvea-Souza, C.P.; Boldrini-França, J.; Gomes, V.M.; Cherene, M.B.; Galdino Alves, N.E.; Vasconcelos, C.M. Dry heat and pressure favor bioactive compounds preservation and peptides formation in sorghum [Sorghum bicolor (L.) Moench]. Curr. Res. Food Sci. 2022, 5, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Labuschagne, M.T. A review of cereal grain proteomics and its potential for sorghum improvement. J. Cereal Sci. 2018, 84, 151–158. [Google Scholar] [CrossRef]
- Zhou, G.; Chen, L.; Zheng, T.; Zhou, Z.; Yuan, H. Potential of anaerobic codigestion of vinegar residue with different ratios of pig and chicken manure. J. Agro-Environ. Sci. 2019, 38, 1357–1364. [Google Scholar] [CrossRef]
- Gao, J.G.; Heslop-Harrison, P.; Liu, P.L.; Zhang, R.G. Panicum virgatum (Poaceae). Trends Genet. 2021, 37, 771–772. [Google Scholar] [CrossRef]
- PN-R-04032; Soil and Mineral Materials—Sampling and Determination of Particle Size Distribution. Polish Committee for Standardizations: Warsaw, Poland, 1998.
- ISO 10390; Soil Quality—Determination of pH. International Organization for Standardization: Geneva, Switzerland, 2005.
- Klute, A. Methods of soil analysis. In Agronomy Monograph; American Society of Agronomy: Madison, WI, USA, 1996; Volume 9. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Method of Soil Analysis: Chemical Methods; Sparks, D.L., Ed.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 1201–1229. [Google Scholar]
- ISO 11261; Soil Quality—Determination of Total Nitrogen—Modified Kjeldahl Method. International Organization for Standardization: Geneva, Switzerland, 1995.
- Egner, H.; Riehm, H.; Domingo, W.R. Untersuchun-gen über die chemische Bodenanalyse als Grundlage für die Beurteilung des Nährstoffzustandes der Böden. II. Chemische Extractionsmethoden zur Phospor-und Kaliumbestimmung. K. Lantbr. Ann. 1960, 26, 199–215. [Google Scholar]
- Schlichting, E.; Blume, H.P.; Stahr, K. Bodenkundliches Praktikum. Pareys Studientexte 81; Blackwell Wissenschafts: Berlin, Germany, 1995. [Google Scholar]
- Öhlinger, R. Dehydrogenase activity with the substrate TTC. In Methods in Soil Biology; Schinner, F., Ohlinger, R., Kandler, E., Margesin, R., Eds.; Springer: Berlin, Germany, 1996; pp. 241–243. [Google Scholar]
- Methods in Applied Soil Microbiology and Biochemistry; Alef, K.; Nannipieri, P. (Eds.) Academic: London, UK, 1998; pp. 316–365. [Google Scholar]
- Wyszkowska, J.; Borowik, A.; Kucharski, M.; Kucharski, J. Applicability of biochemical indices to quality assessment of soil polluted with heavy metals. J. Elementol. 2013, 18, 733–756. [Google Scholar] [CrossRef]
- Mulder, E.G.; Antheumisse, J. Morphologie, physiologie et ecologie des Arthrobacter. Ann. L’Inst. Pasteur 1963, 105, 46–74. [Google Scholar]
- Jayan, N.; Bhatlu, M.L.D. Isolation and studies on zinc removal using microorganism from contaminated soil. Mater. Today Proc. 2021, 44, 1892–1897. [Google Scholar] [CrossRef]
- Bunt, J.S.; Rovira, A.D. Microbiological studies of some subantarctic soil. J. Soil Sci. 1955, 6, 119–128. [Google Scholar] [CrossRef]
- Parkinson, D.; Gray, F.R.G.; Williams, S.T. Methods for Studying the Ecology of Soil Microorganism; IBP Handbook 19; Blackwell Scientific Publication: Oxford, UK, 1971. [Google Scholar]
- Martin, J. Use of acid rose bengal and streptomycin in the plate method for estimating soil fungi. Soil Sci. 1950, 69, 215–233. [Google Scholar] [CrossRef]
- De Leji, F.A.M.; Whipps, M.; Lynch, J.M. The use for colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 1993, 27, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil con-tamination with Ekodiesel Ultra fuel. Environ. Sci. Pollut. Res. 2017, 24, 24346–24363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Xu, J.; Dong, F.; Liu, X.; Zhang, Y.; Wu, X.; Zheng, Y. Effect of tetraconazole application on the soil microbial community. Environ. Sci. Pollut. Res. 2014, 21, 8323–8332. [Google Scholar] [CrossRef]
- Statsoft Inc. Data Analysis Software System. Version 12.0. 2016. Available online: http://www.statsoft.com (accessed on 10 April 2022).
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 10 April 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 10 April 2022).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, M.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data. R Package Version 2.17.0. 2020. Available online: https://CRAN.R-project.org/package=gplots (accessed on 10 April 2022).
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]




| Object | TI of Aerial Parts of Plants | TI of Roots |
|---|---|---|
| Sorghum Moench (S) | ||
| B | 66.094 a | 34.721 c |
| Zn | 9.620 g | 18.440 d |
| BP_Zn | 7.494 g | 2.330 h |
| Sorghum Moench (S) with humic acid (H) | ||
| B | 51.301 d | 43.461 b |
| Zn | 2.705 h | 14.434 ef |
| BP_Zn | 4.946 h | 11.244 fg |
| Panicum virgatum (P) | ||
| B | 54.680 c | 43.461 b |
| Zn | 20.015 e | 17.624 e |
| B_Zn | 17.777 e | 11.244 efg |
| Panicum virgatum (P) with humic acid (H) | ||
| B | 63.075 b | 56.439 a |
| Zn | 15.459 f | 10.775 g |
| B_Zn | 17.067 f | 14.520 efg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaborowska, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Effect of Separate and Combined Toxicity of Bisphenol A and Zinc on the Soil Microbiome. Int. J. Mol. Sci. 2022, 23, 5937. https://doi.org/10.3390/ijms23115937
Zaborowska M, Wyszkowska J, Borowik A, Kucharski J. Effect of Separate and Combined Toxicity of Bisphenol A and Zinc on the Soil Microbiome. International Journal of Molecular Sciences. 2022; 23(11):5937. https://doi.org/10.3390/ijms23115937
Chicago/Turabian StyleZaborowska, Magdalena, Jadwiga Wyszkowska, Agata Borowik, and Jan Kucharski. 2022. "Effect of Separate and Combined Toxicity of Bisphenol A and Zinc on the Soil Microbiome" International Journal of Molecular Sciences 23, no. 11: 5937. https://doi.org/10.3390/ijms23115937






