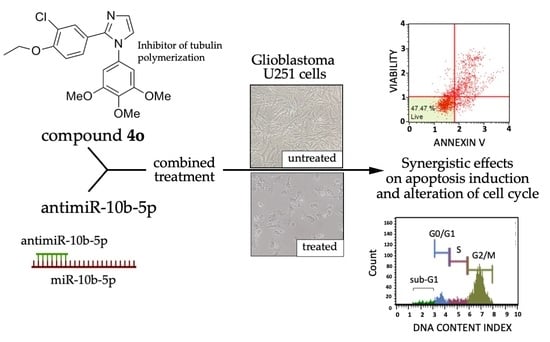
Synergistic Effects of A Combined Treatment of Glioblastoma U251 Cells with An Anti-miR-10b-5p Molecule and An AntiCancer Agent Based on 1-(3′,4′,5′-Trimethoxyphenyl)-2-Aryl-1H-Imidazole Scaffold
Abstract
:1. Introduction
2. Results
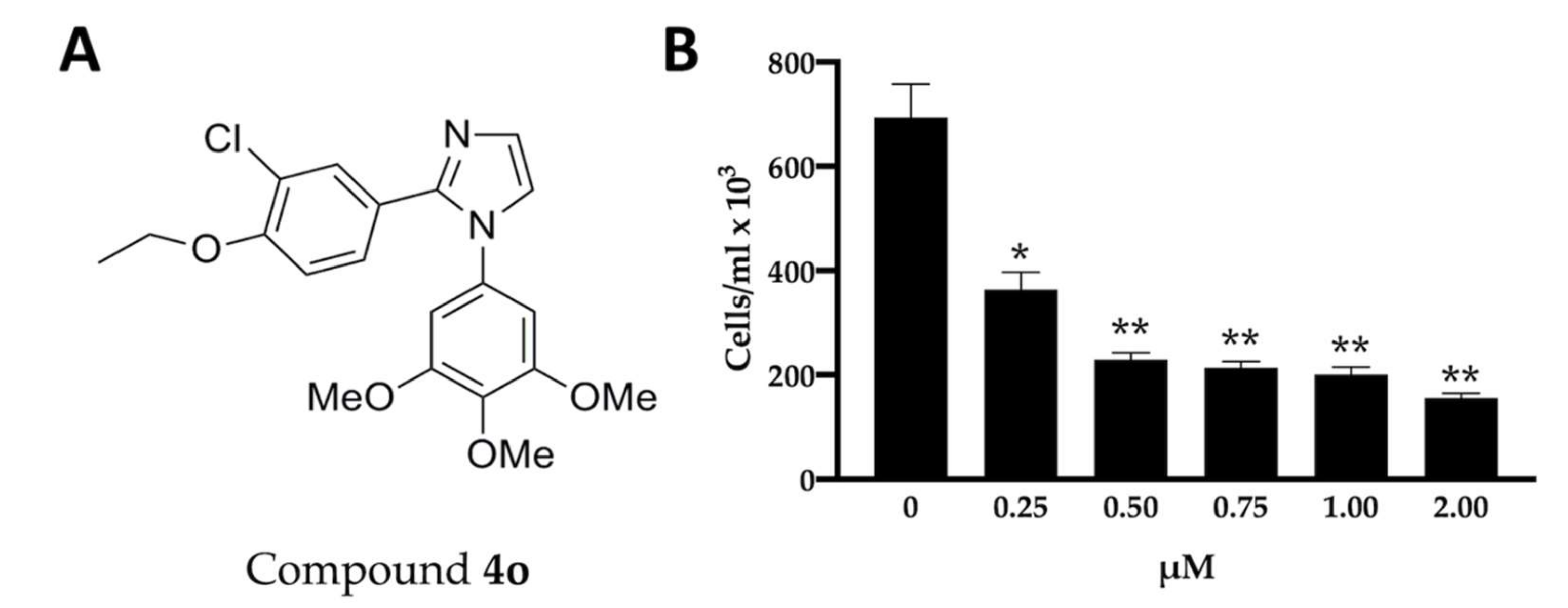
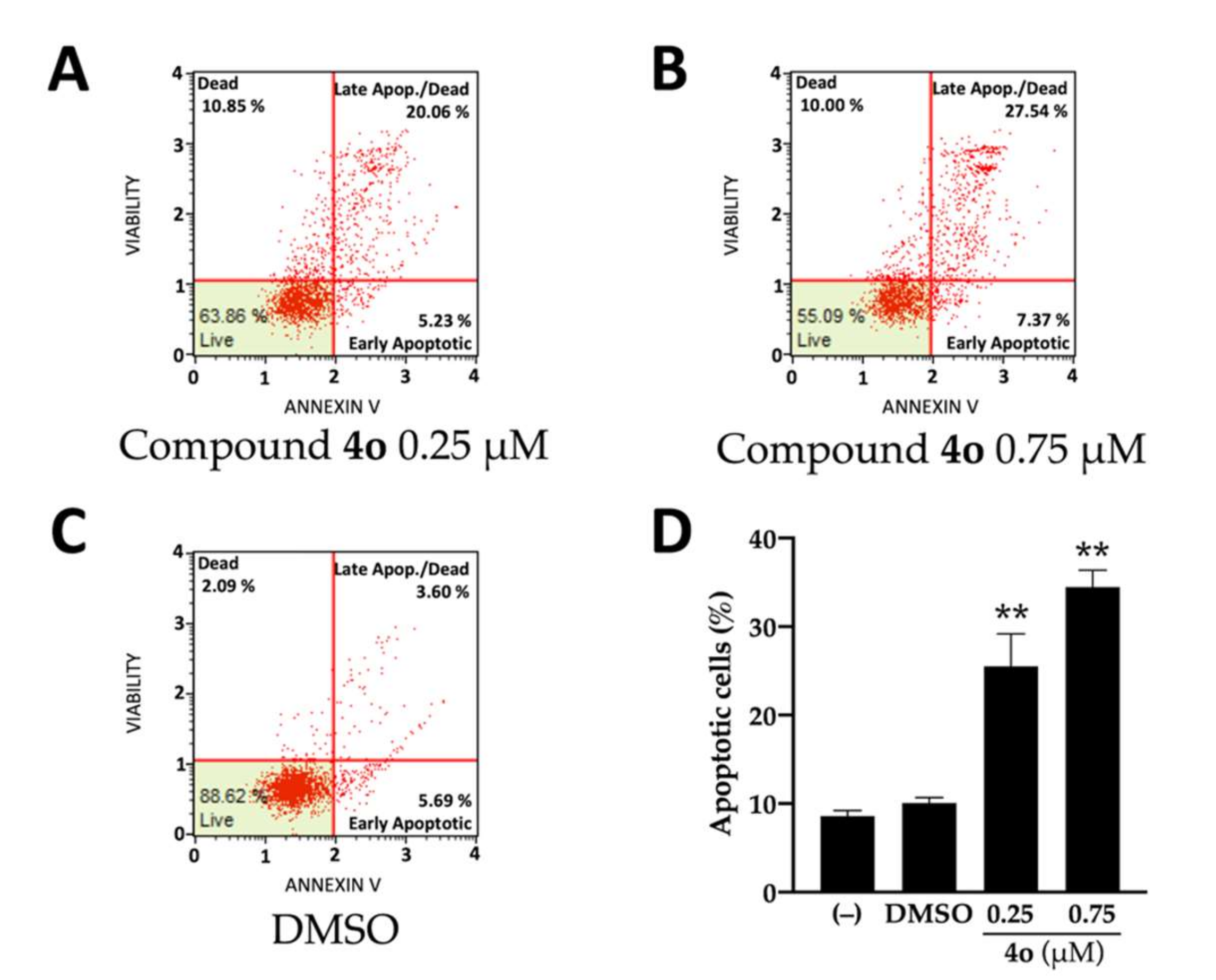
2.1. Effects of Compound 4o on U251 Cell Growth and Apoptosis
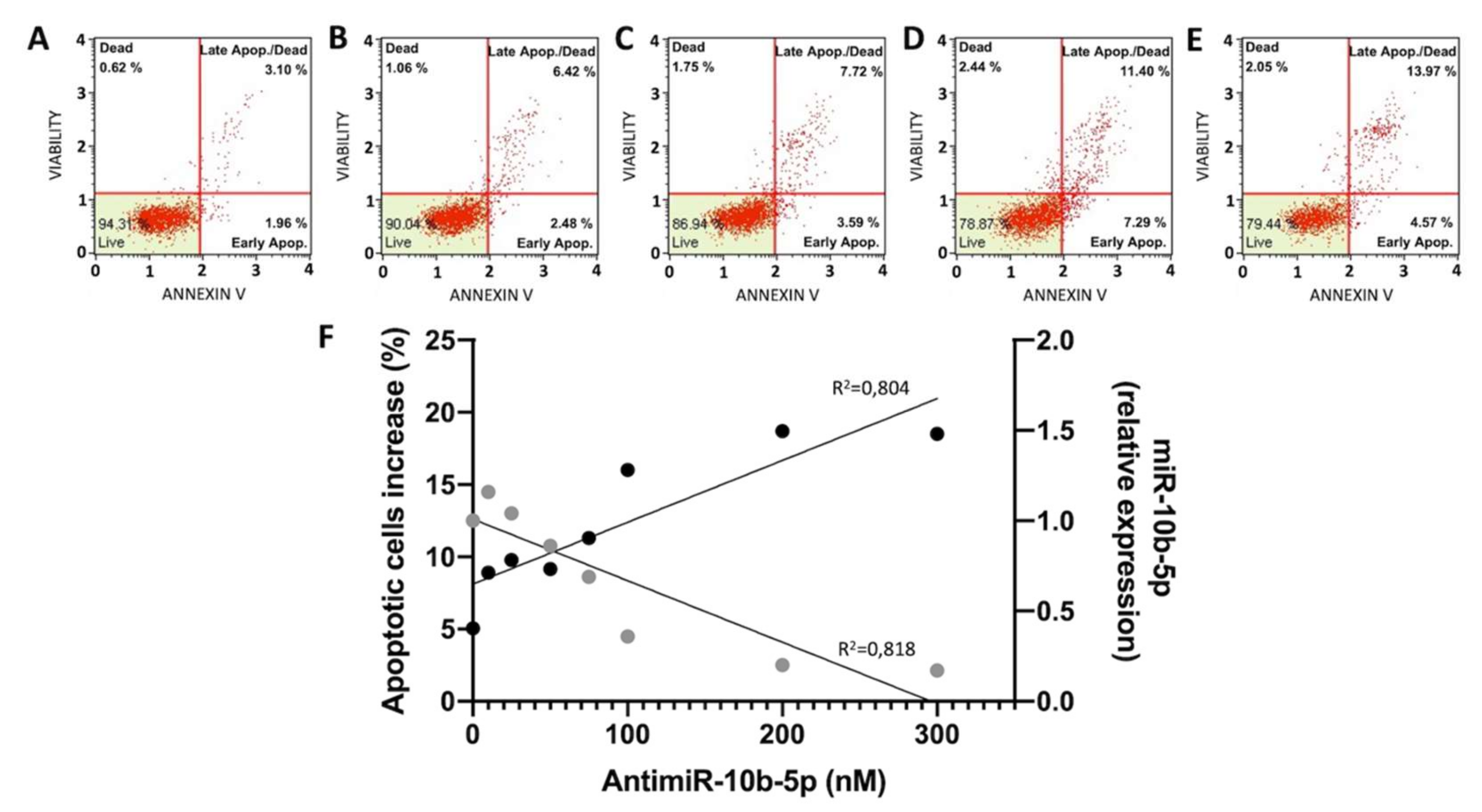
2.2. Effects of Anti-miR-10b-5p on U251 Apoptosis
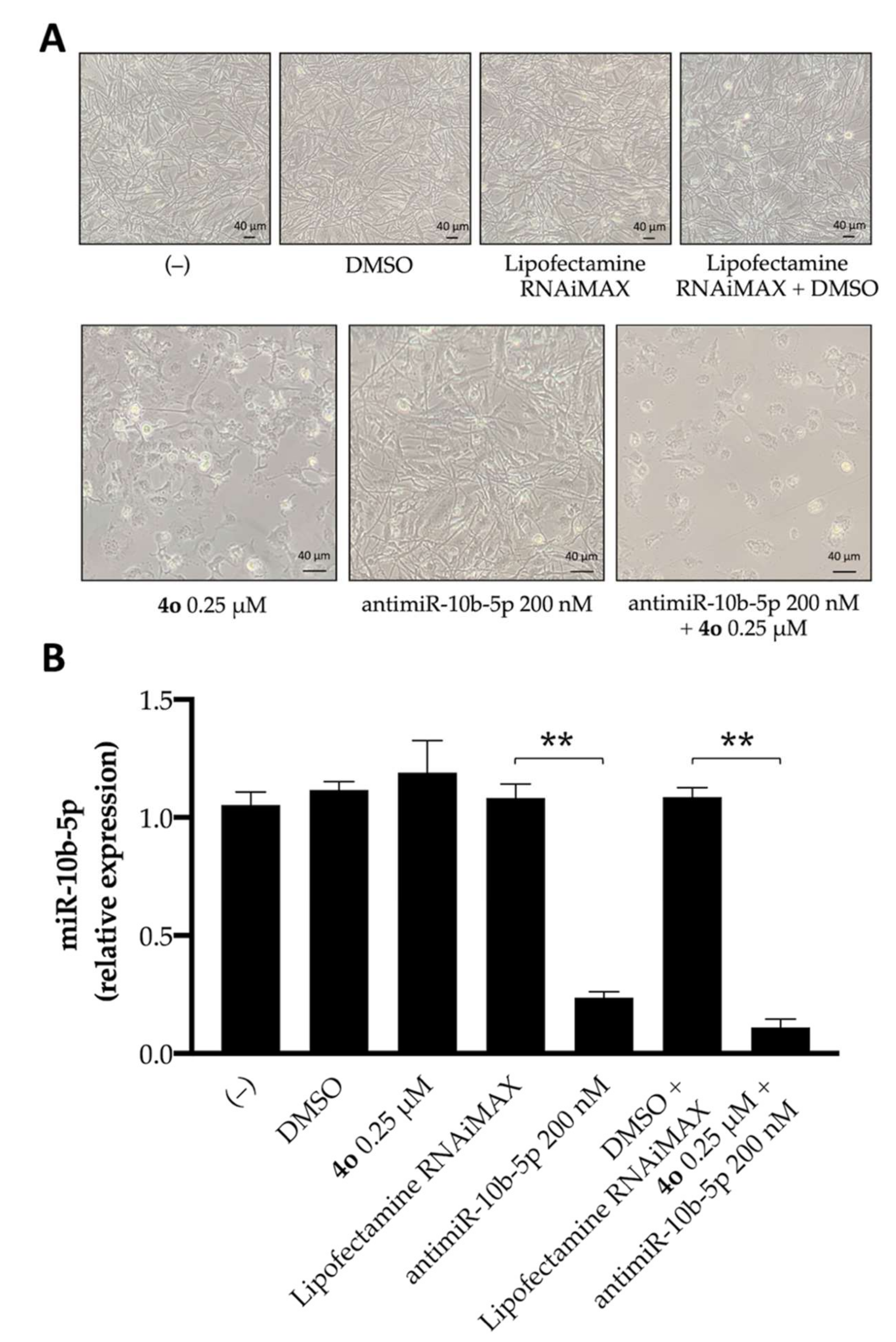
2.3. Effects of Compound 4o on miR-10b-5p Expression and Combined Effects with Anti-miR-10b-5p Transfection
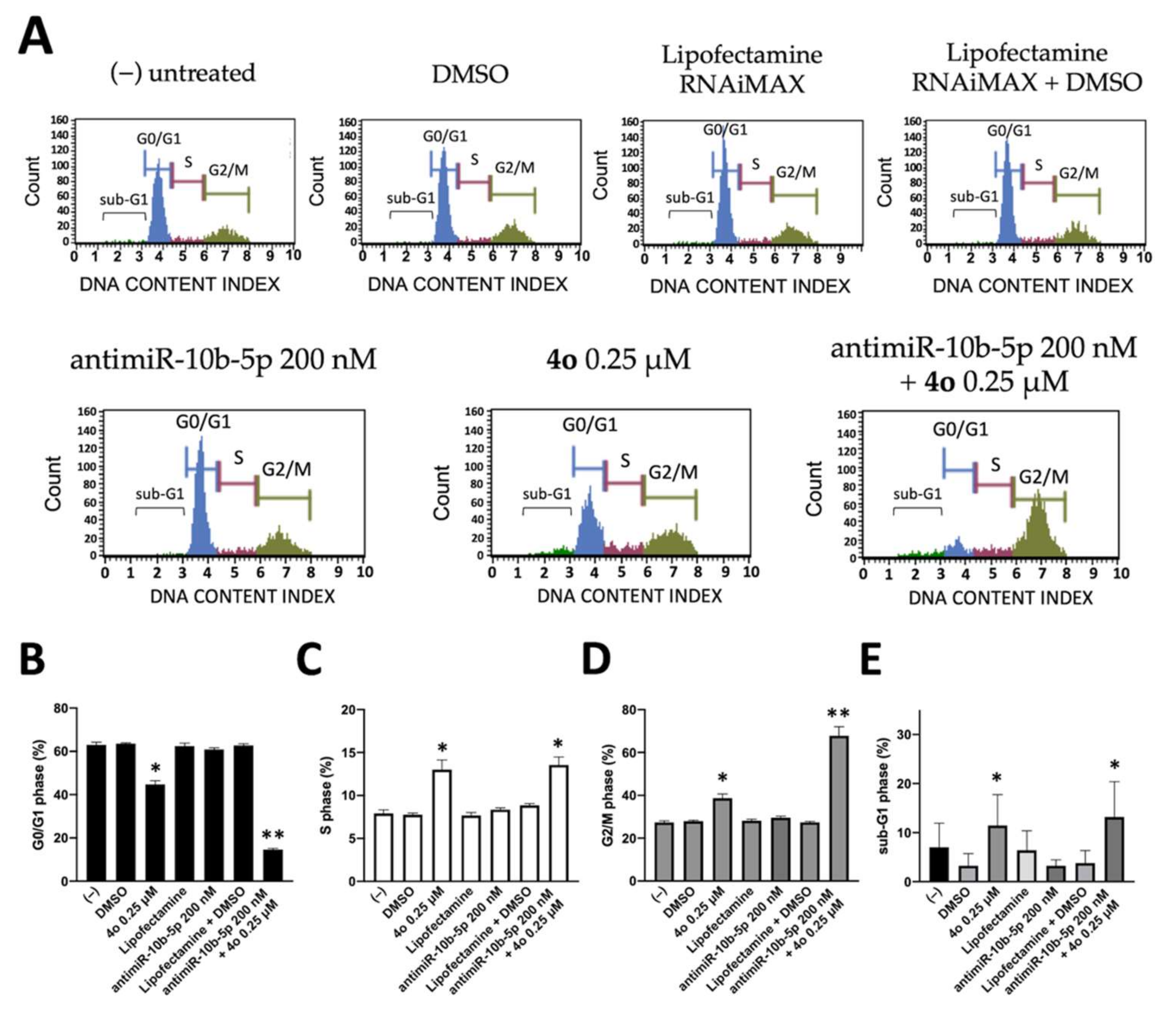
2.4. Co-Treatment of U251 Cells with Compound 4o and AntimiR-10b-5p: Effects on Cell Cycle
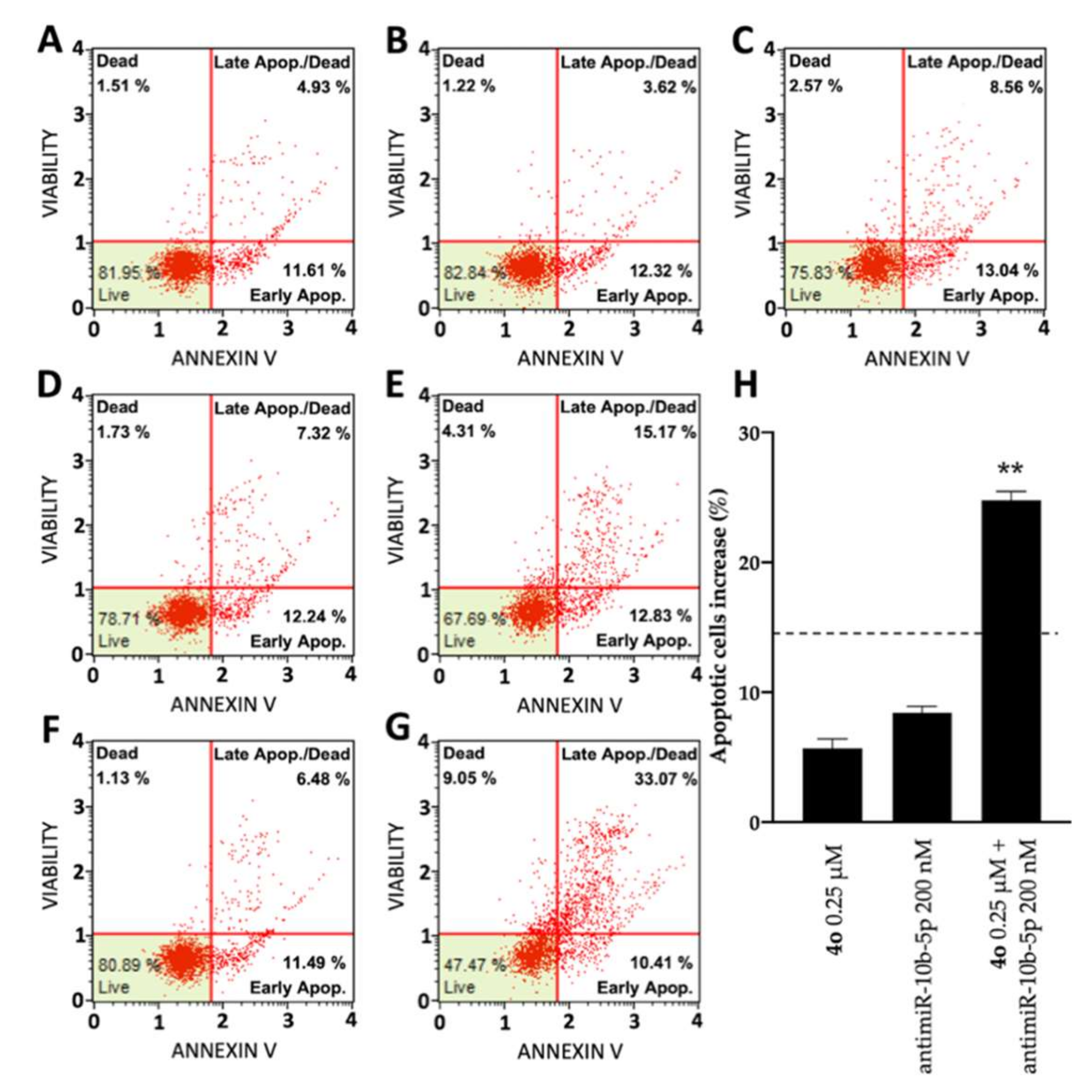
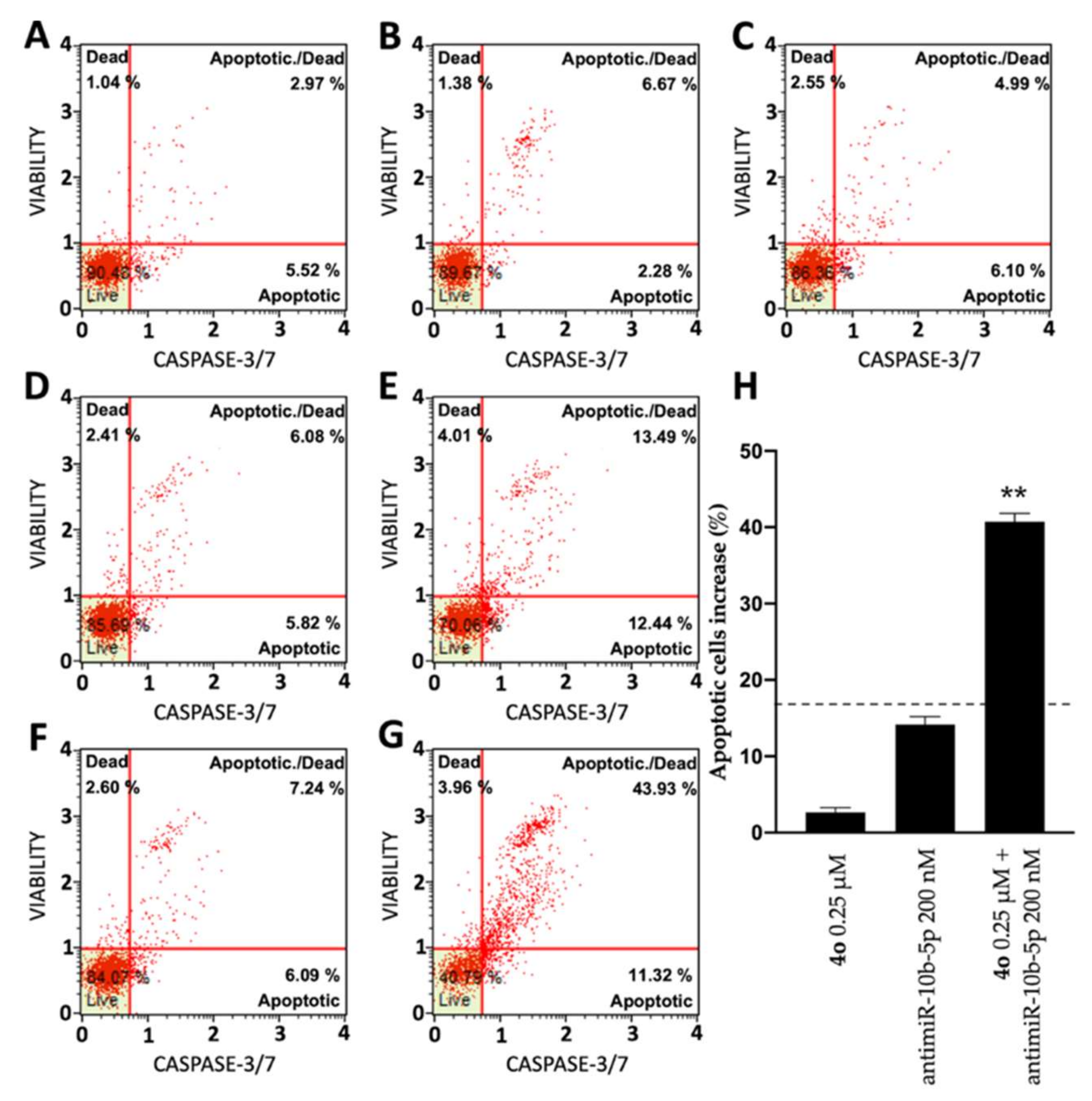
2.5. Co-Treatment of U251 Cells with Compound 4o and AntimiR-10b-5p: Effects on Apoptosis
3. Discussion
4. Materials and Methods
4.1. Chemistry and Reagents
4.2. Cell Lines, Cell Growth Conditions, Antiproliferative Assay
4.3. Morphological Analysis
4.4. AntimiRNA Transfection
4.5. RNA Extraction
4.6. Quantitative Analyses of miRNAs
4.7. Analysis of Caspase-3 mRNA by RT-qPCR
4.8. Effects on the Cell Cycle
4.9. Cell Apoptosis Assays
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Von Neubeck, C.; Seidlitz, A.; Kitzler, H.H.; Beuthien-Baumann, B.; Krause, M. Glioblastoma multiforme: Emerging treatments and stratification markers beyond new drugs. Br. J. Radiol. 2015, 88, 20150354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buczkowicz, P.; Hawkins, C. Pathology, Molecular Genetics, and Epigenetics of Diffuse Intrinsic Pontine Glioma. Front. Oncol. 2015, 5, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, A.; Dirven, L.; Koekkoek, J.A.F.; Golla, H.; Fleming, J.; Rudà, R.; Marosi, C.; Le Rhun, E.; Grant, R.; Oliver, K.; et al. European Association for Neuro-Oncology (EANO) guidelines for palliative care in adults with glioma. Lancet Oncol. 2017, 18, e330–e340. [Google Scholar] [CrossRef] [Green Version]
- Anjum, K.; Shagufta, B.I.; Abbas, S.Q.; Patel, S.; Khan, I.; Shah, S.A.A.; Akhter, N.; Hassan, S.S.U. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: A review. Biomed. Pharmacother. 2017, 92, 681–689. [Google Scholar] [CrossRef]
- Santangelo, A.; Rossato, M.; Lombardi, G.; Benfatto, S.; Lavezzari, D.; De Salvo, G.L.; Indraccolo, S.; Dechecchi, M.C.; Prandini, P.; Gambari, R.; et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro-Oncology 2020, 23, 264–276. [Google Scholar] [CrossRef]
- Touat, M.; Idbaih, A.; Sanson, M.; Ligon, K.L. Glioblastoma targeted therapy: Updated approaches from recent biological insights. Ann. Oncol. 2017, 28, 1457–1472. [Google Scholar] [CrossRef]
- Tai, S.-H.; Lin, Y.-W.; Huang, T.-Y.; Chang, C.-C.; Chao, L.-C.; Wu, T.-S.; Lee, E.-J. Cinnamophilin enhances temozolomide-induced cytotoxicity against malignant glioma: The roles of ROS and cell cycle arrest. Transl. Cancer Res. 2021, 10, 3906–3920. [Google Scholar] [CrossRef]
- Xu, P.; Wang, H.; Pan, H.; Chen, J.; Deng, C. Anlotinib combined with temozolomide suppresses glioblastoma growth via mediation of JAK2/STAT3 signaling pathway. Cancer Chemother. Pharmacol. 2022, 89, 183–196. [Google Scholar] [CrossRef]
- Dongpo, S.; Zhengyao, Z.; Xiaozhuo, L.; Qing, W.; Mingming, F.; Fengqun, M.; Mei, L.; Qian, H.; Tong, C. Efficacy and Safety of Bevacizumab Combined with Other Therapeutic Regimens for Treatment of Recurrent Glioblastoma: A Network Meta-analysis. World Neurosurg. 2021, 160, e61–e79. [Google Scholar] [CrossRef]
- Pająk, B.; Siwiak-Niedbalska, E.; Jaśkiewicz, A.; Sołtyka, M.; Zieliński, R.; Domoradzki, T.; Fokt, I.; Skóra, S.; Priebe, W. Synergistic Anticancer Effect of Glycolysis and Histone Deacetylases Inhibitors in a Glioblastoma Model. Biomedicines 2021, 9, 1749. [Google Scholar] [CrossRef]
- Hsieh, T.-H.; Liang, M.-L.; Zheng, J.-H.; Lin, Y.-C.; Yang, Y.-C.; Vo, T.-H.; Liou, J.-P.; Yen, Y.; Chen, C.-H. Combining an Autophagy Inhibitor, MPT0L145, with Abemaciclib Is a New Therapeutic Strategy in GBM Treatment. Cancers 2021, 13, 6117. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Novello, S.; Guclu, S.Z.; Bentsion, D.; Zvirbule, Z.; Szilasi, M.; Bernabe, R.; Syrigos, K.; Byers, L.A.; Clingan, P.; et al. Veliparib in Combination With Platinum-Based Chemotherapy for First-Line Treatment of Advanced Squamous Cell Lung Cancer: A Randomized, Multicenter Phase III Study. J. Clin. Oncol. 2021, 39, 3633–3644. [Google Scholar] [CrossRef] [PubMed]
- Gasparello, J.; Papi, C.; Zurlo, M.; Gambari, L.; Rozzi, A.; Manicardi, A.; Corradini, R.; Gambari, R.; Finotti, A. Treatment of Human Glioblastoma U251 Cells with Sulforaphane and a Peptide Nucleic Acid (PNA) Targeting miR-15b-5p: Synergistic Effects on Induction of Apoptosis. Molecules 2022, 27, 1299. [Google Scholar] [CrossRef]
- Miroshnichenko, S.; Patutina, O. Enhanced Inhibition of Tumorigenesis Using Combinations of miRNA-Targeted Therapeutics. Front. Pharmacol. 2019, 10, 488. [Google Scholar] [CrossRef]
- Gajda, E.; Godlewska, M.; Mariak, Z.; Nazaruk, E.; Gawel, D. Combinatory Treatment with miR-7-5p and Drug-Loaded Cubosomes Effectively Impairs Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5039. [Google Scholar] [CrossRef]
- Sun, X.; Xu, H.; Huang, T.; Zhang, C.; Wu, J.; Luo, S. Simultaneous delivery of anti-miRNA and docetaxel with supramolecular self-assembled “chitosome” for improving chemosensitivity of triple negative breast cancer cells. Drug Deliv. Transl. Res. 2021, 11, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Ghasabi, M.; Majidi, J.; Mansoori, B.; Mohammadi, A.; Shomali, N.; Shirafkan, N.; Baghbani, E.; Kazemi, T.; Baradaran, B. The effect of combined miR-200c replacement and cisplatin on apoptosis induction and inhibition of gastric cancer cell line migration. J. Cell. Physiol. 2019, 234, 22581–22592. [Google Scholar] [CrossRef]
- Gasparello, J.; Gambari, L.; Papi, C.; Rozzi, A.; Manicardi, A.; Corradini, R.; Gambari, R.; Finotti, A. High Levels of Apoptosis Are Induced in the Human Colon Cancer HT-29 Cell Line by Co-Administration of Sulforaphane and a Peptide Nucleic Acid Targeting miR-15b-5p. Nucleic Acid Ther. 2020, 30, 164–174. [Google Scholar] [CrossRef]
- Zurlo, M.; Romagnoli, R.; Oliva, P.; Gasparello, J.; Finotti, A.; Gambari, R. Synergistic effects of the combined treatment of U251 and T98G glioma cells with an anti-tubulin tetrahydrothieno[2,3-c]pyridine derivative and a peptide nucleic acid targeting miR-221-3p. Int. J. Oncol. 2021, 59, 1–13. [Google Scholar] [CrossRef]
- Sontheimer, E.J.; Carthew, R.W. Silence from within: Endogenous siRNAs and miRNAs. Cell 2005, 122, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Ivan, M.; Cimmino, A.; Negrini, M.; Calin, G.A. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin. Biol. Ther. 2007, 7, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Schiemann, W.P. Therapeutic opportunities for targeting microRNAs in cancer. Mol. Cell. Ther. 2014, 2, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambari, R.; Brognara, E.; Spandidos, D.; Fabbri, E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology (Review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Sasayama, T.; Nishihara, M.; Kondoh, T.; Hosoda, K.; Kohmura, E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer 2009, 125, 1407–1413. [Google Scholar] [CrossRef]
- Gabriely, G.; Yi, M.; Narayan, R.S.; Niers, J.M.; Wurdinger, T.; Imitola, J.; Ligon, K.L.; Kesari, S.; Esau, C.; Stephens, R.M.; et al. Human Glioma Growth Is Controlled by MicroRNA-10b. Cancer Res. 2011, 71, 3563–3572. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Zhao, X.; Ming, J.; Liu, X.; Liu, D.; Jiang, C. Stepwise detection and evaluation reveal miR-10b and miR-222 as a remarkable prognostic pair for glioblastoma. Oncogene 2019, 38, 6142–6157. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Wei, F.; Xia, H.; Liu, H.; Dong, X.; Zhang, Y.; Luo, Q.; Liu, Y.; Li, Y. MicroRNA-10b mediates TGF-β1-regulated glioblastoma proliferation, migration and epithelial-mesenchymal transition. Int. J. Oncol. 2017, 50, 1739–1748. [Google Scholar] [CrossRef] [Green Version]
- Guessous, F.; Alvarado-Velez, M.; Marcinkiewicz, L.; Zhang, Y.; Kim, J.; Heister, S.; Kefas, B.; Godlewski, J.; Schiff, D.; Purow, B.; et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J. Neuro-Oncol. 2013, 112, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Junior, L.G.D.; Baroni, M.; Lira, R.C.P.; Teixeira, S.; Fedatto, P.F.; Silveira, V.S.; Suazo, V.K.; Veronez, L.C.; Panepucci, R.A.; Antônio, D.S.M.; et al. High-throughput microRNA profile in adult and pediatric primary glioblastomas: The role of miR-10b-5p and miR-630 in the tumor aggressiveness. Mol. Biol. Rep. 2020, 47, 6949–6959. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Subramanian, S.; Uhlmann, E.J.; Krichevsky, A.M. Genome Editing Reveals Glioblastoma Addiction to MicroRNA-10b. Mol. Ther. 2017, 25, 368–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.G.; Wu, W.K.; Feng, S.Y.; Wang, X.J.; Shao, J.F.; Qiao, J. Co-inhibition of microRNA-10b and microRNA-21 exerts synergistic inhibition on the proliferation and invasion of human glioma cells. Int. J. Oncol. 2012, 41, 1005–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teplyuk, N.M.; Uhlmann, E.J.; Wong, A.H.-K.; Karmali, P.; Basu, M.; Gabriely, G.; Jain, A.; Wang, Y.; Chiocca, E.A.; Stephens, R.; et al. MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget 2015, 6, 3770–3783. [Google Scholar] [CrossRef] [Green Version]
- Ananta, J.S.; Paulmurugan, R.; Massoud, T.F. Tailored Nanoparticle Codelivery of antimiR-21 and antimiR-10b Augments Glioblastoma Cell Kill by Temozolomide: Toward a “Personalized” Anti-microRNA Therapy. Mol. Pharm. 2016, 13, 3164–3175. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting micro RNA -10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef]
- Uckun, F.M. Rationally Designed Anti-mitotic Agents with Pro-Apoptotic Activity. Curr. Pharm. Des. 2001, 7, 1627–1639. [Google Scholar] [CrossRef]
- Katsetos, C.D. Tubulins as Therapeutic Targets in Cancer: From Bench to Bedside. Curr. Pharm. Des. 2012, 18, 2778–2792. [Google Scholar] [CrossRef]
- Zottel, A.; Jovčevska, I.; Šamec, N.; Komel, R. Cytoskeletal proteins as glioblastoma biomarkers and targets for therapy: A systematic review. Crit. Rev. Oncol. 2021, 160, 103283. [Google Scholar] [CrossRef]
- Bordji, K.; Grandval, A.; Cuhna-Alves, L.; Lechapt-Zalcman, E.; Bernaudin, M. Hypoxia-inducible factor-2α (HIF-2α), but not HIF-1α, is essential for hypoxic induction of class III β-tubulin expression in human glioblastoma cells. FEBS J. 2014, 281, 5220–5236. [Google Scholar] [CrossRef]
- Horne, E.A.; Diaz, P.; Cimino, P.J.; Jung, E.; Xu, C.; Hamel, E.; Wagenbach, M.; Kumasaka, D.; Wageling, N.B.; Azorín, D.D.; et al. A brain-penetrant microtubule-targeting agent that disrupts hallmarks of glioma tumorigenesis. Neuro-Oncol. Adv. 2020, 3, vdaa165. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fu, Q.; Lu, Y.; Zhang, W.; Yu, P.; Liu, Z.; Sun, X. Anti-tubulin agent vinorelbine inhibits metastasis of cancer cells by regulating epithelial-mesenchymal transition. Eur. J. Med. Chem. 2020, 200, 112332. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, R.; Prencipe, F.; Oliva, P.; Cacciari, B.; Balzarini, J.; Liekens, S.; Hamel, E.; Brancale, A.; Ferla, S.; Manfredini, S.; et al. Synthesis and Biological Evaluation of New Antitubulin Agents Containing 2-(3′,4′,5′-trimethoxyanilino)-3,6-disubstituted-4,5,6,7-tetrahydrothieno[2,3-c]pyridine Scaffold. Molecules 2020, 25, 1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Liu, W.; Gong, Z.; Huang, Y.; Li, Y.; Peng, Z. Design, synthesis, biological evaluation and molecular docking studies of new chalcone derivatives containing diaryl ether moiety as potential anticancer agents and tubulin polymerization inhibitors. Bioorg. Chem. 2019, 95, 103565. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yu, L.-Z.; Diao, P.-C.; Jian, X.-E.; Zhou, M.-F.; Jiang, C.-S.; You, W.-W.; Ma, W.-F.; Zhao, P.-L. Novel [1,2,4]triazolo[1,5-a]pyrimidine derivatives as potent antitubulin agents: Design, multicomponent synthesis and antiproliferative activities. Bioorg. Chem. 2019, 92, 103260. [Google Scholar] [CrossRef]
- Döbber, A.; Phoa, A.F.; Abbassi, R.H.; Stringer, B.W.; Day, B.W.; Johns, T.G.; Abadleh, M.; Peifer, C.; Munoz, L. Development and Biological Evaluation of a Photoactivatable Small Molecule Microtubule-Targeting Agent. ACS Med. Chem. Lett. 2017, 8, 395–400. [Google Scholar] [CrossRef] [Green Version]
- Altinoz, M.A.; Topcu, G.; Hacimuftuoglu, A.; Ozpinar, A.; Ozpinar, A.; Hacker, E.; Elmaci, I. Noscapine, a Non-addictive Opioid and Microtubule-Inhibitor in Potential Treatment of Glioblastoma. Neurochem. Res. 2019, 44, 1796–1806. [Google Scholar] [CrossRef]
- Romagnoli, R.; Baraldi, P.G.; Prencipe, F.; Oliva, P.; Baraldi, S.; Tabrizi, M.A.; Lopez-Cara, L.C.; Ferla, S.; Brancale, A.; Hamel, E.; et al. Design and Synthesis of Potent in Vitro and in Vivo Anticancer Agents Based on 1-(3′,4′,5′-Trimethoxyphenyl)-2-Aryl-1H-Imidazole. Sci. Rep. 2016, 6, 26602. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Ye, W.; Xu, J.-P.; Wang, H.-T.; Li, X.-F.; Wang, W.-Y.; Zhou, Z.-Z. Discovery of novel trimethoxyphenylbenzo[d]oxazoles as dual tubulin/PDE4 inhibitors capable of inducing apoptosis at G2/M phase arrest in glioma and lung cancer cells. Eur. J. Med. Chem. 2021, 224, 113700. [Google Scholar] [CrossRef]
- Lin, J.; Teo, S.; Lam, D.H.; Jeyaseelan, K.; Wang, S. MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis. 2012, 3, e398. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, L.; Wang, Z.; Cheng, Y.; Zhang, P.; Wang, X.; Wen, W.; Yang, H.; Liu, H.; Jin, W.; et al. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget 2016, 8, 19244–19254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, C.; Li, M.; Ouyang, Y.; Tan, Z.; Jiang, Y. MiR-424 functions as a tumor suppressor in glioma cells and is down-regulated by DNA methylation. J. Neuro-Oncol. 2017, 133, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Hübner, M.; Moellhoff, N.; Effinger, D.; Hinske, C.L.; Hirschberger, S.; Wu, T.; Müller, M.B.; Strauß, G.; Kreth, F.-W.; Kreth, S. MicroRNA-93 acts as an “anti-inflammatory tumor suppressor” in glioblastoma. Neuro-Oncol. Adv. 2020, 2. [Google Scholar] [CrossRef] [PubMed]
- Kajstura, M.; Halicka, H.D.; Pryjma, J.; Darzynkiewicz, Z. Discontinuous fragmentation of nuclear DNA during apoptosis revealed by discrete “sub-G1” peaks on DNA content histograms. Cytom. Part A 2007, 71A, 125–131. [Google Scholar] [CrossRef]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Xiong, Q.; Tian, X.; Ru, Q. MicroRNA-10b-5p downregulation inhibits the invasion of glioma cells via modulating homeobox B3 expression. Exp. Ther. Med. 2019, 17, 4577–4585. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Liu, X.; Zheng, J.; Xue, Y.; Liu, L.; Ma, J.; Wang, P.; Yang, C.; Wang, D.; Shao, L.; et al. Interaction of BACH2 with FUS promotes malignant progression of glioma cells via the TSLNC8–miR-10b-5p–WWC3 pathway. Mol. Oncol. 2020, 14, 2936–2959. [Google Scholar] [CrossRef]
- Mokhtari, R.B.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H. Combination therapy in combating cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar] [CrossRef] [Green Version]
- Torsvik, A.; Stieber, D.; Enger, P.; Golebiewska, A.; Molven, A.; Svendsen, A.; Westermark, B.; Niclou, S.P.; Olsen, T.K.; Enger, M.C.; et al. U-251 revisited: Genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med. 2014, 3, 812–824. [Google Scholar] [CrossRef]
- Brognara, E.; Fabbri, E.; Bazzoli, E.; Montagner, G.; Ghimenton, C.; Eccher, A.; Cantù, C.; Manicardi, A.; Bianchi, N.; Finotti, A.; et al. Uptake by human glioma cell lines and biological effects of a peptide-nucleic acids targeting miR-221. J. Neuro-Oncol. 2014, 118, 19–28. [Google Scholar] [CrossRef]

| miRNA Name | Assay ID (Applied Biosystems by Thermo Fisher Scientific, Inc., Waltham, MA, USA) |
|---|---|
| hsa-miR-10b-5p | 002218 |
| hsa-U6 snRNA | 001973 |
| hsa-let-7c-5p | 000379 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zurlo, M.; Romagnoli, R.; Oliva, P.; Gasparello, J.; Finotti, A.; Gambari, R. Synergistic Effects of A Combined Treatment of Glioblastoma U251 Cells with An Anti-miR-10b-5p Molecule and An AntiCancer Agent Based on 1-(3′,4′,5′-Trimethoxyphenyl)-2-Aryl-1H-Imidazole Scaffold. Int. J. Mol. Sci. 2022, 23, 5991. https://doi.org/10.3390/ijms23115991
Zurlo M, Romagnoli R, Oliva P, Gasparello J, Finotti A, Gambari R. Synergistic Effects of A Combined Treatment of Glioblastoma U251 Cells with An Anti-miR-10b-5p Molecule and An AntiCancer Agent Based on 1-(3′,4′,5′-Trimethoxyphenyl)-2-Aryl-1H-Imidazole Scaffold. International Journal of Molecular Sciences. 2022; 23(11):5991. https://doi.org/10.3390/ijms23115991
Chicago/Turabian StyleZurlo, Matteo, Romeo Romagnoli, Paola Oliva, Jessica Gasparello, Alessia Finotti, and Roberto Gambari. 2022. "Synergistic Effects of A Combined Treatment of Glioblastoma U251 Cells with An Anti-miR-10b-5p Molecule and An AntiCancer Agent Based on 1-(3′,4′,5′-Trimethoxyphenyl)-2-Aryl-1H-Imidazole Scaffold" International Journal of Molecular Sciences 23, no. 11: 5991. https://doi.org/10.3390/ijms23115991
APA StyleZurlo, M., Romagnoli, R., Oliva, P., Gasparello, J., Finotti, A., & Gambari, R. (2022). Synergistic Effects of A Combined Treatment of Glioblastoma U251 Cells with An Anti-miR-10b-5p Molecule and An AntiCancer Agent Based on 1-(3′,4′,5′-Trimethoxyphenyl)-2-Aryl-1H-Imidazole Scaffold. International Journal of Molecular Sciences, 23(11), 5991. https://doi.org/10.3390/ijms23115991