Generation of Matrix Degradation Products Using an In Vitro MMP Cleavage Assay
Abstract
:1. Introduction
2. Results
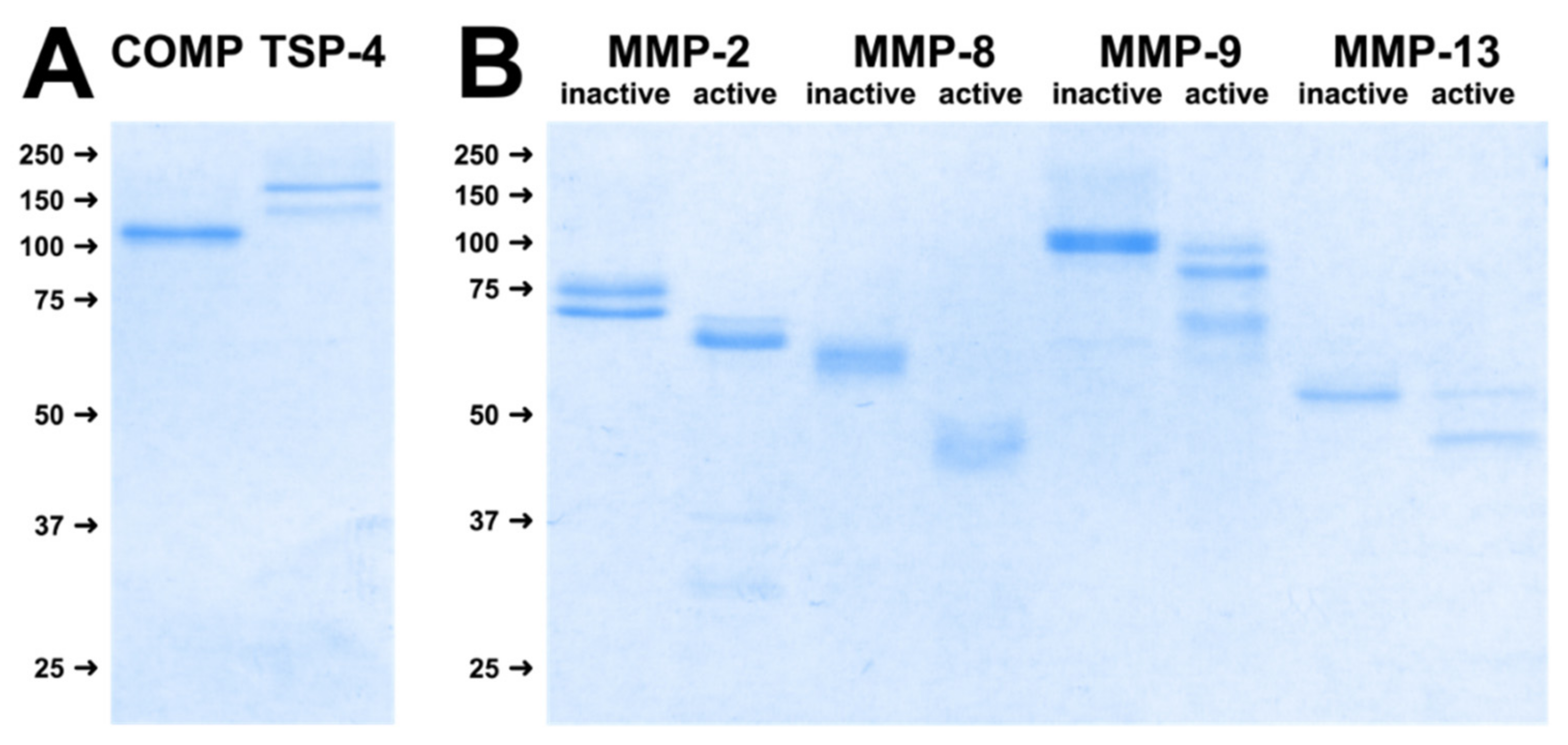
2.1. Substrates and MMPs Used for Digestion Assays
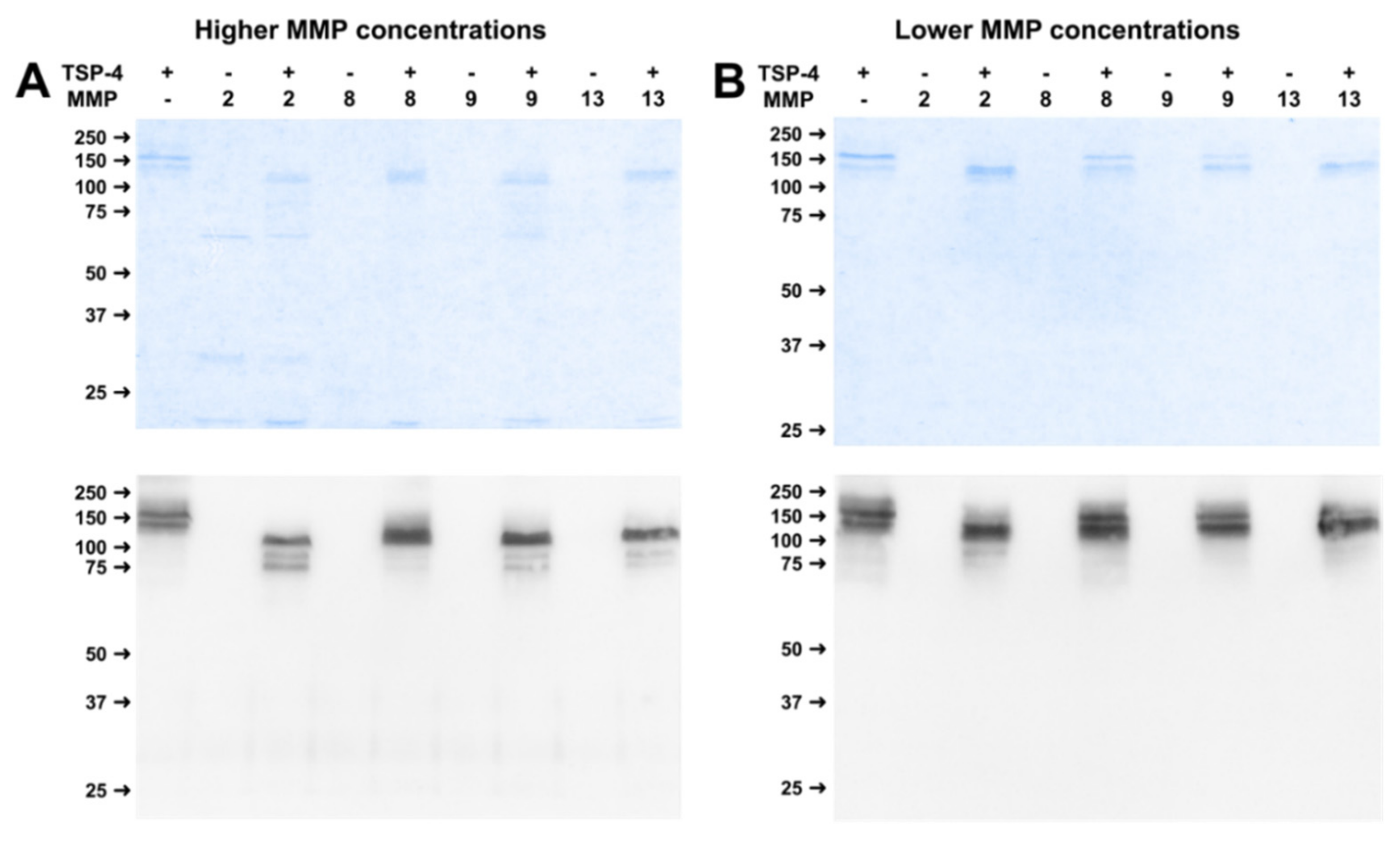
2.2. MMPs Cleave COMP and TSP-4 in a Concentration-Dependent Manner
2.3. Kinetics of COMP- and TSP-4-Cleavage by MMPs
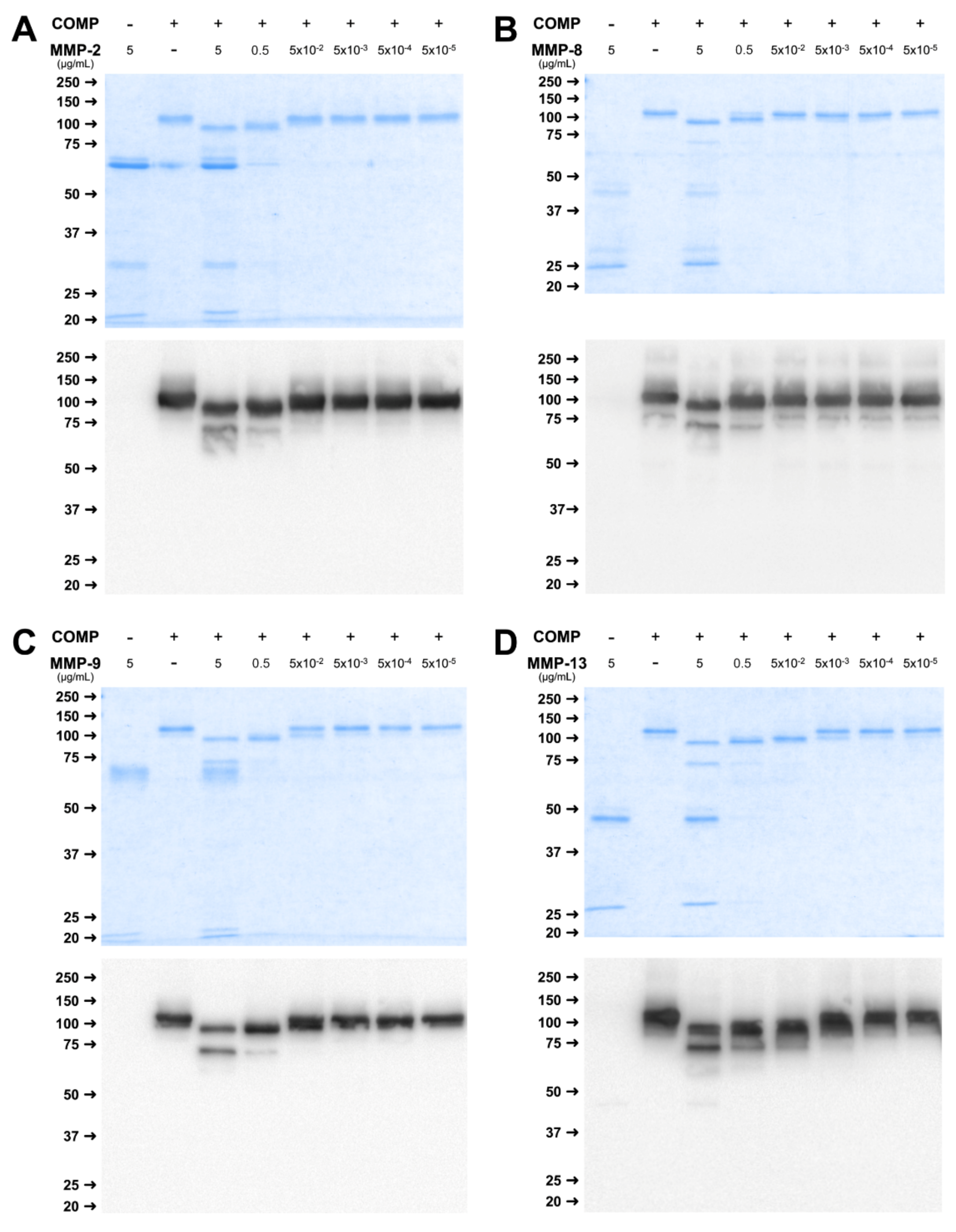
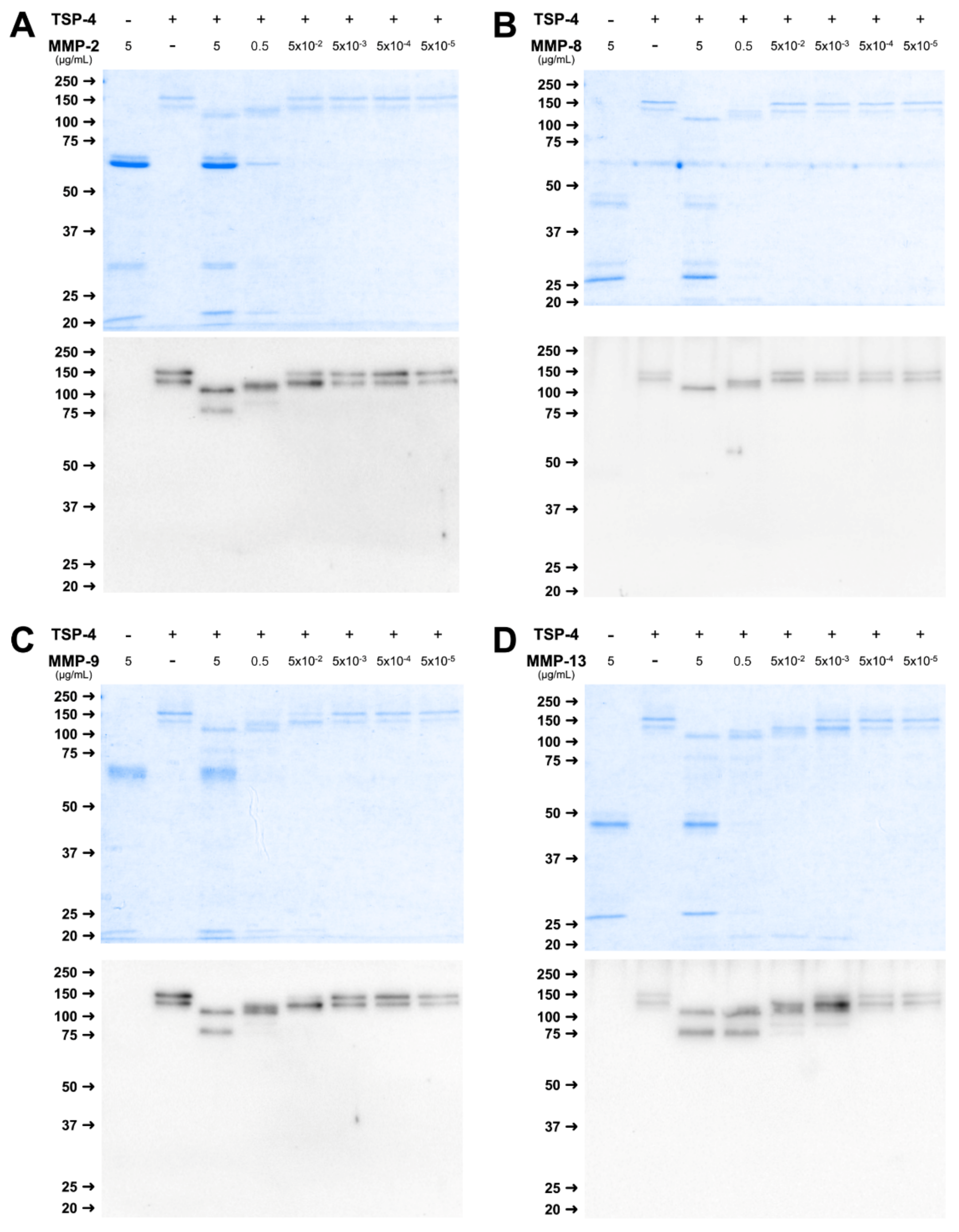
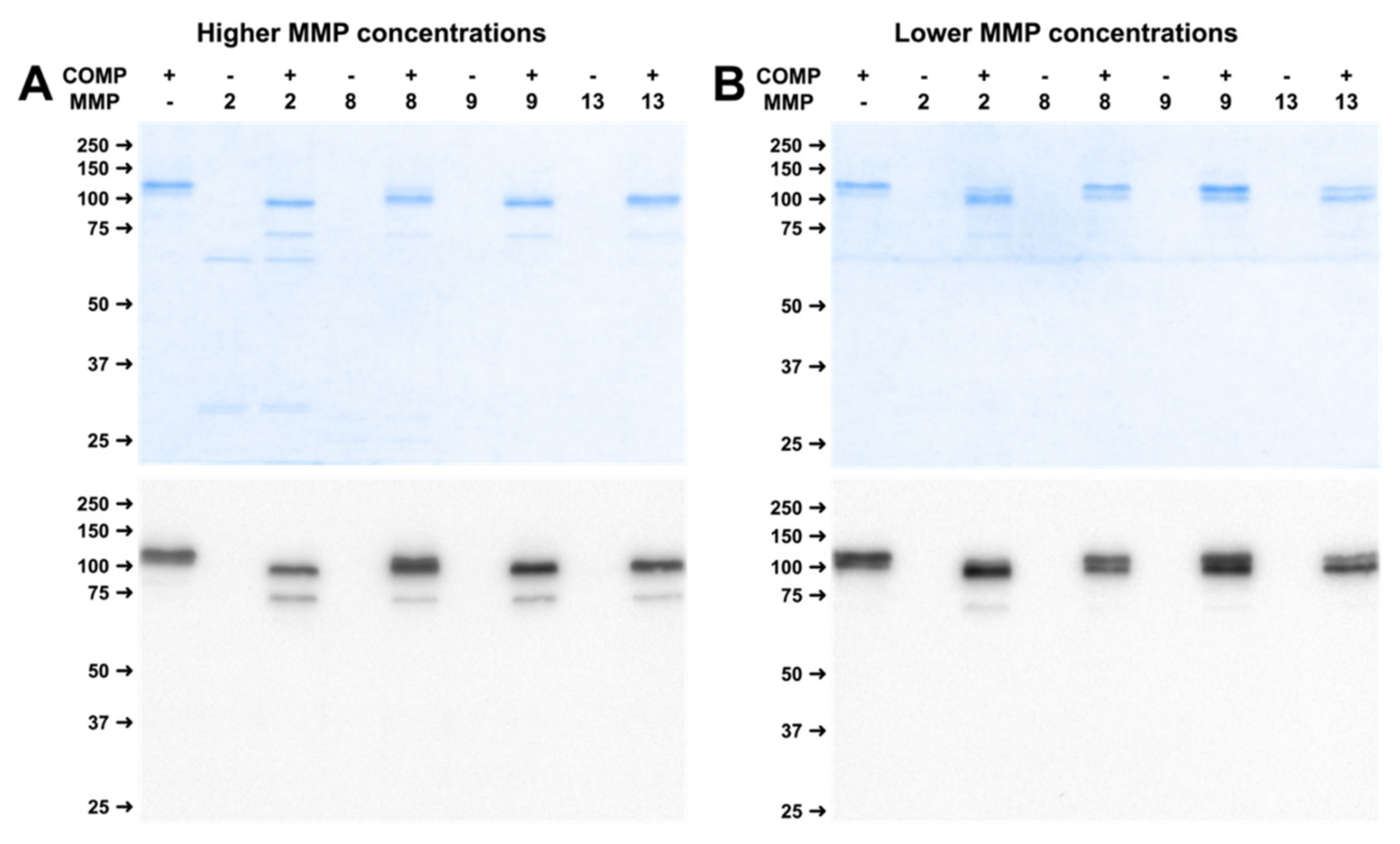
2.4. Physiological Concentrations of MMP Lead to Stable Cleavage Products in Long-Term Digestion Assay
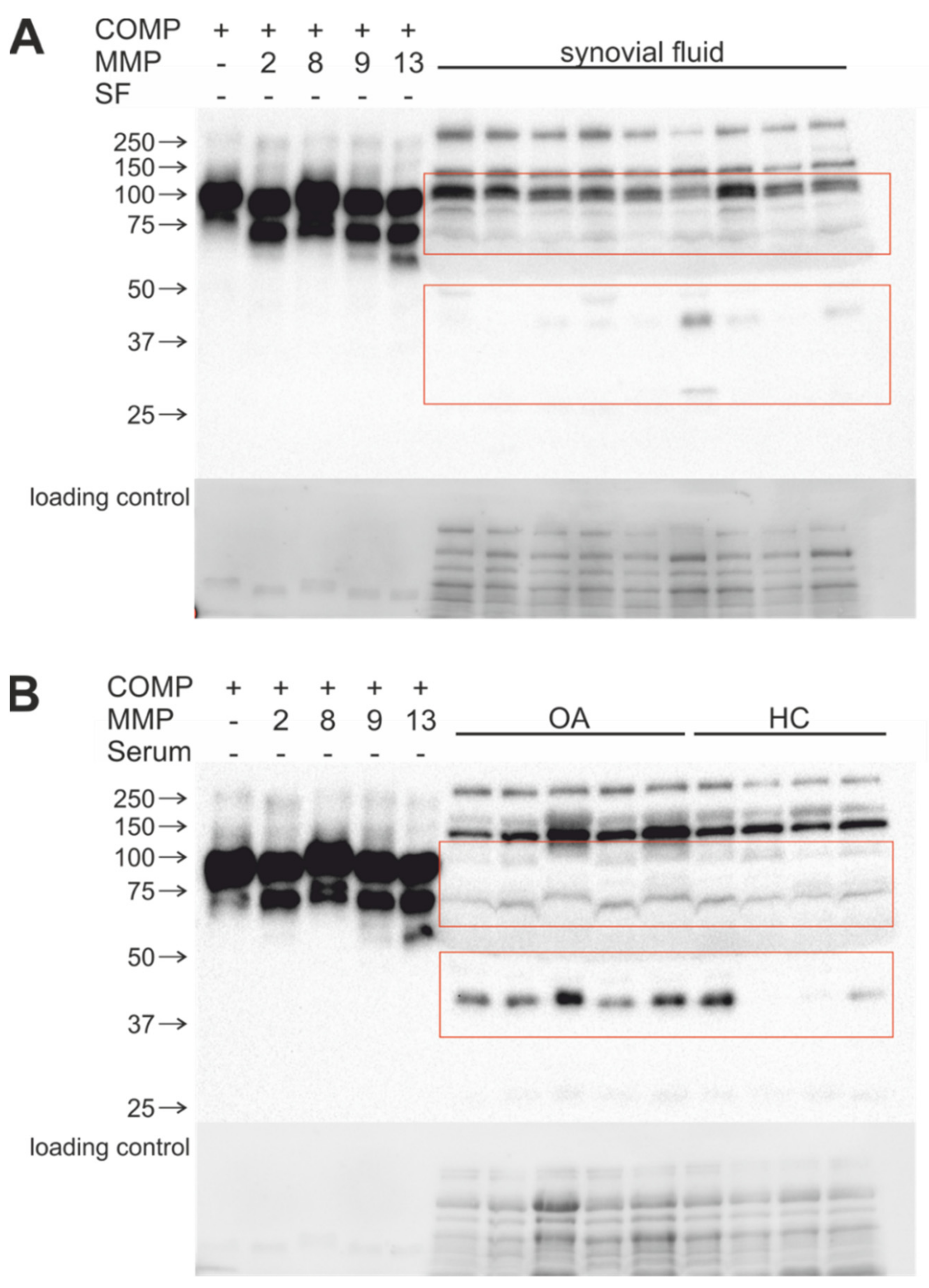
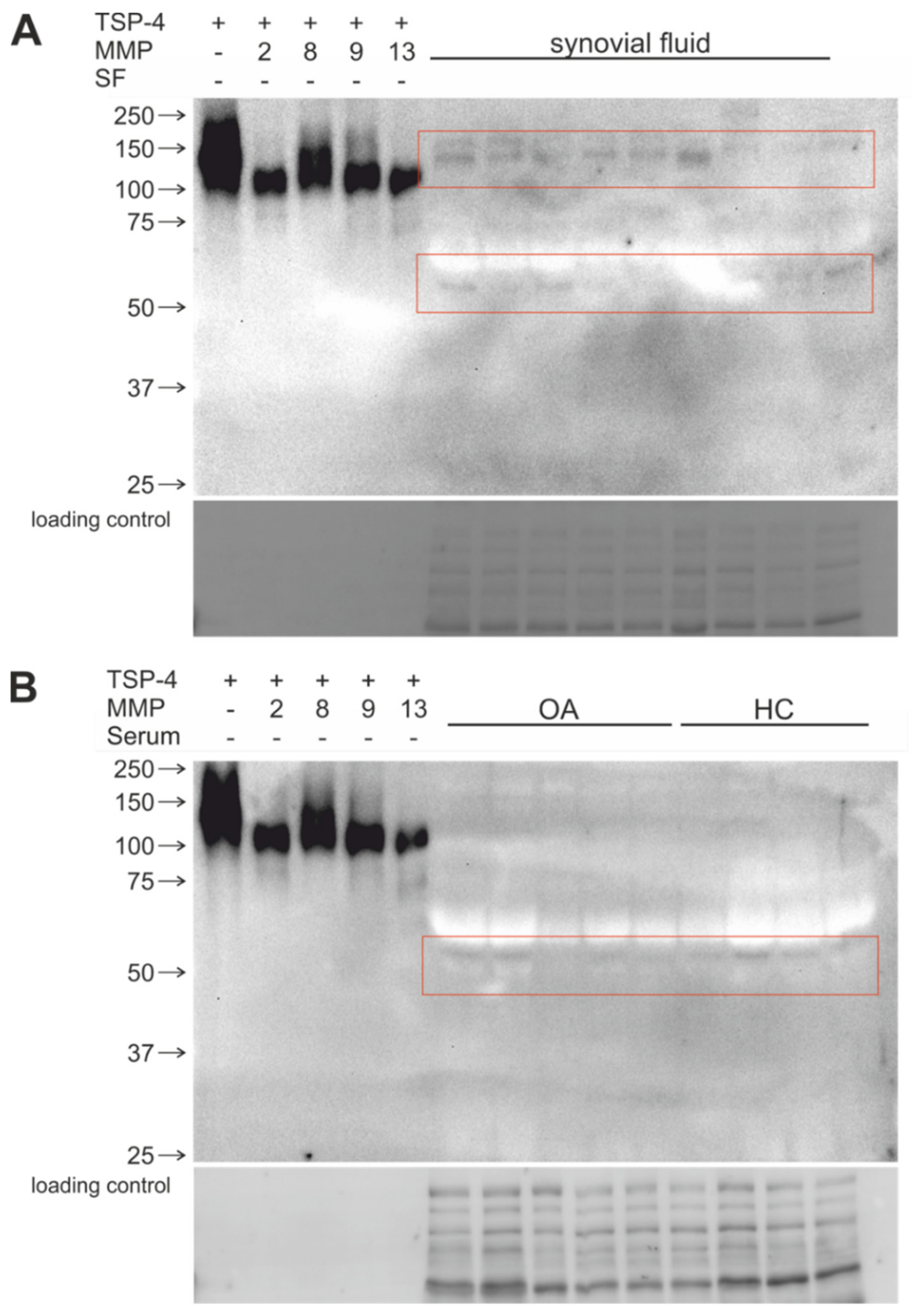
2.5. Stable COMP and TSP-4 Cleavage Products Could Be Detected in Human Synovial Fluid and Serum Samples
3. Discussion
4. Materials and Methods
4.1. Recombinant Expression and Protein Purification
4.2. In Vitro Activation of MMPs
4.3. Physiological MMP Concentration Ranges in Synovial Fluid
4.4. Digestion of COMP and TSP-4 with MMPs
4.5. Harvesting of Synovial Fluid and Serum Samples
4.6. SDS-Polyacrylamide Gel Electrophoresis and Immunoblot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAMTS | A disintegrin and metalloprotease with thrombospondin motifs |
| APMA | 4-aminophenylmercuric Acetate |
| COMP | Cartilage oligomeric matrix protein |
| DMEM/F-12 | Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 |
| ECM | Extracellular Matrix |
| ELISA | Enzyme-linked Immunosorbent Assay |
| FBS | Fetal bovine serum |
| HC | Healthy Individuals/Controls |
| HEK293 EBNA | Human Embryonic Kidney 293 Epstein-Barr Virus Nuclear Antigen |
| HRP | Horseradish peroxidase |
| IL-1β | Interleukin-1-beta |
| kDa | Kilo Dalton |
| LDS | Lithium Dodecyl Sulfate |
| MMP | Matrix metalloproteinase |
| NEM | N-Ethylmaleimide |
| OA | Osteoarthritis |
| PMSF | Phenylmethylsulfonylfluorid |
| PVDF | Polyvinylidene Fluoride |
| R | Concentration ranges |
| RT | Room temperature |
| SDS | Sodium dodecyl sulfate |
| SF | Synovial fluid |
| TBS | Tris-buffered saline |
| TBST | Tris-buffered saline with 0.1% 0Tween20 |
| TGF-β | Transforming growth factor beta |
| TNFα | Tumor necrosis factor alpha |
| TSP-4 | Thrombospondin-4 |
| TSP-5 | Thrombospondin-5 |
| TLR-2 | Toll-like receptor 2 |
References
- Stöcker, W.; Bode, W. Structural Features of a Superfamily of Zinc-Endopeptidases: The Metzincins. Curr. Opin. Struct. Biol. 1995, 5, 383–390. [Google Scholar] [CrossRef]
- Tokito, A.; Jougasaki, M. Matrix Metalloproteinases in Non-Neoplastic Disorders. Int. J. Mol. Sci 2016, 17, 1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, A.; Prado, A.F.; Antonio, R.C.; Issa, J.P.; Gerlach, R.F. Matrix Metalloproteinases Are Involved in Cardiovascular Diseases. Basic Clin. Pharmacol. Toxicol. 2014, 115, 301–314. [Google Scholar] [CrossRef]
- Wu, T.C.; Leu, H.B.; Lin, W.T.; Lin, C.P.; Lin, S.J.; Chen, J.W. Plasma Matrix Metalloproteinase-3 Level Is an Independent Prognostic Factor in Stable Coronary Artery Disease. Eur. J. Clin. Investig. 2005, 35, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Opincariu, D.; Nora, R.; Benedek, I. The Role of Matrix Metalloproteinases in the Progression and Vulnerabilization of Coronary Atherosclerotic Plaques. J. Cardiovasc. Emerg. 2021, 7, 9–16. [Google Scholar] [CrossRef]
- Muir, E.M.; Adcock, K.H.; Morgenstern, D.A.; Clayton, R.; von Stillfried, N.; Rhodes, K.; Ellis, C.; Fawcett, J.W.; Rogers, J.H. Matrix Metalloproteases and Their Inhibitors Are Produced by Overlapping Populations of Activated Astrocytes. Brain Res. Mol. Brain Res. 2002, 100, 103–117. [Google Scholar] [CrossRef]
- Deb, S.; Wenjun Zhang, J.; Gottschall, P.E. Beta-Amyloid Induces the Production of Active, Matrix-Degrading Proteases in Cultured Rat Astrocytes. Brain Res. 2003, 970, 205–213. [Google Scholar] [CrossRef]
- Łukaszewicz-Zając, M.; Mroczko, B.; Słowik, A. Matrix Metalloproteinases (MMPs) and Their Tissue Inhibitors (TIMPs) in Amyotrophic Lateral Sclerosis (ALS). J. Neural. Transm. 2014, 121, 1387–1397. [Google Scholar] [CrossRef] [Green Version]
- Mehana, E.-S.E.; Khafaga, A.F.; El-Blehi, S.S. The Role of Matrix Metalloproteinases in Osteoarthritis Pathogenesis: An Updated Review. Life Sci. 2019, 234, 116786. [Google Scholar] [CrossRef]
- Dimas, G.; Iliadis, F.; Grekas, D. Matrix Metalloproteinases, Atherosclerosis, Proteinuria and Kidney Disease: Linkage-Based Approaches. Hippokratia 2013, 17, 292–297. [Google Scholar]
- Conlon, G.A.; Murray, G.I. Recent Advances in Understanding the Roles of Matrix Metalloproteinases in Tumour Invasion and Metastasis. J. Pathol. 2019, 247, 629–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatti, O.; Vehviläinen, P.; Lehti, K.; Keski-Oja, J. MT1-MMP Releases Latent TGF-Beta1 from Endothelial Cell Extracellular Matrix via Proteolytic Processing of LTBP-1. Exp. Cell Res. 2008, 314, 2501–2514. [Google Scholar] [CrossRef] [PubMed]
- Van Lint, P.; Libert, C. Chemokine and Cytokine Processing by Matrix Metalloproteinases and Its Effect on Leukocyte Migration and Inflammation. J. Leukoc. Biol. 2007, 82, 1375–1381. [Google Scholar] [CrossRef] [Green Version]
- Van Den Steen, P.E.; Wuyts, A.; Husson, S.J.; Proost, P.; Van Damme, J.; Opdenakker, G. Gelatinase B/MMP-9 and Neutrophil Collagenase/MMP-8 Process the Chemokines Human GCP-2/CXCL6, ENA-78/CXCL5 and Mouse GCP-2/LIX and Modulate Their Physiological Activities. Eur. J. Biochem. 2003, 270, 3739–3749. [Google Scholar] [CrossRef]
- Veidal, S.S.; Larsen, D.V.; Chen, X.; Sun, S.; Zheng, Q.; Bay-Jensen, A.-C.; Leeming, D.J.; Nawrocki, A.; Larsen, M.R.; Schett, G.; et al. MMP Mediated Type V Collagen Degradation (C5M) Is Elevated in Ankylosing Spondylitis. Clin. Biochem. 2012, 45, 541–546. [Google Scholar] [CrossRef]
- Genovese, F.; Karsdal, M.A. Protein Degradation Fragments as Diagnostic and Prognostic Biomarkers of Connective Tissue Diseases: Understanding the Extracellular Matrix Message and Implication for Current and Future Serological Biomarkers. Expert Rev. Proteom. 2016, 13, 213–225. [Google Scholar] [CrossRef]
- Bay-Jensen, A.-C.; Liu, Q.; Byrjalsen, I.; Li, Y.; Wang, J.; Pedersen, C.; Leeming, D.J.; Dam, E.B.; Zheng, Q.; Qvist, P.; et al. Enzyme-Linked Immunosorbent Assay (ELISAs) for Metalloproteinase Derived Type II Collagen Neoepitope, CIIM--Increased Serum CIIM in Subjects with Severe Radiographic Osteoarthritis. Clin. Biochem. 2011, 44, 423–429. [Google Scholar] [CrossRef]
- Maquart, F.-X.; Pasco, S.; Ramont, L.; Hornebeck, W.; Monboisse, J.-C. An Introduction to Matrikines: Extracellular Matrix-Derived Peptides Which Regulate Cell Activity. Crit. Rev. Oncol./Hematol. 2004, 49, 199–202. [Google Scholar] [CrossRef]
- Pérez-García, S.; Carrión, M.; Gutiérrez-Cañas, I.; Villanueva-Romero, R.; Castro, D.; Martínez, C.; González-Álvaro, I.; Blanco, F.J.; Juarranz, Y.; Gomariz, R.P. Profile of Matrix-Remodeling Proteinases in Osteoarthritis: Impact of Fibronectin. Cells 2019, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Ricard-Blum, S.; Salza, R. Matricryptins and Matrikines: Biologically Active Fragments of the Extracellular Matrix. Exp. Dermatol. 2014, 23, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Heinegård, D.; Saxne, T. The Role of the Cartilage Matrix in Osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, Y.; Nakamura, H.; Obata, K.; Yamada, H.; Hayakawa, T.; Fujikawa, K.; Okada, Y. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Synovial Fluids from Patients with Rheumatoid Arthritis or Osteoarthritis. Ann. Rheum. Dis. 2000, 59, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Ecker, M. Overview of MMP-13 as a Promising Target for the Treatment of Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 1742. [Google Scholar] [CrossRef]
- Milaras, C.; Lepetsos, P.; Dafou, D.; Potoupnis, M.; Tsiridis, E. Association of Matrix Metalloproteinase (MMP) Gene Polymorphisms With Knee Osteoarthritis: A Review of the Literature. Cureus 2021, 13, e18607. [Google Scholar] [CrossRef] [PubMed]
- Heard, B.J.; Martin, L.; Rattner, J.B.; Frank, C.B.; Hart, D.A.; Krawetz, R. Matrix Metalloproteinase Protein Expression Profiles Cannot Distinguish between Normal and Early Osteoarthritic Synovial Fluid. BMC Musculoskelet. Disord. 2012, 13, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulejová, H.; Baresová, V.; Klézl, Z.; Polanská, M.; Adam, M.; Senolt, L. Increased Level of Cytokines and Matrix Metalloproteinases in Osteoarthritic Subchondral Bone. Cytokine 2007, 38, 151–156. [Google Scholar] [CrossRef]
- Tetlow, L.C.; Adlam, D.J.; Woolley, D.E. Matrix Metalloproteinase and Proinflammatory Cytokine Production by Chondrocytes of Human Osteoarthritic Cartilage: Associations with Degenerative Changes. Arthritis Rheum. 2001, 44, 585–594. [Google Scholar] [CrossRef]
- Ganu, V.; Goldberg, R.; Peppard, J.; Rediske, J.; Melton, R.; Hu, S.I.; Wang, W.; Duvander, C.; Heinegård, D. Inhibition of Interleukin-1alpha-Induced Cartilage Oligomeric Matrix Protein Degradation in Bovine Articular Cartilage by Matrix Metalloproteinase Inhibitors: Potential Role for Matrix Metalloproteinases in the Generation of Cartilage Oligomeric Matrix Protein Fragments in Arthritic Synovial Fluid. Arthritis Rheum. 1998, 41, 2143–2151. [Google Scholar] [CrossRef]
- Maly, K.; Schaible, I.; Riegger, J.; Brenner, R.E.; Meurer, A.; Zaucke, F. The Expression of Thrombospondin-4 Correlates with Disease Severity in Osteoarthritic Knee Cartilage. Int. J. Mol. Sci. 2019, 20, 447. [Google Scholar] [CrossRef] [Green Version]
- Maly, K.; Andres Sastre, E.; Farrell, E.; Meurer, A.; Zaucke, F. COMP and TSP-4: Functional Roles in Articular Cartilage and Relevance in Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 2242. [Google Scholar] [CrossRef] [PubMed]
- Koelling, S.; Clauditz, T.S.; Kaste, M.; Miosge, N. Cartilage Oligomeric Matrix Protein Is Involved in Human Limb Development and in the Pathogenesis of Osteoarthritis. Arthritis Res. Ther. 2006, 8, R56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernotiene, E.; Bagdonas, E.; Kirdaite, G.; Bernotas, P.; Kalvaityte, U.; Uzieliene, I.; Thudium, C.S.; Hannula, H.; Lorite, G.S.; Dvir-Ginzberg, M.; et al. Emerging Technologies and Platforms for the Immunodetection of Multiple Biochemical Markers in Osteoarthritis Research and Therapy. Front. Med. 2020, 7, 572977. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Meulenbelt, I.; Chassaing, E.; Arden, N.K.; Bierma-Zeinstra, S.; Hart, D.; Hofman, A.; Karsdal, M.; Kloppenburg, M.; Kroon, H.M.; et al. Large Scale Meta-Analysis of Urinary C-Terminal Telopeptide, Serum Cartilage Oligomeric Protein and Matrix Metalloprotease Degraded Type II Collagen and Their Role in Prevalence, Incidence and Progression of Osteoarthritis. Osteoarthr. Cartil. 2014, 22, 683–689. [Google Scholar] [CrossRef] [Green Version]
- Luan, Y.; Kong, L.; Howell, D.R.; Ilalov, K.; Fajardo, M.; Bai, X.-H.; Di Cesare, P.E.; Goldring, M.B.; Abramson, S.B.; Liu, C.-J. Inhibition of ADAMTS-7 and ADAMTS-12 Degradation of Cartilage Oligomeric Matrix Protein by Alpha-2-Macroglobulin. Osteoarthr. Cartil. 2008, 16, 1413–1420. [Google Scholar] [CrossRef] [Green Version]
- Lees, S.; Golub, S.B.; Last, K.; Zeng, W.; Jackson, D.C.; Sutton, P.; Fosang, A.J. Bioactivity in an Aggrecan 32-Mer Fragment Is Mediated via Toll-like Receptor 2. Arthritis Rheumatol. 2015, 67, 1240–1249. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Chen, C.T.; Bhargava, M.; Torzilli, P.A. A Comparative Study of Fibronectin Cleavage by MMP-1, -3, -13, and -14. Cartilage 2012, 3, 267–277. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.E.; Ishihara, S.; Tran, P.B.; Golub, S.B.; Last, K.; Miller, R.J.; Fosang, A.J.; Malfait, A.-M. An Aggrecan Fragment Drives Osteoarthritis Pain through Toll-like Receptor 2. JCI Insight 2018, 3, 95704. [Google Scholar] [CrossRef]
- Hwang, H.S.; Lee, M.H.; Kim, H.A. The TLR-2/TonEBP Signaling Pathway Regulates 29-KDa Fibronectin Fragment-Dependent Expression of Matrix Metalloproteinases. Sci. Rep. 2021, 11, 8891. [Google Scholar] [CrossRef]
- Thur, J.; Rosenberg, K.; Nitsche, D.P.; Pihlajamaa, T.; Ala-Kokko, L.; Heinegård, D.; Paulsson, M.; Maurer, P. Mutations in Cartilage Oligomeric Matrix Protein Causing Pseudoachondroplasia and Multiple Epiphyseal Dysplasia Affect Binding of Calcium and Collagen I, II, and IX. J. Biol. Chem. 2001, 276, 6083–6092. [Google Scholar] [CrossRef] [Green Version]
- Rübenhagen, R.; Schüttrumpf, J.P.; Stürmer, K.M.; Frosch, K.-H. Interleukin-7 Levels in Synovial Fluid Increase with Age and MMP-1 Levels Decrease with Progression of Osteoarthritis. Acta Orthop. 2012, 83, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Li, M.; Zeng, J.; Feng, Z.; Yang, J.; Shen, B.; Zeng, Y. Differential Expression of Renin-Angiotensin System-Related Components in Patients with Rheumatoid Arthritis and Osteoarthritis. Am. J. Med. Sci. 2020, 359, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Choi, H.M.; Lee, Y.-A.; Choi, I.A.; Lee, S.-H.; Hong, S.-J.; Yang, H.-I.; Yoo, M.C. Expression Levels and Association of Gelatinases MMP-2 and MMP-9 and Collagenases MMP-1 and MMP-13 with VEGF in Synovial Fluid of Patients with Arthritis. Rheumatol. Int. 2011, 31, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Shi, Q.; Chen, J.; Wang, H.; Wu, W.; Zha, Z. Expression and Significance of MMPs in Synovial Fluid, Serum and PBMC Culture Supernatant Stimulated by LPS in Osteoarthritis Patients With or Without Diabetes. Exp. Clin. Endocrinol. Diabetes 2019, 127, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhen, E.Y.; Brittain, I.J.; Laska, D.A.; Mitchell, P.G.; Sumer, E.U.; Karsdal, M.A.; Duffin, K.L. Characterization of Metalloprotease Cleavage Products of Human Articular Cartilage. Arthritis Rheum. 2008, 58, 2420–2431. [Google Scholar] [CrossRef] [PubMed]
- Neidhart, M.; Hauser, N.; Paulsson, M.; DiCesare, P.E.; Michel, B.A.; Häuselmann, H.J. Small Fragments of Cartilage Oligomeric Matrix Protein in Synovial Fluid and Serum as Markers for Cartilage Degradation. Br. J. Rheumatol. 1997, 36, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Firner, S.; Zaucke, F.; Heilig, J.; Marées, M.; Willwacher, S.; Brüggemann, G.; Niehoff, A. Impact of Knee Joint Loading on Fragmentation of Serum Cartilage Oligomeric Matrix Protein. J. Orthop. Res. 2020, 38, 1710–1718. [Google Scholar] [CrossRef] [Green Version]
- Lin, E.A.; Liu, C.-J. The Emerging Roles of ADAMTS-7 and ADAMTS-12 Matrix Metalloproteinases. Open Access Rheumatol. 2009, 1, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Gebauer, J.M.; Köhler, A.; Dietmar, H.; Gompert, M.; Neundorf, I.; Zaucke, F.; Koch, M.; Baumann, U. COMP and TSP-4 Interact Specifically with the Novel GXKGHR Motif Only Found in Fibrillar Collagens. Sci. Rep. 2018, 8, 17187. [Google Scholar] [CrossRef]
- Calamia, V.; Fernández-Puente, P.; Lourido, L.; Camacho, M.; González, L.; Blanco, F.J.; Ruiz-Romero, C. Development of a Multiplexed Assay for the Targeted Analysis of Cartilage Endogenous Peptides in Synovial Fluid and Serum from Osteoarthritis Patients. Osteoarthr. Cartil. 2016, 24, S160. [Google Scholar] [CrossRef] [Green Version]
- Li, K.-W.; Kim, D.-S.; Zaucke, F.; Luo, Z.D. Trigeminal Nerve Injury-Induced Thrombospondin-4 up-Regulation Contributes to Orofacial Neuropathic Pain States in a Rat Model. Eur. J. Pain 2014, 18, 489–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosby, N.D.; Zaucke, F.; Kras, J.V.; Dong, L.; Luo, Z.D.; Winkelstein, B.A. Thrombospondin-4 and Excitatory Synaptogenesis Promote Spinal Sensitization after Painful Mechanical Joint Injury. Exp. Neurol. 2015, 264, 111–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-S.; Li, K.-W.; Boroujerdi, A.; Peter Yu, Y.; Zhou, C.-Y.; Deng, P.; Park, J.; Zhang, X.; Lee, J.; Corpe, M.; et al. Thrombospondin-4 Contributes to Spinal Sensitization and Neuropathic Pain States. J. Neurosci. 2012, 32, 8977–8987. [Google Scholar] [CrossRef] [PubMed]
- Bay-Jensen, A.-C.; Andersen, T.L.; Charni-Ben Tabassi, N.; Kristensen, P.W.; Kjaersgaard-Andersen, P.; Sandell, L.; Garnero, P.; Delaissé, J.-M. Biochemical Markers of Type II Collagen Breakdown and Synthesis Are Positioned at Specific Sites in Human Osteoarthritic Knee Cartilage. Osteoarthr. Cartil. 2008, 16, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Kuhi, L.; Tamm, A.E.; Tamm, A.O.; Kisand, K. Risk Assessment of the Progression of Early Knee Osteoarthritis by Collagen Neoepitope C2C: A Longitudinal Study of an Estonian Middle-Aged Cohort. Diagnostics 2021, 11, 1236. [Google Scholar] [CrossRef]
- Xie, D.L.; Hui, F.; Meyers, R.; Homandberg, G.A. Cartilage Chondrolysis by Fibronectin Fragments Is Associated with Release of Several Proteinases: Stromelysin Plays a Major Role in Chondrolysis. Arch. Biochem. Biophys. 1994, 311, 205–212. [Google Scholar] [CrossRef]
- Klatt, A.R.; Klinger, G.; Paul-Klausch, B.; Kühn, G.; Renno, J.H.; Wagener, R.; Paulsson, M.; Schmidt, J.; Malchau, G.; Wielckens, K. Matrilin-3 Activates the Expression of Osteoarthritis-Associated Genes in Primary Human Chondrocytes. FEBS Lett. 2009, 583, 3611–3617. [Google Scholar] [CrossRef] [Green Version]
- Andrés Sastre, E.; Zaucke, F.; Witte-Bouma, J.; van Osch, G.J.V.M.; Farrell, E. Cartilage Oligomeric Matrix Protein–Derived Peptides Secreted by Cartilage Do Not Induce Responses Commonly Observed during Osteoarthritis. Cartilage 2021, 13, 1229S–1236S. [Google Scholar] [CrossRef]
- Kohfeldt, E.; Maurer, P.; Vannahme, C.; Timpl, R. Properties of the Extracellular Calcium Binding Module of the Proteoglycan Testican. FEBS Lett. 1997, 414, 557–561. [Google Scholar] [CrossRef]
- Smyth, N.; Odenthal, U.; Merkl, B.; Paulsson, M. Eukaryotic Expression and Purification of Recombinant Extracellular Matrix Proteins Carrying the Strep II Tag. In Extracellular Matrix Protocols; Even-Ram, S., Artym, V., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 522, pp. 63–72. ISBN 978-1-58829-984-0. [Google Scholar]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a Human Cell Line Transformed by DNA from Human Adenovirus Type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Liu, M.; Wang, S.; Zhang, H.; Zhao, Y. Cloning, Expression, Purification, and Characterization of the Catalytic Domain of Sika Deer MMP-13. Protein Expr. Purif. 2016, 127, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Ladner-Keay, C.L.; Turner, R.J.; Edwards, R.A. Fluorescent Protein Visualization Immediately After Gel Electrophoresis Using an In-Gel Trichloroethanol Photoreaction with Tryptophan. In Protein Gel Detection and Imaging; Kurien, B.T., Scofield, R.H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1853, pp. 179–190. ISBN 978-1-4939-8744-3. [Google Scholar]
- Gürtler, A.; Kunz, N.; Gomolka, M.; Hornhardt, S.; Friedl, A.A.; McDonald, K.; Kohn, J.E.; Posch, A. Stain-Free Technology as a Normalization Tool in Western Blot Analysis. Anal. Biochem. 2013, 433, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, E.T.; Zaucke, F.; Clegg, D.O. Thrombospondin-4 and Matrix Three-Dimensionality in Axon Outgrowth and Adhesion in the Developing Retina. Exp. Eye Res. 2007, 84, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Spitznagel, L.; Nitsche, D.P.; Paulsson, M.; Maurer, P.; Zaucke, F. Characterization of a Pseudoachondroplasia-Associated Mutation (His587 Arg) in the C-Terminal, Collagen-Binding Domain of Cartilage Oligomeric Matrix Protein (COMP). Biochem. J. 2004, 377, 479–487. [Google Scholar] [CrossRef] [Green Version]

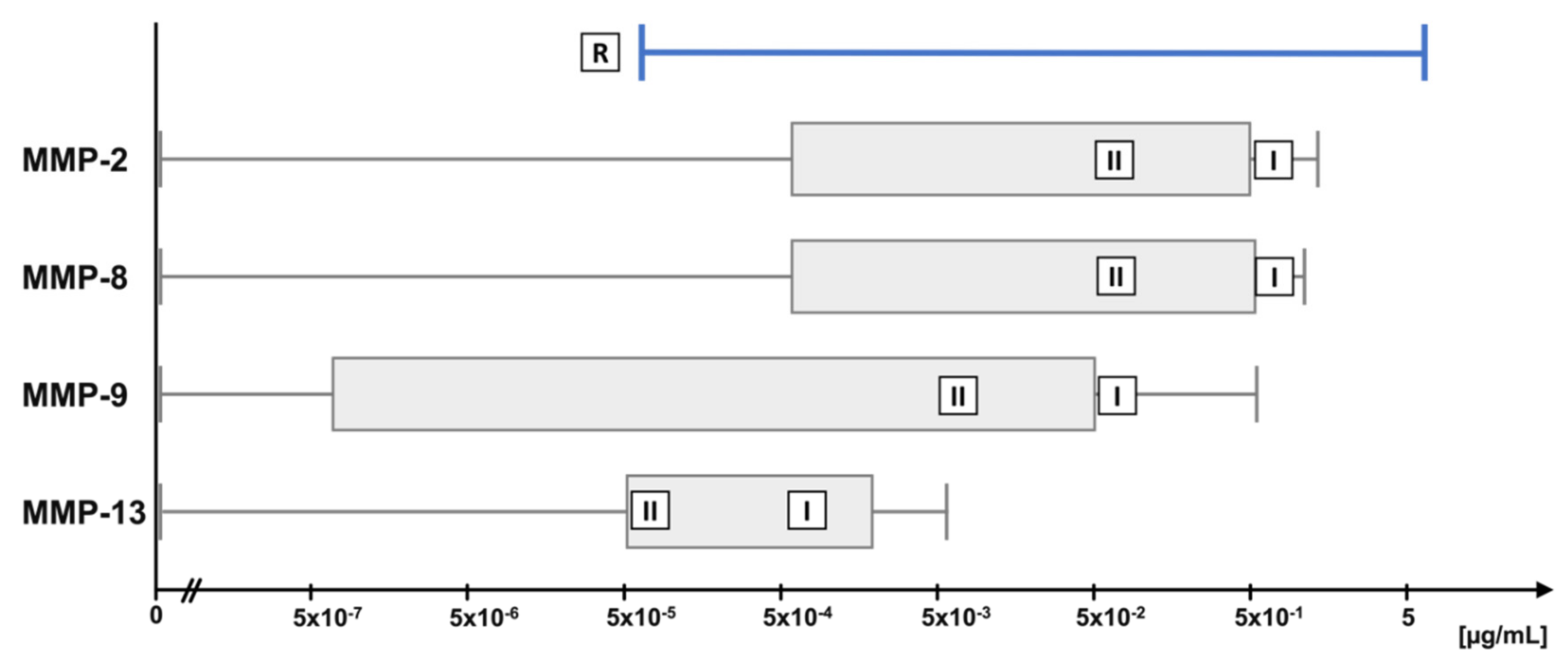
| MMP | Low Concentration (µg/mL) | High Concentration (µg/mL) |
|---|---|---|
| MMP-2 | 5 × 10−2 | 5 × 10−1 |
| MMP-8 | 5 × 10−2 | 5 × 10−1 |
| MMP-9 | 5 × 10−3 | 5 × 10−2 |
| MMP-13 | 5 × 10−5 | 5 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, N.; Rapp, A.E.; Braun, S.; Ehnert, M.; Imhof, T.; Koch, M.; Jenei-Lanzl, Z.; Zaucke, F.; Meurer, A. Generation of Matrix Degradation Products Using an In Vitro MMP Cleavage Assay. Int. J. Mol. Sci. 2022, 23, 6245. https://doi.org/10.3390/ijms23116245
Wagner N, Rapp AE, Braun S, Ehnert M, Imhof T, Koch M, Jenei-Lanzl Z, Zaucke F, Meurer A. Generation of Matrix Degradation Products Using an In Vitro MMP Cleavage Assay. International Journal of Molecular Sciences. 2022; 23(11):6245. https://doi.org/10.3390/ijms23116245
Chicago/Turabian StyleWagner, Niklas, Anna E. Rapp, Sebastian Braun, Markus Ehnert, Thomas Imhof, Manuel Koch, Zsuzsa Jenei-Lanzl, Frank Zaucke, and Andrea Meurer. 2022. "Generation of Matrix Degradation Products Using an In Vitro MMP Cleavage Assay" International Journal of Molecular Sciences 23, no. 11: 6245. https://doi.org/10.3390/ijms23116245






