Role of Nucleolin in Endometrial Precancerous Hyperplasia and Carcinogenesis: Ex Vivo and In Silico Study
Abstract
:1. Introduction
2. Results
2.1. Demographic Data
2.2. NCL Gene Expression
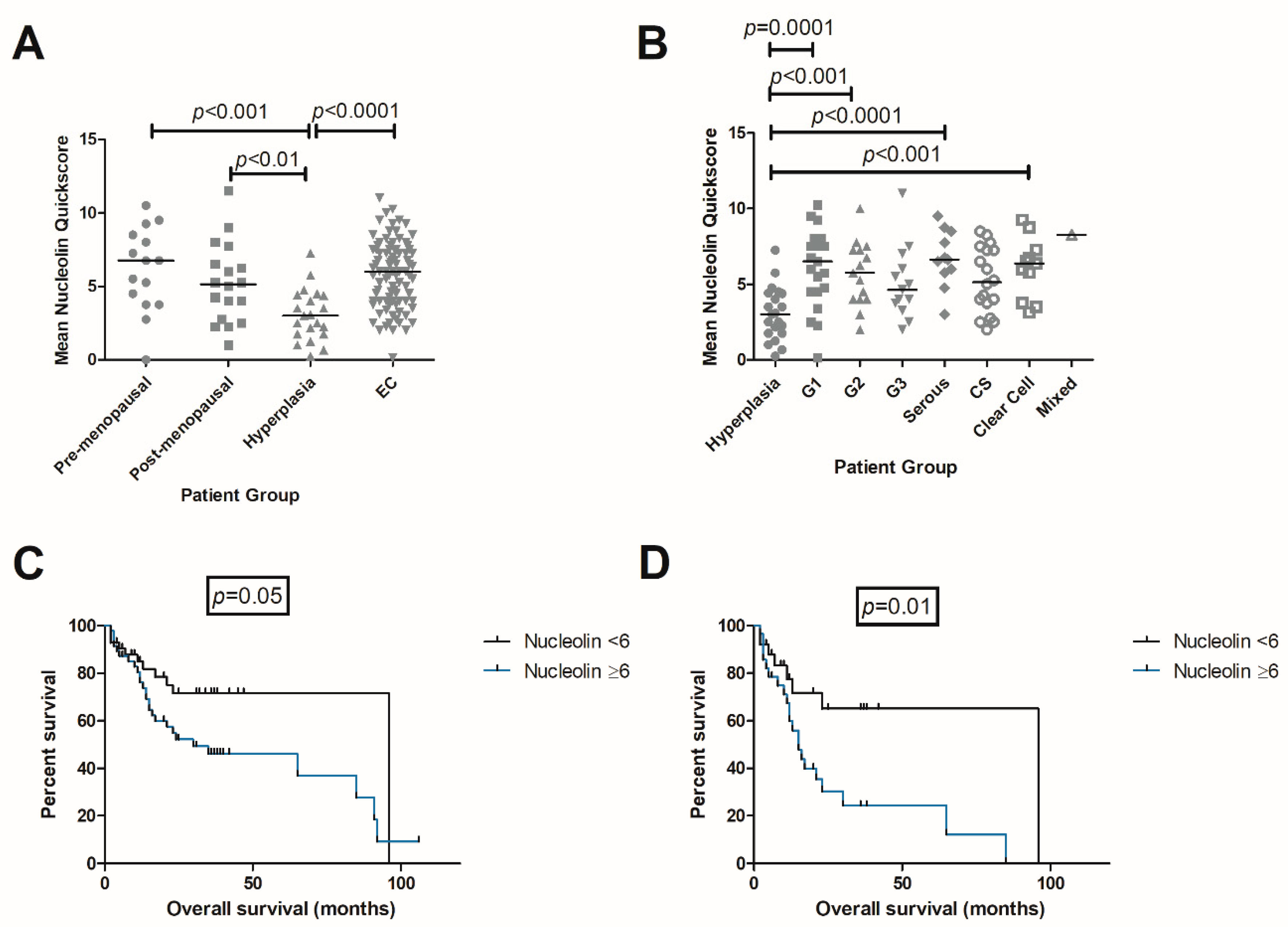
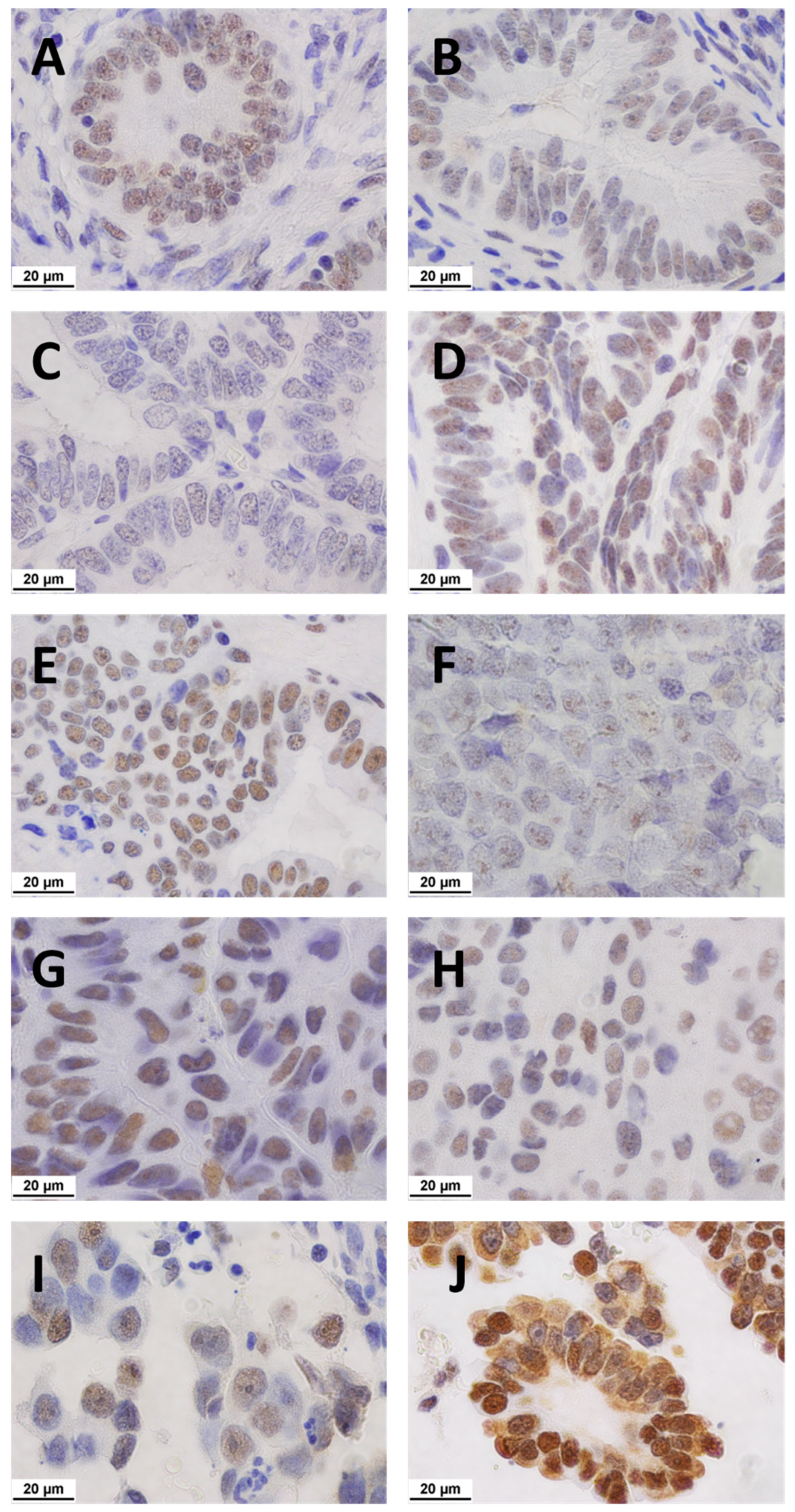
2.3. Immunolocalisation of NCL
2.3.1. Matched Pairs Analysis of NCL Staining
2.3.2. Observer Agreement
2.4. In Silico Analysis
2.4.1. NCL in TCGA Data
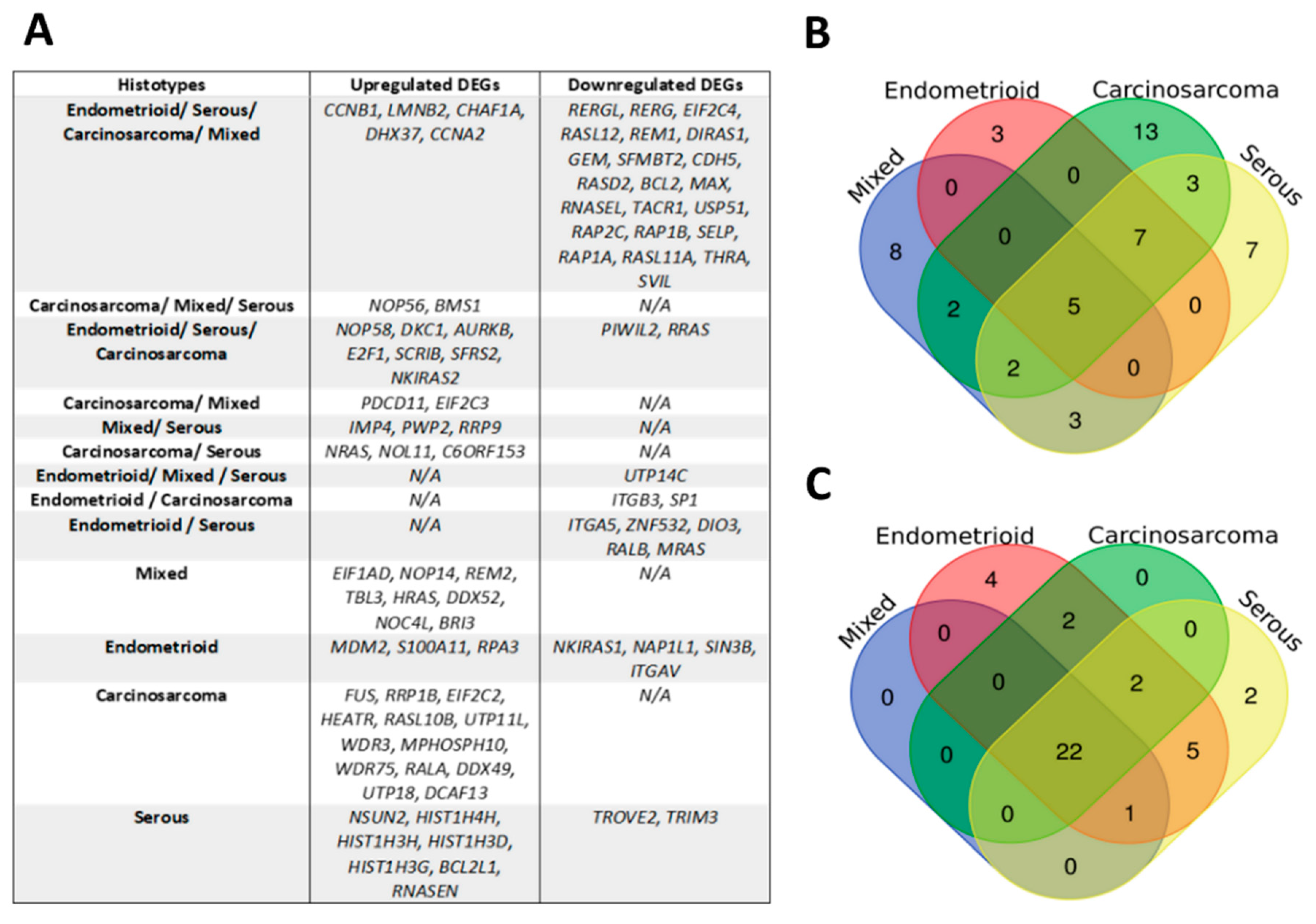
2.4.2. Identification of NAGs and DEGs
2.4.3. Identification of Prognostic DEGs
2.4.4. External Validation of Differentially Expressed NAGs
2.4.5. Biological Processes and Pathway Analyses
3. Discussion
4. Materials and Methods
4.1. Endometrial Tissue Samples
4.2. Real-Time qPCR
4.3. Immunohistochemistry
4.4. In Silico Study
4.4.1. Identification of Nucleolin Associated Genes (NAGs)
4.4.2. TCGA Cohort
4.4.3. Identification of Differentially Expressed Genes in TCGA Dataset
4.4.4. Biological Processes and Pathway Analyses
4.4.5. Validation of Differentially Expressed NAGs
4.4.6. STRING Network of Known Molecular Markers for EC and NAGs
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Gene Name | Sequences (5′ ≥ 3′) | Product Size (bp) | Efficiency (%) |
|---|---|---|---|
| IPO8 | Fwd: AGGATCAGAGGACAGCACTGCA | 102 | 97 |
| Rev: AGGTGAAGCCTCCCTGTTGTTC | |||
| PPIA | Fwd: AGACAAGGTCCCAAAGAC | 118 | 100 |
| Rev: ACCACCCTGACACATAAA | |||
| MRPL19 | Fwd: CAGGAAGAGGACTTGGAGCTAC | 137 | 101.5 |
| Rev: GCTATCATCCAGCCGTTTCTCTA | |||
| NCL | https://www.bio-rad.com/en-uk/prime-pcr-assays/assay/qhsaced0043572-primepcr-sybr-green-assay-ncl-human | 160 | 94 |
References
- Uterine Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/uterine-cancer (accessed on 10 May 2021).
- Ovarian Cancer Survival Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/ovarian-cancer (accessed on 19 May 2021).
- Cervical Cancer Survival Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/cervical-cancer (accessed on 19 May 2021).
- Mongelard, F.; Bouvet, P. Nucleolin: A multiFACeTed protein. Trends Cell Biol. 2007, 17, 80–86. [Google Scholar] [CrossRef]
- Salvetti, A.; Couté, Y.; Epstein, A.; Arata, L.; Kraut, A.; Navratil, V.; Bouvet, P.; Greco, A. Nuclear Functions of Nucleolin through Global Proteomics and Interactomic Approaches. J. Proteome Res. 2016, 15, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, X. Roles of nucleolin. Focus on cancer and anti-cancer therapy. Saudi Med. J. 2016, 37, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Zhou, F.; Zhang, Q.; Sun, X.; Shi, X.; Liang, Y.; Wang, X.; Yue, L. Overexpression of nucleolin and different expression sites both related to the prognosis of gastric cancer. Apmis 2013, 121, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Liang, J.; Wang, H.; Song, X.; Rashid, A.; Gomez, H.F.; Corley, L.J.; Abbruzzese, J.L.; Fleming, J.B.; Evans, D.B.; et al. High levels of nucleolar expression of nucleolin are associated with better prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2010, 16, 3734–3742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, J.; Li, H.; Liu, N.; Xing, Y.; Zhou, G.; Wu, Y.; Liu, Y.; Chen, W.; Yue, J.; Han, B.; et al. The implications and mechanisms of the extra-nuclear nucleolin in the esophageal squamous cell carcinomas. Med. Oncol. 2015, 32, 45. [Google Scholar] [CrossRef]
- Hammoudi, A.; Song, F.; Reed, K.R.; Jenkins, R.E.; Meniel, V.S.; Watson, A.J.; Pritchard, D.M.; Clarke, A.R.; Jenkins, J.R. Proteomic profiling of a mouse model of acute intestinal Apc deletion leads to identification of potential novel biomarkers of human colorectal cancer (CRC). Biochem. Biophys. Res. Commun. 2013, 440, 364–370. [Google Scholar] [CrossRef]
- Guo, X.; Xiong, L.; Yu, L.; Li, R.; Wang, Z.; Ren, B.; Dong, J.; Li, B.; Wang, D. Increased level of nucleolin confers to aggressive tumor progression and poor prognosis in patients with hepatocellular carcinoma after hepatectomy. Diagn. Pathol. 2014, 9, 175. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci. 2017, 186, 1–10. [Google Scholar] [CrossRef]
- Hapangama, D.K.; Turner, M.A.; Drury, J.A.; Quenby, S.; Hart, A.; Maddick, M.; Martin-Ruiz, C.; von Zglinicki, T. Sustained replication in endometrium of women with endometriosis occurs without evoking a DNA damage response. Hum. Reprod. 2009, 24, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Ma, X.; Hu, S.; Li, R.; Wei, X.; Han, B.; Ma, Y.; Liu, P.; Pang, Y. Overexpression of Nucleolin is a Potential Prognostic Marker in Endometrial Carcinoma. Cancer Manag. Res. 2021, 13, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, C.J.S.; Song, F.; Reed, K.R.; Al-Khafaji, N.; Clarke, A.R.; Vimalachandran, D.; Miyajima, F.; Pritchard, D.M.; Jenkins, J.R. NAP1L1: A Novel Human Colorectal Cancer Biomarker Derived From Animal Models of Apc Inactivation. Front. Oncol. 2020, 10, 1565. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Sugihara, K.; Nakayama, J.; Akama, T.O.; Wong, S.M.; Kawashima, H.; Zhang, J.; Smith, D.F.; Ohyama, C.; Fukuda, M.; et al. Identification of mRNA splicing factors as the endothelial receptor for carbohydrate-dependent lung colonization of cancer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 3095–3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; Toomer, K.H.; Chen, Z.; Cai, X. Genome-wide analysis of alternative transcripts in human breast cancer. Breast Cancer Res. Treat. 2015, 151, 295–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S.; Young, R.H. WHO Classification of Tumours of Female Reproductive Organs, 4th ed.; IARC: Lyon, France, 2014. [Google Scholar]
- Lacey, J.V.; Chia, V.M. Endometrial hyperplasia and the risk of progression to carcinoma. Maturitas 2009, 63, 39–44. [Google Scholar] [CrossRef]
- Morrison, J.; Balega, J.; Buckley, L.; Clamp, A.; Crosbie, E.; Drew, Y.; Durrant, L.; Forrest, J.; Fotopoulou, C.; Gajjar, K.; et al. British Gynaecological Cancer Society (BGCS) uterine cancer guidelines: Recommendations for practice. Eur. J. Obstet. Gynecol. Reprod. Biol. 2022, 270, 50–89. [Google Scholar] [CrossRef]
- Cusimano, M.C.; Simpson, A.N.; Han, A.; Hayeems, R.; Bernardini, M.Q.; Robertson, D.; Kives, S.L.; Satkunaratnam, A.; Baxter, N.N.; Ferguson, S.E. Barriers to care for women with low-grade endometrial cancer and morbid obesity: A qualitative study. BMJ Open 2019, 9, e026872. [Google Scholar] [CrossRef] [Green Version]
- Fortelny, N.; Overall, C.M.; Pavlidis, P.; Freue, G.V.C. Can we predict protein from mRNA levels? Nature 2017, 547, E19–E20. [Google Scholar] [CrossRef]
- Mehra, A.; Lee, K.H.; Hatzimanikatis, V. Insights into the relation between mRNA and protein expression patterns: I. Theoretical considerations. Biotechnol. Bioeng. 2003, 84, 822–833. [Google Scholar] [CrossRef]
- Mao, W.; Wei, S.; Yang, H.; Yu, Q.; Xu, M.; Guo, J.; Gao, L. Clinicopathological study of organ metastasis in endometrial cancer. Future Oncol. 2020, 16, 525–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Call, M.E.; Wucherpfennig, K.W. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol. Immunol. 2004, 40, 1295–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodges, E.; Krishna, M.T.; Pickard, C.; Smith, J.L. Diagnostic role of tests for T cell receptor (TCR) genes. J. Clin. Pathol. 2003, 56, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Cantrell, L.A.; Blank, S.V.; Duska, L.R. Uterine carcinosarcoma: A review of the literature. Gynecol. Oncol. 2015, 137, 581–588. [Google Scholar] [CrossRef]
- Milde-Langosch, K.; Bamberger, A.M.; Goemann, C.; Rössing, E.; Rieck, G.; Kelp, B.; Löning, T. Expression of cell-cycle regulatory proteins in endometrial carcinomas: Correlations with hormone receptor status and clinicopathologic parameters. J. Cancer Res. Clin. Oncol. 2001, 127, 537–544. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, T.; Miao, H.; Liang, B. The Calcium Binding Protein S100A11 and Its Roles in Diseases. Front. Cell Dev. Biol. 2021, 9, 693262. [Google Scholar] [CrossRef]
- Gabril, M.; Girgis, H.; Scorilas, A.; Rotondo, F.; Wala, S.; Bjarnason, G.A.; Ding, Q.; Evans, A.; Tawedrous, E.; Pasic, M.; et al. S100A11 is a potential prognostic marker for clear cell renal cell carcinoma. Clin. Exp. Metastasis 2016, 33, 63–71. [Google Scholar] [CrossRef]
- Rehman, I.; Azzouzi, A.R.; Cross, S.S.; Deloulme, J.C.; Catto, J.W.; Wylde, N.; Larre, S.; Champigneuille, J.; Hamdy, F.C. Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum. Pathol. 2004, 35, 1385–1391. [Google Scholar] [CrossRef]
- Cross, S.S.; Hamdy, F.C.; Deloulme, J.C.; Rehman, I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology 2005, 46, 256–269. [Google Scholar] [CrossRef]
- Sobolewski, C.; Abegg, D.; Berthou, F.; Dolicka, D.; Calo, N.; Sempoux, C.; Fournier, M.; Maeder, C.; Ay, A.S.; Clavien, P.A.; et al. S100A11/ANXA2 belongs to a tumour suppressor/oncogene network deregulated early with steatosis and involved in inflammation and hepatocellular carcinoma development. Gut 2020, 69, 1841–1854. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Sang, L.; Wang, X. S100 Calcium Binding Protein A11 (S100A11) Promotes the Proliferation, Migration and Invasion of Cervical Cancer Cells, and Activates Wnt/β-Catenin Signaling. Onco Targets Ther. 2019, 12, 8675–8685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Han, X.; Gao, B. Knockdown of S100A11 expression suppresses ovarian cancer cell growth and invasion. Exp. Ther. Med. 2015, 9, 1460–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.M.; Ding, G.L.; Jiang, Y.; Pan, H.J.; Zhang, D.; Wang, T.T.; Zhang, R.J.; Shu, J.; Sheng, J.Z.; Huang, H.F. Down-regulation of S100A11, a calcium-binding protein, in human endometrium may cause reproductive failure. J. Clin. Endocrinol. Metab. 2012, 97, 3672–3683. [Google Scholar] [CrossRef] [Green Version]
- Burke, B.; Stewart, C.L. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 2013, 14, 13–24. [Google Scholar] [CrossRef]
- Su, X.; Feng, X.; Gao, C.; Wang, G.; Liu, L. ROR promotes the proliferation and migration of esophageal cancer through regulating miR-145/LMNB2 signal axis. Am. J. Transl. Res. 2020, 12, 7223–7235. [Google Scholar]
- Zhao, C.C.; Chen, J.; Zhang, L.Y.; Liu, H.; Zhang, C.G.; Liu, Y. Lamin B2 promotes the progression of triple negative breast cancer via mediating cell proliferation and apoptosis. Biosci. Rep. 2021, 41. [Google Scholar] [CrossRef]
- Dong, C.H.; Jiang, T.; Yin, H.; Song, H.; Zhang, Y.; Geng, H.; Shi, P.C.; Xu, Y.X.; Gao, H.; Liu, L.Y.; et al. LMNB2 promotes the progression of colorectal cancer by silencing p21 expression. Cell Death Dis. 2021, 12, 331. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, Z.; Wang, Z.; Hu, L.; Wang, H.; Fang, Z. Supervillin promotes tumor angiogenesis in liver cancer. Oncol. Rep. 2020, 44, 674–684. [Google Scholar] [CrossRef]
- Cheuk, I.W.; Siu, M.T.; Ho, J.C.; Chen, J.; Shin, V.Y.; Kwong, A. ITGAV targeting as a therapeutic approach for treatment of metastatic breast cancer. Am. J. Cancer Res. 2020, 10, 211–223. [Google Scholar]
- Fry, S.A.; Sinclair, J.; Timms, J.F.; Leathem, A.J.; Dwek, M.V. A targeted glycoproteomic approach identifies cadherin-5 as a novel biomarker of metastatic breast cancer. Cancer Lett. 2013, 328, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Pecorelli, S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int. J. Gynaecol. Obstet. 2009, 105, 103–104. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, B.; Innes, B.A.; Bulmer, J.N.; Otun, H.A.; Chadwick, T.J.; Robson, S.C.; Lash, G.E. Localization of angiogenic growth factors and their receptors in the human placental bed throughout normal human pregnancy. Placenta 2009, 30, 79–87. [Google Scholar] [CrossRef]
- STRING Version 11.0. Available online: https://string-db.org/ (accessed on 19 April 2021).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- QIAGEN. Ingenuity Pathway Analysis (IPA). Available online: https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ (accessed on 30 April 2021).
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Science, W.I.o. GeneCards. Available online: https://www.genecards.org/ (accessed on 28 May 2021).
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Bradfield, A.; Button, L.; Drury, J.; Green, D.C.; Hill, C.J.; Hapangama, D.K. Investigating the Role of Telomere and Telomerase Associated Genes and Proteins in Endometrial Cancer. Methods Protoc. 2020, 3, 63. [Google Scholar] [CrossRef]
- iDEP.91. Available online: http://bioinformatics.sdstate.edu/idep/ (accessed on 15 May 2021).
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [Green Version]
- Bioinformatics & Evolutionary Genomics. Available online: https://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 21 May 2021).
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 2 June 2021).
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Enrichr. Available online: https://maayanlab.cloud/Enrichr/ (accessed on 11 June 2021).
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]




| Study Group | Number of Patients | Age (Years) Median (Range) | BMI (kg/m2) Median (Range) |
|---|---|---|---|
| Healthy Control | 38 | 56.5 (30–85) | 26.8 (20–52.2) |
| Pre-menopausal | 15 | 44 (30–47) | 27.8 (21.6–52.2) |
| Post-menopausal | 23 | 62 (52–85) | 25.2 (20–39.6) |
| Endometrial Hyperplasia | 21 | 57 (37–74) | 37.8 (23.6–63.1) |
| Endometrial Cancer | 98 | 67 (33–89) | 30 (20.2–51.4) |
| Low Grade EC | 39 | 60 (33–88) | 30.6 (21.6–48.4) |
| Grade 1 Endometrioid Carcinoma | 22 | 58 (33–88) | 31.2 (21.6–47.6) |
| Grade 2 Endometrioid Carcinoma | 17 | 64 (41–79) | 28.9 (41–48.4) |
| High Grade EC | 59 | 73 (52–89) | 29.6 (20.2–51.4) |
| Grade 3 Endometrioid Carcinoma | 14 | 70 (54–83) | 28.6 (23.9–42.7) |
| Serous Carcinoma | 13 | 78 (60–87) | 29.6 (23.4–38.7) |
| Carcinosarcoma | 20 | 78 (58–89) | 26 (20.2–51.4) |
| Clear Cell Carcinoma | 11 | 64 (52–82) | 30.3 (26.6–39) |
| Mixed Endometrioid and Clear Cell | 1 | 83 | 26.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barzilova, V.D.; Drury, J.; Rogers, B.; Thomas, E.; Ahmed, F.; Bradfield, A.; Al-Lamee, H.; Hapangama, D.K. Role of Nucleolin in Endometrial Precancerous Hyperplasia and Carcinogenesis: Ex Vivo and In Silico Study. Int. J. Mol. Sci. 2022, 23, 6228. https://doi.org/10.3390/ijms23116228
Barzilova VD, Drury J, Rogers B, Thomas E, Ahmed F, Bradfield A, Al-Lamee H, Hapangama DK. Role of Nucleolin in Endometrial Precancerous Hyperplasia and Carcinogenesis: Ex Vivo and In Silico Study. International Journal of Molecular Sciences. 2022; 23(11):6228. https://doi.org/10.3390/ijms23116228
Chicago/Turabian StyleBarzilova, Vanya D., Josephine Drury, Bryony Rogers, Emily Thomas, Fareen Ahmed, Alice Bradfield, Hannan Al-Lamee, and Dharani K. Hapangama. 2022. "Role of Nucleolin in Endometrial Precancerous Hyperplasia and Carcinogenesis: Ex Vivo and In Silico Study" International Journal of Molecular Sciences 23, no. 11: 6228. https://doi.org/10.3390/ijms23116228
APA StyleBarzilova, V. D., Drury, J., Rogers, B., Thomas, E., Ahmed, F., Bradfield, A., Al-Lamee, H., & Hapangama, D. K. (2022). Role of Nucleolin in Endometrial Precancerous Hyperplasia and Carcinogenesis: Ex Vivo and In Silico Study. International Journal of Molecular Sciences, 23(11), 6228. https://doi.org/10.3390/ijms23116228







