Systematic Investigations on the Metabolic and Transcriptomic Regulation of Lactate in the Human Colon Epithelial Cells
Abstract
:1. Introduction
2. Results
2.1. The Effect of Lactate on Caco-2 Cells Growth
2.2. Metabolite Assignments for the 1H NMR Spectroscopy
2.3. Significantly Changed Metabolites Detected by High-Resolution 1H NMR
2.4. Lactate Modulated Transcriptome Signatures in Human Colon Epithelial Cells
3. Discussion
4. Materials and Methods
4.1. Caco-2 Cells Culture and the Growth Curve
4.2. RNA Sequencing (RNA-Seq)
4.3. Metabolism Sample Collection and Preparation
4.4. NMR Experiment and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Caco-2 | Human colon epithelial cells; |
| NMR | Nuclear Magnetic Resonance; |
| HDAC | Histone deacetylase activity; |
| GPR81 | G protein-coupled receptor 81; |
| TNF | Tumor Necrosis Factor; |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; |
| PCA | Principal component analysis; |
| OPLS-DA | Orthogonal projection to the latent structures discriminant analysis; |
| SAM | S-adenosyl-methionine; |
| ROS | Reactive oxygen species; |
| ETC | Electron transport chain; |
| NADH | Nicotinamide adenine dinucleotide; |
| MAT | Methionine adenosyltransferase; |
| DMNT | DNA methyltransferase; |
| UHRF2 | E3 ubiquitin-protein ligase; |
| SLCs | Solute carriers; |
| DEGs | Differentially expressed genes; |
| NOESY | Nuclear Overhauser effect spectroscopy; |
| JRES | J-resolved spectroscopy; |
| 1H−1H COSY | 1H−1H correlation spectroscopy; |
| 1H−1H TOCSY | 1H−1H total correlation spectroscopy; |
| 1H-13C HSQC | 1H-13C heteronuclear single quantum coherence spectroscopy; |
| 1H-13C HMBC | 1H-13C heteronuclear multiple bond correlation spectroscopy; |
| OXPHOS | Oxidative phosphorylation; |
| PA | Pantothenate; |
| Ile | Isoleucine; |
| Leu | Leucine; |
| Val | Valine; |
| α-KIV | α-Keto-isovalerate; |
| α-KMV | α-keto-beta-methyl-valerate; |
| Lac | Lactate; |
| Ala | Alanine; |
| Thr | Threonine; |
| Glu | Glutamate; |
| Gln | Glutamine; |
| NAG | N-Acetyl-Glutamine; |
| Suc | Succinate; |
| Met | Methionine; |
| DMG | Dimethylglycine; |
| Cre | Creatine; |
| Cho | Choline; |
| PC | Phosphocholine; |
| GPC | Glycerophosphocholine; |
| Tau | Taurine; |
| GSH | Glutathione; |
| Glc | Glucose; |
| Fru | Fructose; |
| Gal-1-P | Galactose-1-phosphate; |
| Tyr | Tyrosine; |
| Phe | Phenylalanine; |
| Fum | Fumaric acid; |
| NAD+ | Nicotinamide adenine dinucleotide; |
| FA | Formate; |
| His | Histidine; |
| AMP | Adenosine monophosphate; |
| ATP | Adenosine triphosphate; |
| UMP | Uridine 5′-monophosphate; |
| UDP | Uridine-5′-diphosphate; |
| UDPG | UDP Glucuronate; |
| UDP-N-Ace | UDP-N-acetylglucosamine. |
References
- Gladden, L. Lactate metabolism: A new paradigm for the third millennium. J. Physiol. 2004, 558, 5–30. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, M.; Albertsen, J.; Rentsch, M.; Juel, C. Lactate and force production in skeletal muscle. J. Physiol. 2005, 562, 521–526. [Google Scholar] [CrossRef]
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [PubMed]
- Husain, Z.; Huang, Y.; Seth, P.; Sukhatme, V.P. Tumor-derived lactate modifies antitumor immune response: Effect on myeloid-derived suppressor cells and NK cells. J. Immunol. Res. 2013, 191, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Heim, C.E.; Bosch, M.E.; Yamada, K.J.; Aldrich, A.L.; Chaudhari, S.S.; Klinkebiel, D.; Gries, C.M.; Alqarzaee, A.A.; Li, Y.; Thomas, V.C. Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol. 2020, 5, 1271–1284. [Google Scholar] [CrossRef] [PubMed]
- Romero-Garcia, S.; Moreno-Altamirano, M.M.B.; Prado-Garcia, H.; Sánchez-García, F.J. Lactate contribution to the tumor microenvironment: Mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol. 2016, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Korol, D.L.; Gardner, R.S.; Tunur, T.; Gold, P.E. Involvement of lactate transport in two object recognition tasks that require either the hippocampus or striatum. Behav. Neurosci. 2019, 133, 176. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Sattler, U.G.; Mueller-Klieser, W. Lactate: A metabolic key player in cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [Green Version]
- Brown, T.P.; Bhattacharjee, P.; Ramachandran, S.; Sivaprakasam, S.; Ristic, B.; Sikder, M.O.F.; Ganapathy, V. The lactate receptor GPR81 promotes breast cancer growth via a paracrine mechanism involving antigen-presenting cells in the tumor microenvironment. Oncogene 2020, 39, 3292–3304. [Google Scholar]
- Matsuki, T.; Pédron, T.; Regnault, B.; Mulet, C.; Hara, T.; Sansonetti, P.J. Epithelial cell proliferation arrest induced by lactate and acetate from Lactobacillus casei and Bifidobacterium breve. PLoS ONE 2013, 8, e63053. [Google Scholar]
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.; Martínez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.T.; Lacroix, C.; Braegger, C.P.; Chassard, C. Lactate-utilizing community is associated with gut microbiota dysbiosis in colicky infants. Sci. Rep. 2017, 7, 11176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, L.T. Breast and gut: The relationship between lactating mammary function and neonatal gastrointestinal function. Proc. Nutr. Soc. 1992, 51, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, B.; Ai, Z.; Zeng, S.; Song, Y.; Song, J.; Zeng, Q.; Liao, Z.; Wang, T.; Huang, C.; Su, D. Gut microbiome-derived lactate promotes to anxiety-like behaviors through GPR81 receptor-mediated lipid metabolism pathway. Psychoneuroendocrinology 2020, 117, 104699. [Google Scholar] [CrossRef] [PubMed]
- Phypers, B.; Pierce, J.T. Lactate physiology in health and disease. Contin. Educ. Anaesth. Crit. Care Pain 2006, 6, 128–132. [Google Scholar]
- Garrote, G.L.; Abraham, A.G.; Rumbo, M. Is lactate an undervalued functional component of fermented food products? Front. Microbiol. 2015, 6, 629. [Google Scholar] [CrossRef]
- Latham, T.; Mackay, L.; Sproul, D.; Karim, M.; Culley, J.; Harrison, D.J.; Hayward, L.; Langridge-Smith, P.; Gilbert, N.; Ramsahoye, B.H. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012, 40, 4794–4803. [Google Scholar] [CrossRef] [Green Version]
- Brown, T.P.; Ganapathy, V. Lactate/GPR81 signaling and proton motive force in cancer: Role in angiogenesis, immune escape, nutrition, and Warburg phenomenon. Pharmacol. Ther. 2020, 206, 107451. [Google Scholar] [CrossRef]
- Roland, C.L.; Arumugam, T.; Deng, D.; Liu, S.H.; Philip, B.; Gomez, S.; Burns, W.R.; Ramachandran, V.; Wang, H.; Cruz-Monserrate, Z. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res. 2014, 74, 5301–5310. [Google Scholar] [CrossRef] [Green Version]
- Ippolito, L.; Morandi, A.; Giannoni, E.; Chiarugi, P. Lactate: A metabolic driver in the tumour landscape. Trends Biochem. Sci. 2019, 44, 153–166. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.A. The science and translation of lactate shuttle theory. Cell Metab. 2018, 27, 757–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villamil, M.F.; Ordieres, A.M.; Blanco, P.T. Immobilized enzyme electrode for the determination of L-lactate in food samples. Anal. Chim. Acta 1997, 345, 37–43. [Google Scholar] [CrossRef]
- Shapiro, F.; Silanikove, N. Rapid and accurate determination of D-and L-lactate, lactose and galactose by enzymatic reactions coupled to formation of a fluorochromophore: Applications in food quality control. Food Chem. 2010, 119, 829–833. [Google Scholar] [CrossRef]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef] [Green Version]
- Solligård, E.; Juel, I.S.; Spigset, O.; Romundstad, P.; Grønbech, J.E.; Aadahl, P. Gut luminal lactate measured by microdialysis mirrors permeability of the intestinal mucosa after ischemia. Shock 2008, 29, 245–251. [Google Scholar] [CrossRef]
- Ogawa, K.; Ben, R.A.; Pons, S.; de Paolo, M. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J. Pediatr. Gastroenterol. Nutr. 1992, 15, 248–252. [Google Scholar] [CrossRef]
- Vernia, P.; Caprilli, R.; Latella, G.; Barbetti, F.; Magliocca, F.M.; Cittadini, M. Fecal lactate and ulcerative colitis. Gastroenterology 1988, 95, 1564–1568. [Google Scholar] [CrossRef]
- Hove, H.; Nordgaard-Andersen, I.; Mortensen, P.B. Faecal DL-lactate concentration in 100 gastrointestinal patients. Scand. J. Gastroenterol. 1994, 29, 255–259. [Google Scholar] [CrossRef]
- Samuvel, D.J.; Sundararaj, K.P.; Nareika, A.; Lopes-Virella, M.F.; Huang, Y. Lactate boosts TLR4 signaling and NF-κB pathway-mediated gene transcription in macrophages via monocarboxylate transporters and MD-2 up-regulation. J. Immunol. Res. 2009, 182, 2476–2484. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Li, H.; Chen, J.; Qian, Q. Lactic acid: No longer an inert and end-product of glycolysis. Physiology 2017, 32, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Dienel, G.A. Brain lactate metabolism: The discoveries and the controversies. J. Cereb. Blood Flow Metab. 2012, 32, 1107–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, G.; Hassan, I.; Dix, C.; Williamson, I.; Shah, R.; Mackay, M.; Artursson, P. Transport and permeability properties of human Caco-2 cells: An in vitro model of the intestinal epithelial cell barrier. J. Control. Release 1990, 11, 25–40. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borgan, E.; Sitter, B.; Lingjærde, O.C.; Johnsen, H.; Lundgren, S.; Bathen, T.F.; Sørlie, T.; Børresen-Dale, A.-L.; Gribbestad, I.S. Merging transcriptomics and metabolomics-advances in breast cancer profiling. BMC Cancer 2010, 10, 628. [Google Scholar] [CrossRef] [Green Version]
- Viguerie, N.; Poitou, C.; Cancello, R.; Stich, V.; Clement, K.; Langin, D. Transcriptomics applied to obesity and caloric restriction. Biochimie 2005, 87, 117–123. [Google Scholar] [CrossRef]
- Kuehnbaum, N.L.; Britz-McKibbin, P. New advances in separation science for metabolomics: Resolving chemical diversity in a post-genomic era. Chem. Rev. 2013, 113, 2437–2468. [Google Scholar] [CrossRef]
- Manson, J.M.; Rauch, M.; Gilmore, M.S. The commensal microbiology of the gastrointestinal tract. GI Microbiota Regul. Immune Syst. 2008, 635, 15–28. [Google Scholar]
- Sumei, S.; Xiangyun, K.; Fenrong, C.; Xueguang, S.; Sijun, H.; Bin, B.; Xiaolei, S.; Yongjiu, T.; Kaichun, W.; Qingchuan, Z. Hypermethylation of DHRS3 as a Novel Tumor Suppressor Involved in Tumor Growth and Prognosis in Gastric Cancer. Front. Cell Dev. Biol 2021, 11. [Google Scholar] [CrossRef]
- Fraga, M.; Perelmuter, K.; Delucchi, L.; Cidade, E.; Zunino, P. Vaginal lactic acid bacteria in the mare: Evaluation of the probiotic potential of native Lactobacillus spp. and Enterococcus spp. strains. Antonie Van Leeuwenhoek 2008, 93, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Dietl, K.; Renner, K.; Dettmer, K.; Timischl, B.; Eberhart, K.; Dorn, C.; Hellerbrand, C.; Kastenberger, M.; Kunz-Schughart, L.A.; Oefner, P.J. Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J. Immunol. Res. 2010, 184, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- Belenguer, A.; Duncan, S.H.; Holtrop, G.; Anderson, S.E.; Lobley, G.E.; Flint, H.J. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl. Environ. Microbiol. 2007, 73, 6526–6533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra-and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Li, T.; Wernersson, R.; Hansen, R.B.; Horn, H.; Mercer, J.; Slodkowicz, G.; Workman, C.T.; Rigina, O.; Rapacki, K.; Stærfeldt, H.H. A scored human protein–protein interaction network to catalyze genomic interpretation. Nat. Methods 2017, 14, 61–64. [Google Scholar] [CrossRef] [Green Version]
- Zhao, R.Z.; Jiang, S.; Zhang, L.; Yu, Z.B. Mitochondrial electron transport chain, ROS generation and uncoupling. Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Tauffenberger, A.; Fiumelli, H.; Almustafa, S.; Magistretti, P.J. Lactate and pyruvate promote cellular stress resistance and longevity through ROS signaling. Cell Death Dis. 2019, 10, 542316. [Google Scholar] [CrossRef]
- Zelenka, J.; Dvořák, A.; Alán, L. L-lactate protects skin fibroblasts against aging-associated mitochondrial dysfunction via mitohormesis. Oxid. Med. Cell. Longev. 2015, 2015, 351698. [Google Scholar] [CrossRef] [Green Version]
- Galardo, M.N.; Regueira, M.; Riera, M.F.; Pellizzari, E.H.; Cigorraga, S.B.; Meroni, S.B. Lactate regulates rat male germ cell function through reactive oxygen species. PLoS ONE 2014, 9, e88024. [Google Scholar] [CrossRef] [Green Version]
- Vinogradov, A.D.; Grivennikova, V.G. Oxidation of NADH and ROS production by respiratory complex I. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 863–871. [Google Scholar] [CrossRef]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef] [PubMed]
- Balestrino, M.; Adriano, E. Creatine as a candidate to prevent statin myopathy. Biomolecules 2019, 9, 496. [Google Scholar] [CrossRef] [Green Version]
- Das, B.; Srivastava, S.K. Activation of aldose reductase from human tissues. Diabetes 1985, 34, 1145–1151. [Google Scholar] [CrossRef]
- González, R.G.; Barnett, P.; Aguayo, J.; Cheng, H.-M.; Chylack, L. Direct measurement of polyol pathway activity in the ocular lens. Diabetes 1984, 33, 196–199. [Google Scholar] [CrossRef] [Green Version]
- Liemburg-Apers, D.C.; Willems, P.H.; Koopman, W.J.; Grefte, S. Interactions between mitochondrial reactive oxygen species and cellular glucose metabolism. Arch. Toxicol. 2015, 89, 1209–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.J. Redox imbalance stress in diabetes mellitus: Role of the polyol pathway. Anim. Models Exp. Med. 2018, 1, 7–13. [Google Scholar] [CrossRef]
- Schenz, J.; Heilig, L.; Lohse, T.; Tichy, L.; Bomans, K.; Büttner, M.; Weigand, M.A.; Uhle, F. Extracellular Lactate Acts as a Metabolic Checkpoint and Shapes Monocyte Function Time Dependently. Front. Immunol. 2021, 12, 729209. [Google Scholar] [CrossRef]
- Hwang, J.J.; Jiang, L.; Hamza, M.; Dai, F.; Belfort-DeAguiar, R.; Cline, G.; Rothman, D.L.; Mason, G.; Sherwin, R.S. The human brain produces fructose from glucose. JCI Insight 2017, 2, e90508. [Google Scholar] [CrossRef] [PubMed]
- Barnett, P.A.; González, R.G.; Chylack, L.T., Jr.; Cheng, H.-M. The effect of oxidation on sorbitol pathway kinetics. Diabetes 1986, 35, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Chandra, D.; Jackson, E.B.; Ramana, K.V.; Kelley, R.; Srivastava, S.K.; Bhatnagar, A. Nitric oxide prevents aldose reductase activation and sorbitol accumulation during diabetes. Diabetes 2002, 51, 3095–3101. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, J.H.; Merola, L.O.; Satoh, K.; Dikmak, E. Osmotic changes caused by the accumulation of dulcitol in the lenses of rats fed with galactose. Nature 1962, 194, 1085–1087. [Google Scholar] [PubMed]
- Tang, W.; Martin, K.A.; Hwa, J. Aldose reductase, oxidative stress, and diabetic mellitus. Front. Pharmacol. 2012, 3, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyama, T.; Miyasita, Y.; Watanabe, H.; Shirai, K. The role of polyol pathway in high glucose-induced endothelial cell damages. Diabetes Res. Clin. Pract. 2006, 73, 227–234. [Google Scholar] [PubMed]
- Clare, C.E.; Brassington, A.H.; Kwong, W.Y.; Sinclair, K.D. One-carbon metabolism: Linking nutritional biochemistry to epigenetic programming of long-term development. Annu. Rev. Anim. Biosci. 2019, 7, 263–287. [Google Scholar] [PubMed]
- Chiang, P.K.; Gordon, R.K.; Tal, J.; Zeng, G.; Doctor, B.; Pardhasaradhi, K.; McCann, P.P. S-Adenosylmetliionine and methylation. FASEB J. 1996, 10, 471–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramani, K.; Mato, J.M.; Lu, S.C. Role of methionine adenosyltransferase genes in hepatocarcinogenesis. Cancers 2011, 3, 1480–1497. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.A.; Weinshilboum, R.M. Rabbit lung indolethylamine N-methyltransferase: cDNA and gene cloning and characterization. J. Biol. Chem. 1998, 273, 34502–34510. [Google Scholar] [CrossRef] [Green Version]
- Jacob, M.S.; Presti, D.E. Endogenous psychoactive tryptamines reconsidered: An anxiolytic role for dimethyltryptamine. Med. Hypotheses 2005, 64, 930–937. [Google Scholar]
- Pichler, G.; Wolf, P.; Schmidt, C.S.; Meilinger, D.; Schneider, K.; Frauer, C.; Fellinger, K.; Rottach, A.; Leonhardt, H. Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J. Cell. Biochem. 2011, 112, 2585–2593. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, B.; Kuang, H.; Korakavi, G.; Lu, L.-Y.; Yu, X. Zinc finger protein 618 regulates the function of UHRF2 (Ubiquitin-like with PHD and ring finger domains 2) as a specific 5-hydroxymethylcytosine reader. J. Biol. Chem. 2016, 291, 13679–13688. [Google Scholar] [CrossRef] [Green Version]
- Engedal, N.; Žerovnik, E.; Rudov, A.; Galli, F.; Olivieri, F.; Procopio, A.D.; Rippo, M.R.; Monsurrò, V.; Betti, M.; Albertini, M.C. From oxidative stress damage to pathways, networks, and autophagy via microRNAs. Oxid. Med. Cell. Longev. 2018, 2018, 4968321. [Google Scholar] [CrossRef] [PubMed]
- Conomos, D.; Reddel, R.R.; Pickett, H.A. NuRD–ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 2014, 21, 760–770. [Google Scholar] [PubMed]
- Schilderink, R.; Verseijden, C.; de Jonge, W.J. Dietary inhibitors of histone deacetylases in intestinal immunity and homeostasis. Front. Immunol. 2013, 4, 226. [Google Scholar]
- Belenguer, A.; Holtrop, G.; Duncan, S.H.; Anderson, S.E.; Calder, A.G.; Flint, H.J.; Lobley, G.E. Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol. Ecol. 2011, 77, 107–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kee, H.J.; Kook, H. Krüppel-like factor 4 mediates histone deacetylase inhibitor-induced prevention of cardiac hypertrophy. J. Mol. Cell. Cardiol. 2009, 47, 770–780. [Google Scholar] [CrossRef]
- Wagner, W.; Ciszewski, W.M.; Kania, K.D. L-and D-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun. Signal. 2015, 13, 36. [Google Scholar]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias-Guiu, X. NF-kB in development and progression of human cancer. Virchows Arch. 2005, 446, 475–482. [Google Scholar]
- Arjunan, P.; Meghil, M.M.; Pi, W.; Xu, J.; Lang, L.; El-Awady, A.; Sullivan, W.; Rajendran, M.; Rabelo, M.S.; Wang, T. Oral pathobiont activates anti-apoptotic pathway, promoting both immune suppression and oncogenic cell proliferation. Sci. Rep. 2018, 8, 16607. [Google Scholar]
- Kamejima, S.; Tatsumi, N.; Anraku, A.; Suzuki, H.; Ohkido, I.; Yokoo, T.; Okabe, M. Gcm1 is involved in cell proliferation and fibrosis during kidney regeneration after ischemia–reperfusion injury. Sci. Rep. 2019, 9, 7883. [Google Scholar]
- Wang, Z.; Reinach, P.S.; Zhang, F.; Vellonen, K.-S.; Urtti, A.; Turner, H.; Wolosin, J.M. DUSP5 and DUSP6 modulate corneal epithelial cell proliferation. Mol. Vis. 2010, 16, 1696. [Google Scholar]
- Hughes, T.K.; Wadsworth, M.H.; Gierahn, T.M.; Do, T.; Weiss, D.; Andrade, P.R.; Ma, F.; de Andrade Silva, B.J.; Shao, S.; Tsoi, L.C. Highly efficient, massively-parallel single-cell RNA-seq reveals cellular states and molecular features of human skin pathology. BioRxiv 2019, 689273. [Google Scholar] [CrossRef] [Green Version]
- Kotolloshi, R.; Hölzer, M.; Gajda, M.; Grimm, M.-O.; Steinbach, D. SLC35F2, a Transporter Sporadically Mutated in the Untranslated Region, Promotes Growth, Migration, and Invasion of Bladder Cancer Cells. Cells 2021, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Thanan, R.; Kaewlert, W.; Sakonsinsiri, C.; Chaiprasert, T.; Armartmuntree, N.; Muengsaen, D.; Techasen, A.; Klanrit, P.; Lert-Itthiporn, W.; Pinlaor, S. Opposing roles of FoxA1 and FoxA3 in intrahepatic cholangiocarcinoma progression. Int. J. Mol. Sci. 2020, 21, 1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Xu, J.; Liang, C.; Meng, Q.; Hua, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Emerging roles of the solute carrier family in pancreatic cancer. Clin. Transl. Med. 2021, 11, e356. [Google Scholar] [CrossRef]
- Schumann, T.; König, J.; Henke, C.; Willmes, D.M.; Bornstein, S.R.; Jordan, J.; Fromm, M.F.; Birkenfeld, A.L. Solute carrier transporters as potential targets for the treatment of metabolic disease. Pharmacol. Rev. 2020, 72, 343–379. [Google Scholar] [CrossRef]
- Pramod, A.B.; Foster, J.; Carvelli, L.; Henry, L.K. SLC6 transporters: Structure, function, regulation, disease association and therapeutics. Mol. Aspects Med. 2013, 34, 197–219. [Google Scholar] [CrossRef] [Green Version]
- Guarch, M.E.; Font-Llitjós, M.; Murillo-Cuesta, S.; Errasti-Murugarren, E.; Celaya, A.M.; Girotto, G.; Vuckovic, D.; Mezzavilla, M.; Vilches, C.; Bodoy, S. Mutations in L-type amino acid transporter-2 support SLC7A8 as a novel gene involved in age-related hearing loss. Elife 2018, 7, e31511. [Google Scholar] [CrossRef]
- Bahuguna, A.; Khan, I.; Bajpai, V.K.; Kang, S.C. MTT assay to evaluate the cytotoxic potential of a drug. Bangladesh J. Pharmacol. 2017, 12, 115–118. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. BioRxiv 2021, 060012. [Google Scholar] [CrossRef] [Green Version]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Luo, Y.; Geng, N.; Zhang, B.; Chen, J.; Zhang, H. Effects of harvesting and extraction methods on metabolite recovery from adherently growing mammalian cells. Anal. Methods 2020, 12, 2491–2498. [Google Scholar] [CrossRef] [PubMed]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Lindgren, F.; Hansen, B.; Karcher, W.; Sjöström, M.; Eriksson, L. Model validation by permutation tests: Applications to variable selection. J. Chemom. 1996, 10, 521–532. [Google Scholar] [CrossRef]
- Cloarec, O.; Dumas, M.E.; Trygg, J.; Craig, A.; Barton, R.H.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal. Chem. 2005, 77, 517–526. [Google Scholar] [CrossRef]


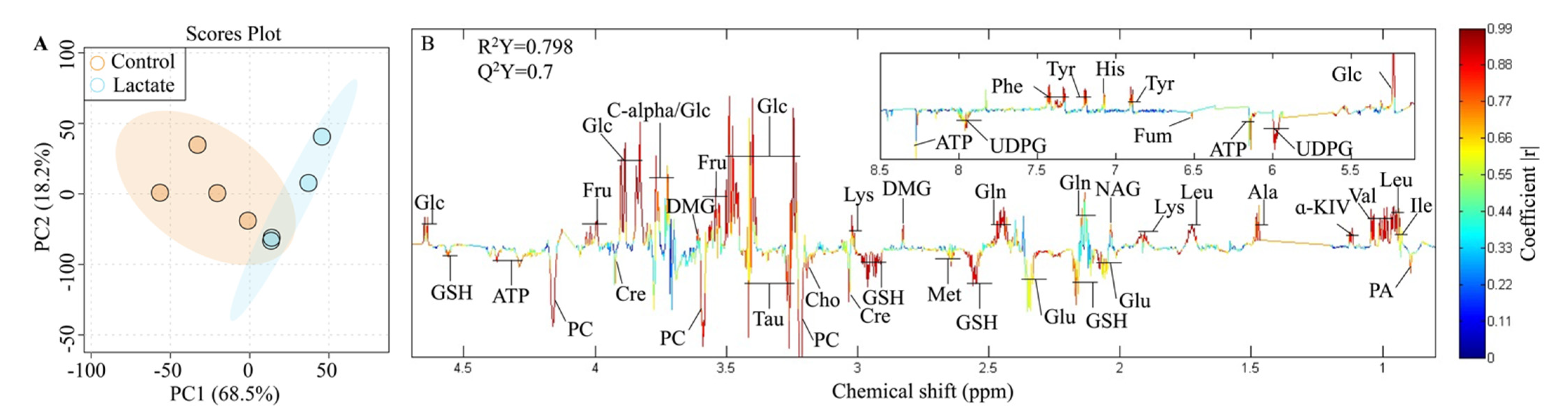
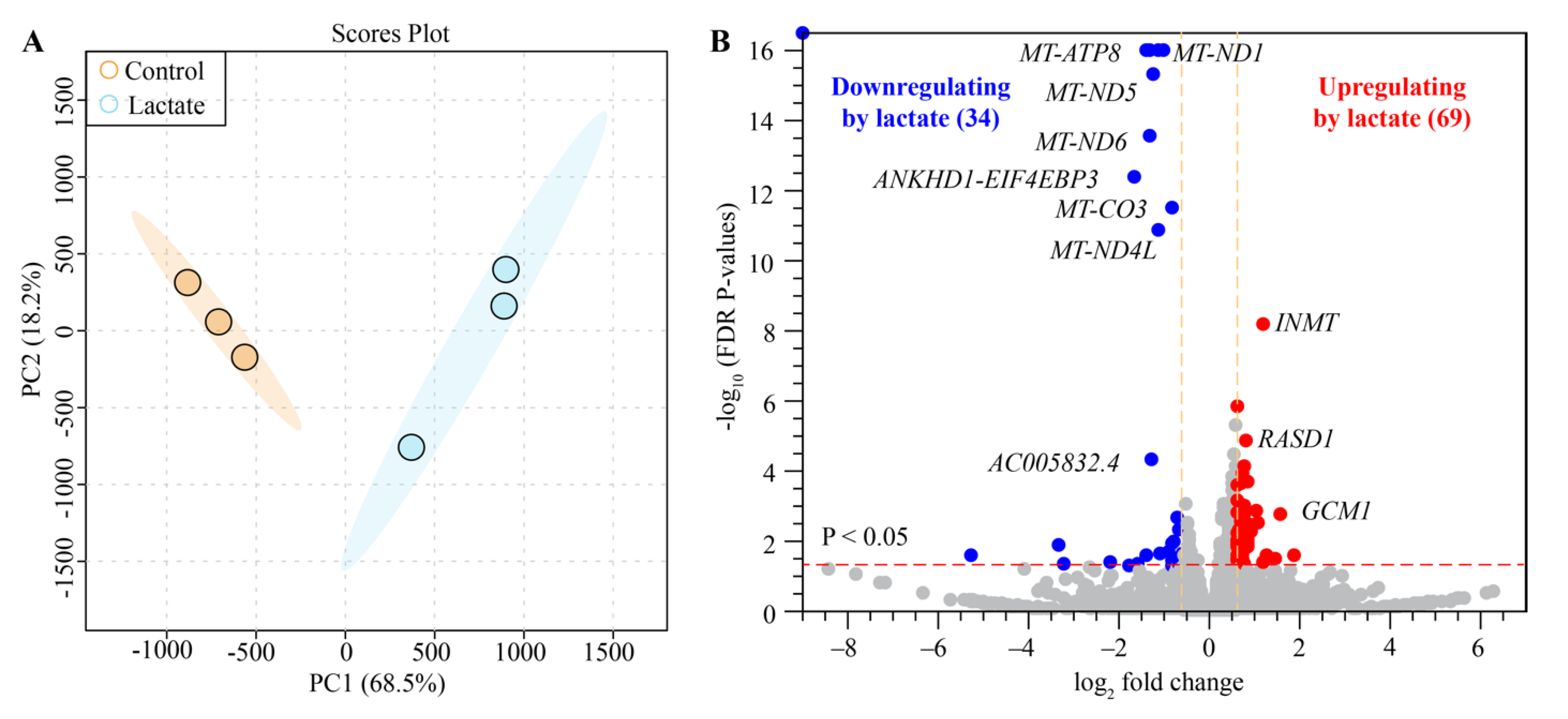

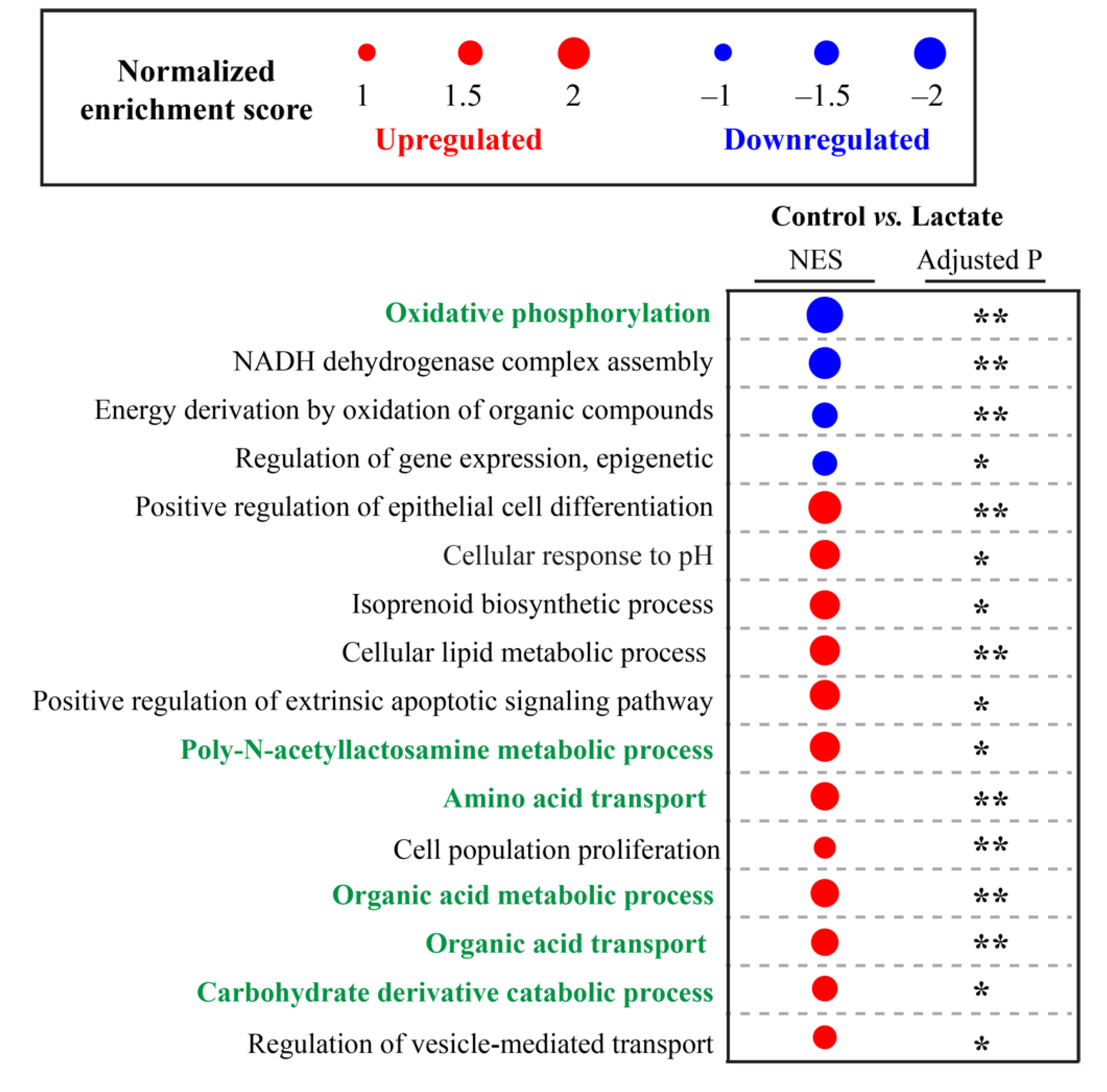

| Number | Metabolites | δ 1H (ppm) and Multiplicity | Abbreviation |
|---|---|---|---|
| 1 | Pantothenate | 0.893 (s), 0.927 (s) | PA |
| 2 | Isoleucine | 0.937 (t), 1.264 (m), 1.46 (m), 1.007 (d), 1.97 (m) | Ile |
| 3 | Leucine | 0.953 (d), 0.964 (d), 1.705 (m), 3.78 (t) | Leu |
| 4 | Valine | 0.989 (d), 1.04 (d), 2.259 (m) | Val |
| 5 | α-Keto-isovalerate | 1.12 (d), 3.01 (m) | α-KIV |
| 6 | α-keto-β-methyl-valerate | 0.883 (t), 1.088 (d) | α-KMV |
| 7 | Lactic acid | 1.324 (d), 4.104 (q) | Lac |
| 8 | Alanine | 1.477 (d), 3.774 (q) | Ala |
| 9 | Threonine | 1.325 (d), 4.247 (dd) | Thr |
| 10 | Glutamate | 2.339 (m), 2.052 (m), 2.117 (m), 3.759 (t) | Glu |
| 11 | Glutamine | 2.453 (m), 2.153 (m), 3.769 (t) | Gln |
| 12 | N-Acetyl-L-Glutamine | 2.037 (s), 2.335 (t) | NAG |
| 13 | Succinate | 2.399 (s) | Suc |
| 14 | Methionine | 2.644 (t) | Met |
| 15 | Dimethylglycine | 2.827 (s) | DMG |
| 16 | Creatine | 3.032 (s), 3.926 (s) | Cre |
| 17 | Choline | 3.193 (s) | Cho |
| 18 | Phosphocholine | 3.218 (s) | PC |
| 19 | Glycerophosphocholine | 3.227 (s) | GPC |
| 20 | Taurine | 3.26 (t), 3.41 (t) | Tau |
| 21 | Glutathione | 2.164 (m), 2.554 (m) 2.931 (dd), 2.975 (dd), 4.55 (m) | GSH |
| 22 | Glucose | 4.644 (d), 5.23 (d) | Glc |
| 23 | Fructose | 4.016 (m), 4.032 (m), 4.06 (m) | Fru |
| 24 | Galactose-1-phosphate | 5.452 (dd) | Gal-1-P |
| 25 | Tyrosine | 6.899 (d), 7.192 (d) | Tyr |
| 26 | Phenylalanine | 7.327 (m), 7.425 (m), 7.373 (m) | Phe |
| 27 | Fumaric acid | 6.515 (s) | Fum |
| 28 | Nicotinamide adenine dinucleotide | 8.428 (s) | NAD+ |
| 29 | Formate | 8.454 (s) | FA |
| 30 | Histidine | 7.073 (s), 7.826 (s) | His |
| 31 | Adenosine monophosphate | 8.607 (s), 8.269 (s), 6.147 (d) | AMP |
| 32 | Adenosine triphosphate | 8.533 (s), 8.269 (s), 6.147 (d) | ATP |
| 33 | Uridine 5’-monophosphate | 8.116 (d) | UMP |
| 34 | Uridine 5’-diphosphate | 7.99 (d), 5.968 (d) | UDP |
| 35 | UDP Glucuronate | 7.952 (d) | UDPG |
| 36 | UDP-N-acetylglucosamine | 7.946 (d) | UDP-N-Ace |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Xu, H.; Zhou, X.; Liu, M.; Li, J.; Liu, C. Systematic Investigations on the Metabolic and Transcriptomic Regulation of Lactate in the Human Colon Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 6262. https://doi.org/10.3390/ijms23116262
Huang C, Xu H, Zhou X, Liu M, Li J, Liu C. Systematic Investigations on the Metabolic and Transcriptomic Regulation of Lactate in the Human Colon Epithelial Cells. International Journal of Molecular Sciences. 2022; 23(11):6262. https://doi.org/10.3390/ijms23116262
Chicago/Turabian StyleHuang, Chongyang, Huanzhou Xu, Xin Zhou, Maili Liu, Jing Li, and Chaoyang Liu. 2022. "Systematic Investigations on the Metabolic and Transcriptomic Regulation of Lactate in the Human Colon Epithelial Cells" International Journal of Molecular Sciences 23, no. 11: 6262. https://doi.org/10.3390/ijms23116262
APA StyleHuang, C., Xu, H., Zhou, X., Liu, M., Li, J., & Liu, C. (2022). Systematic Investigations on the Metabolic and Transcriptomic Regulation of Lactate in the Human Colon Epithelial Cells. International Journal of Molecular Sciences, 23(11), 6262. https://doi.org/10.3390/ijms23116262






