Identification of Structural and Molecular Signatures Mediating Adaptive Changes in the Mouse Kidney in Response to Pregnancy
Abstract
:1. Introduction
2. Results
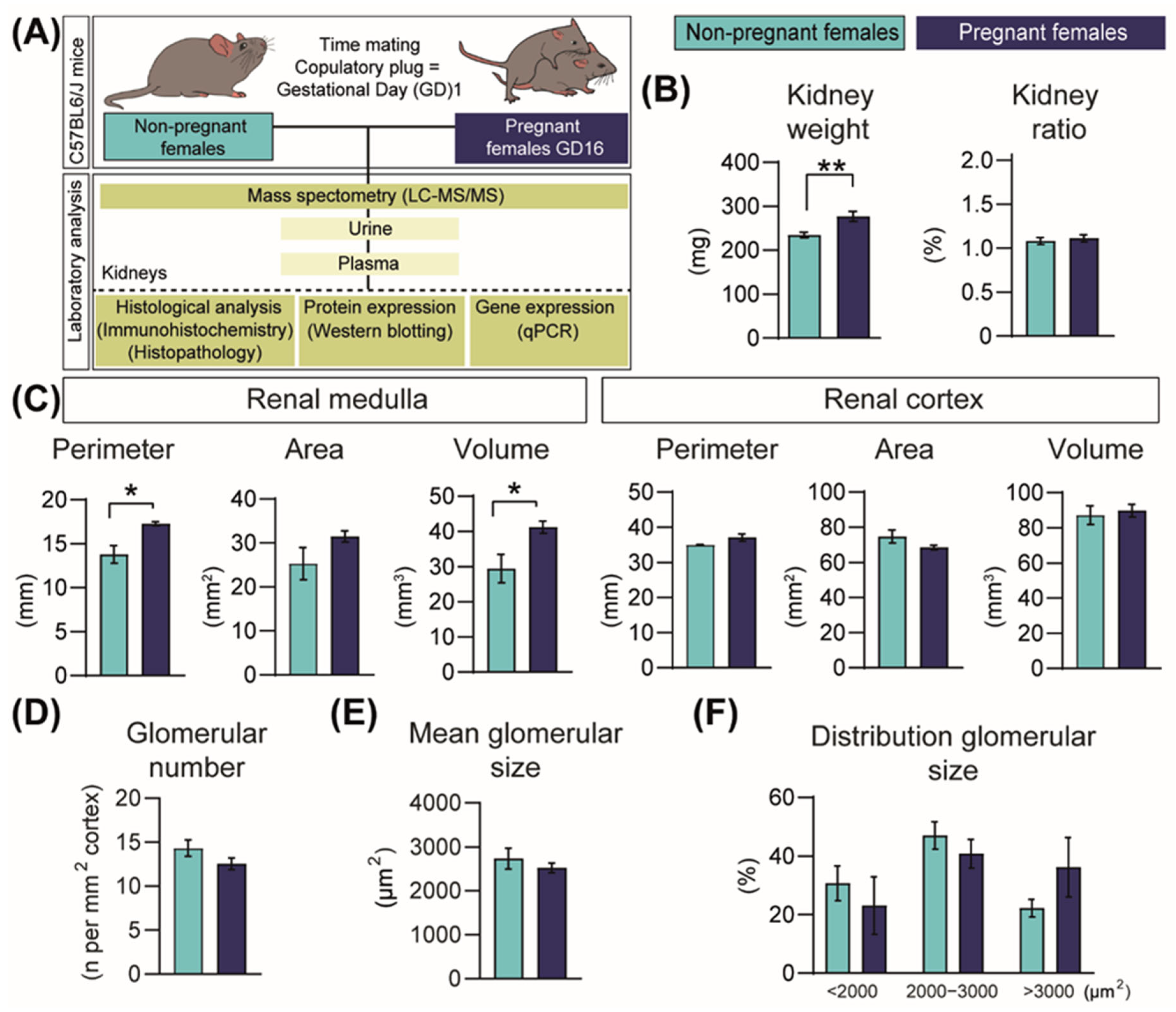
2.1. Pregnancy Increases Kidney Size
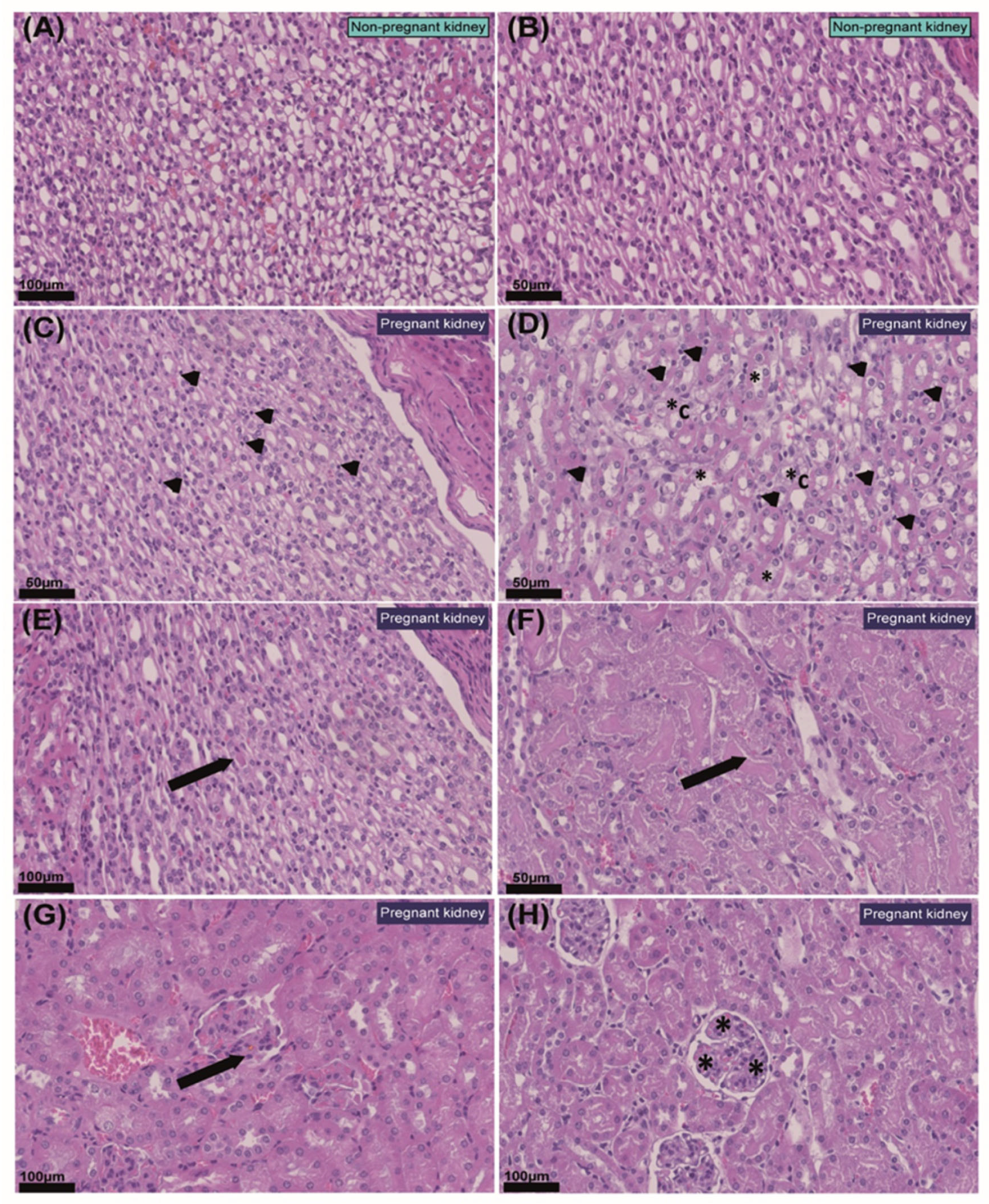
2.2. Pregnancy Modifies Cell Populations in the Kidney
2.3. Pregnancy Increases Cell Proliferation in the Kidney
2.4. Pregnancy Is Associated with Changes in Key Cellular Signalling Pathways and in the mRNA Levels of Cyclin-Dependent Kinases (CDKs)
2.5. Pregnancy Is Associated with Changes in Urine Proteomic Profile
2.6. Pregnancy Is also Associated with Alterations in Plasma Proteomic Profile
3. Discussion
4. Materials and Methods
4.1. Animal Work
4.2. Histology
Immunohistochemical Analysis of Ki67 and TUNEL Staining
4.3. Western Blot Analysis
4.4. RNA Extraction and qPCR
4.5. LC-MS/MS Analysis of Urine and Plasma Samples
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Napso, T.; Yong, H.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. The role of placental hormones in mediating maternal adaptations to support pregnancy and lactation. Front. Physiol. 2018, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Soma-Pillay, P.; Nelson-Piercy, C.; Tolppanen, H.; Mebazaa, A. Physiological changes in pregnancy. Cardiovasc. J. Afr. 2016, 27, 89–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, K.L.; Lafayette, R.A. Renal Physiology of Pregnancy. Adv. Chronic Kidney Dis. 2013, 20, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Beers, K.; Patel, N. Kidney Physiology in Pregnancy. Adv. Chronic Kidney Dis. 2020, 27, 449–454. [Google Scholar] [CrossRef]
- Davison, J.M.; Dunlop, W. Renal hemodynamics and tubular function normal human pregnancy. Kidney Int. 1980, 18, 152–161. [Google Scholar] [CrossRef] [Green Version]
- Odutayo, A.; Hladunewich, M. Obstetric nephrology: Renal hemodynamic and metabolic physiology in normal pregnancy. Clin. J. Am. Soc. Nephrol. 2012, 7, 2073–2080. [Google Scholar] [CrossRef] [Green Version]
- Alhenc-Gelas, F.; Tache, A.; Saint-Andre, J.P.; Milliez, J.; Sureau, C.; Corvol, P.; Menard, J. The renin-angiotensin system in pregnancy and parturition. Adv. Nephrol. Necker Hosp. 1986, 15, 25–33. [Google Scholar]
- Hsueh, W.A.; Luetscher, J.A.; Carlson, E.J.; Grislis, G.; Fraze, E.; McHargue, A. Changes in active and inactive renin throughout pregnancy. J. Clin. Endocrinol. Metab. 1982, 54, 1010–1016. [Google Scholar] [CrossRef]
- Skinner, S.L.; Lumbers, E.R.; Symonds, E.M. Analysis of changes in the renin-angiotensin system during pregnancy. Clin. Sci. 1972, 42, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.; Morganti, A.A.; Zervoudakis, I.; Letcher, R.L.; Romney, B.M.; Von Oeyon, P.; Papera, S.; Sealey, J.E.; Laragh, J.H. Blood pressure, the renin-aldosterone system and sex steroids throughout normal pregnancy. Am. J. Med. 1980, 68, 97–104. [Google Scholar] [CrossRef]
- Xia, Y.; Wen, H.; Prashner, H.R.; Chen, R.; Inagami, T.; Catanzaro, D.F.; Kellems, R.E. Pregnancy-induced changes in renin gene expression in mice. Biol. Reprod. 2002, 66, 135–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, P.E.; Nielsen, F.R. Hydronephrosis during pregnancy: A literature survey. Eur. J. Obstet. Gynecol. Reprod. Biol. 1988, 27, 249–259. [Google Scholar] [CrossRef]
- Fischer, M.J.; Lehnerz, S.D.; Hebert, J.R.; Parikh, C.R. Kidney disease is an independent risk factor for adverse fetal and maternal outcomes in pregnancy. Am. J. Kidney Dis. 2004, 43, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Imbasciati, E.; Gregorini, G.; Cabiddu, G.; Gammaro, L.; Ambroso, G.; Del Giudice, A.; Ravani, P. Pregnancy in CKD stages 3 to 5: Fetal and maternal outcomes. Am. J. Kidney Dis. 2007, 49, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.G.; Cox, S.M.; Harstad, T.W.; Mason, R.A.; Pritchard, J.A. Chronic renal disease and pregnancy outcome. Am. J. Obstet. Gynecol. 1990, 163, 453–459. [Google Scholar] [CrossRef]
- Jones, D.C.; Hayslett, J.P. Outcome of pregnancy in women with moderate or severe renal insufficiency. N. Engl. J. Med. 1996, 335, 226–232. [Google Scholar] [CrossRef]
- Williams, D.; Davison, J. Chronic kidney disease in pregnancy. BMJ 2008, 336, 211–215. [Google Scholar] [CrossRef] [Green Version]
- Wiles, K.S.; Nelson-Piercy, C.; Bramham, K. Reproductive health and pregnancy in women with chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 165–184. [Google Scholar] [CrossRef]
- Powe, C.E.; Thadhani, R. Diabetes and the kidney in pregnancy. Semin. Nephrol. 2011, 31, 59–69. [Google Scholar] [CrossRef]
- Musial, B.; Fernandez-Twinn, D.S.; Vaughan, O.R.; Ozanne, S.E.; Voshol, P.; Sferruzzi-Perri, A.N.; Fowden, A.L. Proximity to Delivery Alters Insulin Sensitivity and Glucose Metabolism in Pregnant Mice. Diabetes 2016, 65, 851–860. [Google Scholar] [CrossRef] [Green Version]
- Han, L.W.; Shi, Y.; Paquette, A.; Wang, L.; Bammler, T.K.; Mao, Q. Key hepatic metabolic pathways are altered in germ-free mice during pregnancy. PLoS ONE 2021, 16, e0248351. [Google Scholar] [CrossRef] [PubMed]
- Paquette, A.; Baloni, P.; Holloman, A.B.; Nigam, S.; Bammler, T.; Mao, Q.; Price, N.D. Temporal transcriptomic analysis of metabolic genes in maternal organs and placenta during murine pregnancy. Biol. Reprod. 2018, 99, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Mazaki-Tovi, S.; Vaisbuch, E.; Tarca, A.L.; Kusanovic, J.P.; Than, N.G.; Chaiworapongsa, T.; Dong, Z.; Hassan, S.S.; Romero, R. Characterization of Visceral and Subcutaneous Adipose Tissue Transcriptome and Biological Pathways in Pregnant and Non-Pregnant Women: Evidence for Pregnancy-Related Regional-Specific Differences in Adipose Tissue. PLoS ONE 2015, 10, e0143779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchanan, T.A.; Metzger, B.E.; Freinkel, N.; Bergman, R.N. Insulin sensitivity and B-cell responsiveness to glucose during late pregnancy in lean and moderately obese women with normal glucose tolerance or mild gestational diabetes. Am. J. Obstet. Gynecol. 1990, 162, 1008–1014. [Google Scholar] [CrossRef]
- Lee, S.; Temple, F.T.; Dawson, P.A. Kidney microRNA profile in pregnant mice reveals molecular insights into kidney adaptation to pregnancy: A pilot study. Mol. Genet. Metab. Rep. 2019, 20, 100486. [Google Scholar] [CrossRef]
- O’Brien, L.L.; Guo, Q.; Lee, Y.; Tran, T.; Benazet, J.-D.; Whitney, P.H.; Valouev, A.; McMahon, A.P. Differential regulation of mouse and human nephron progenitors by the Six family of transcriptional regulators. Development 2016, 143, 595–608. [Google Scholar] [CrossRef] [Green Version]
- Nishino, T.; Sasaki, N.; Nagasaki, K.-I.; Ahmad, Z.; Agui, T. Genetic background strongly influences the severity of glomerulosclerosis in mice. J. Vet. Med. Sci. 2010, 72, 1313–1318. [Google Scholar] [CrossRef] [Green Version]
- Becker, G.J.; Hewitson, T.D. Animal models of chronic kidney disease: Useful but not perfect. Nephrol. Dial. Transplant. 2013, 28, 2432–2438. [Google Scholar] [CrossRef] [Green Version]
- Hemberger, M.; Hanna, C.W.; Dean, W. Mechanisms of early placental development in mouse and humans. Nat. Rev. Genet. 2020, 21, 27–43. [Google Scholar] [CrossRef]
- Carter, A.M. Animal Models of Human Placentation—A Review. Placenta 2007, 28, S41–S47. [Google Scholar] [CrossRef]
- Swanson, A.M.; David, A.L. Animal models of fetal growth restriction: Considerations for translational medicine. Placenta 2015, 36, 623–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, F.; Lee, J.T.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: A target for cancer chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta BBA Mol. Cell Res. 2011, 1813, 1619–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, A.; Du, J. Potential role of Akt signaling in chronic kidney disease. Nephrol. Dial. Transplant. 2015, 30, 385–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Santis, M.C.; Sala, V.; Martini, M.; Ferrero, G.B.; Hirsch, E. PI3K Signaling in Tissue Hyper-Proliferation: From Overgrowth Syndromes to Kidney Cysts. Cancers 2017, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrales, P.; Izquierdo-Lahuerta, A.; Medina-Gómez, G. Maintenance of Kidney Metabolic Homeostasis by PPAR Gamma. Int. J. Mol. Sci. 2018, 19, 2063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juszczak, F.; Caron, N.; Mathew, A.V.; Declèves, A.-E. Critical Role for AMPK in Metabolic Disease-Induced Chronic Kidney Disease. Int. J. Mol. Sci. 2020, 21, 7994. [Google Scholar] [CrossRef]
- Hallows, K.R.; Mount, P.F.; Pastor-Soler, N.M.; Power, D.A. Role of the energy sensor AMP-activated protein kinase in renal physiology and disease. Am. J. Physiol. Renal Physiol. 2010, 298, F1067–F1077. [Google Scholar] [CrossRef] [Green Version]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, L.; Wang, J.; Jin, Q. Urinary proteomic and non-prefractionation quantitative phosphoproteomic analysis during pregnancy and non-pregnancy. BMC Genom. 2013, 14, 777. [Google Scholar] [CrossRef] [Green Version]
- Napso, T.; Zhao, X.; Lligoña, M.I.; Sandovici, I.; Kay, R.G.; George, A.L.; Gribble, F.M.; Reimann, F.; Meek, C.L.; Hamilton, R.S.; et al. Placental secretome characterization identifies candidates for pregnancy complications. Commun. Biol. 2021, 4, 701. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Tuteja, G. TissueEnrich: Tissue-specific gene enrichment analysis. Bioinformatics 2019, 35, 1966–1967. [Google Scholar] [CrossRef]
- Nawata, C.M.; Pannabecker, T.L. Mammalian urine concentration: A review of renal medullary architecture and membrane transporters. J. Comp. Physiol. B 2018, 188, 899–918. [Google Scholar] [CrossRef] [PubMed]
- Lemley, K.V.; Kriz, W. Anatomy of the renal interstitium. Kidney Int. 1991, 39, 370–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovatz, S.; Rathaus, M.; Aderet, N.B.; Bernheim, J. Increased Renal Prostaglandins in Normal Pregnancy and in Pregnancy with Hypertension. Nephron 1982, 32, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Tordera, V.; Sendra, R.; Pérez-Ortín, J.E. The role of histones and their modifications in the informative content of chromatin. Experientia 1993, 49, 780–788. [Google Scholar] [CrossRef]
- Djudjaj, S.; Papasotiriou, M.; Bülow, R.D.; Wagnerova, A.; Lindenmeyer, M.T.; Cohen, C.D.; Strnad, P.; Goumenos, D.S.; Floege, J.; Boor, P. Keratins are novel markers of renal epithelial cell injury. Kidney Int. 2016, 89, 792–808. [Google Scholar] [CrossRef] [Green Version]
- Helenius, T.O.; Antman, C.A.; Asghar, M.N.; Nyström, J.H.; Toivola, D.M. Keratins Are Altered in Intestinal Disease-Related Stress Responses. Cells 2016, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Eckert, R.L.; Efimova, T.; Balasubramanian, S.; Crish, J.F.; Bone, F.; Dashti, S. p38 Mitogen-activated protein kinases on the body surface—A function for p38 delta. J. Investig. Derm. 2003, 120, 823–828. [Google Scholar] [CrossRef]
- Wöll, S.; Windoffer, R.; Leube, R.E. p38 MAPK-dependent shaping of the keratin cytoskeleton in cultured cells. J. Cell Biol. 2007, 177, 795–807. [Google Scholar] [CrossRef]
- Cuenda, A.; Rousseau, S. p38 MAP-Kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta BBA Mol. Cell Res. 2007, 1773, 1358–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomás-Loba, A.; Manieri, E.; González-Terán, B.; Mora, A.; Leiva-Vega, L.; Santamans, A.M.; Romero-Becerra, R.; Rodríguez, E.; Pintor-Chocano, A.; Feixas, F.; et al. p38γ is essential for cell cycle progression and liver tumorigenesis. Nature 2019, 568, 557–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhanumathy, C.D.; Tang, Y.; Monga, S.P.S.; Katuri, V.; Cox, J.A.; Mishra, B.; Mishra, L. Itih-4, a serine protease inhibitor regulated in interleukin-6-dependent liver formation: Role in liver development and regeneration. Dev. Dyn. 2002, 223, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.N.-Y.; Kanazawa, S.; Kado, M.; Okada, K.; Luo, L.; Hayashi, A.; Mizuno, H.; Tanaka, R. Interleukin-6 stimulates Akt and p38 MAPK phosphorylation and fibroblast migration in non-diabetic but not diabetic mice. PLoS ONE 2017, 12, e0178232. [Google Scholar]
- Robertson, S.A.; Christiaens, I.; Dorian, C.L.; Zaragoza, D.B.; Care, A.S.; Banks, A.M.; Olson, D.M. Interleukin-6 is an essential determinant of on-time parturition in the mouse. Endocrinology 2010, 151, 3996–4006. [Google Scholar] [CrossRef] [Green Version]
- Kattah, A. Preeclampsia and Kidney Disease: Deciphering Cause and Effect. Curr. Hypertens. Rep. 2020, 22, 91. [Google Scholar] [CrossRef]
- Saito, S.; Shiozaki, A.; Nakashima, A.; Sakai, M.; Sasaki, Y. The role of the immune system in preeclampsia. Mol. Asp. Med. 2007, 28, 192–209. [Google Scholar] [CrossRef]
- Clark, A.J.; Parikh, S.M. Targeting energy pathways in kidney disease: The roles of sirtuins, AMPK, and PGC1α. Kidney Int. 2021, 99, 828–840. [Google Scholar] [CrossRef]
- Lee, W.H.; Kim, S.G. AMPK-Dependent Metabolic Regulation by PPAR Agonists. PPAR Res. 2010, 2010, 549101. [Google Scholar] [CrossRef]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef]
- Ghosh, S.S.; Gehr, T.W.B.; Ghosh, S.; Fakhry, I.; Sica, D.A.; Lyall, V.; Schoolwerth, A.C. PPARγ ligand attenuates PDGF-induced mesangial cell proliferation: Role of MAP kinase. Kidney Int. 2003, 64, 52–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, T.; Wada, J.; Hida, K.; Eguchi, J.; Hashimoto, I.; Baba, M.; Yasuhara, A.; Shikata, K.; Makino, H. Thiazolidinediones ameliorate diabetic nephropathy via cell cycle-dependent mechanisms. Diabetes 2006, 55, 1666–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grellier, P.; Sabbah, M.; Fouqueray, B.; Woodruff, K.; Yee, D.; Abboud, H.E.; Abboud, S.L. Characterization of insulin-like growth factor binding proteins and regulation of IGFBP3 in human mesangial cells. Kidney Int. 1996, 49, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, K.; Hirata, H.; Majid, S.; Tabatabai, Z.L.; Hinoda, Y.; Dahiya, R. IGFBP-4 activates the Wnt/beta-catenin signaling pathway and induces M-CAM expression in human renal cell carcinoma. Int. J. Cancer 2011, 129, 2360–2369. [Google Scholar] [CrossRef]
- Ning, Y.; Schuller, A.G.P.; Conover, C.A.; Pintar, J.E. Insulin-Like Growth Factor (IGF) Binding Protein-4 Is Both a Positive and Negative Regulator of IGF Activity in Vivo. Mol. Endocrinol. 2008, 22, 1213–1225. [Google Scholar] [CrossRef] [Green Version]
- Karumanchi, S.A.; Maynard, S.E.; Stillman, I.E.; Epstein, F.H.; Sukhatme, V.P. Preeclampsia: A renal perspective. Kidney Int. 2005, 67, 2101–2113. [Google Scholar] [CrossRef] [Green Version]
- Brosens, I.; Pijnenborg, R.; Vercruysse, L.; Romero, R. The ‘Great Obstetrical Syndromes’ are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 2011, 204, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Napso, T.; Lean, S.C.; Lu, M.; Mort, E.J.; Desforges, M.; Moghimi, A.; Bartels, B.; El-Bacha, T.; Fowden, A.L.; Camm, E.J.; et al. Diet-Induced Maternal Obesity Impacts Feto-Placental Growth and Induces Sex-Specific Alterations in Placental Morphology, Mitochondrial Bioenergetics, Dynamics, Lipid Metabolism and Oxidative Stress in Mice. Acta Physiol. 2022, 234, e13795. [Google Scholar] [CrossRef]
- Lopez-Tello, J.; Jimenez-Martinez, M.A.; Herrera, E.A.; Krause, B.J.; Sferruzzi-Perri, A.N. Progressive uterine artery occlusion in the Guinea pig leads to defects in placental structure that relate to fetal growth. Placenta 2018, 72–73, 36–40. [Google Scholar] [CrossRef]
- Aykroyd, B.R.L.; Tunster, S.J.; Sferruzzi-Perri, A.N. Igf2 deletion alters mouse placenta endocrine capacity in a sexually dimorphic manner. J. Endocrinol. 2020, 246, 93–108. [Google Scholar] [CrossRef]
- Waker, C.A.; Kaufman, M.R.; Brown, T.L. Current State of Preeclampsia Mouse Models: Approaches, Relevance, and Standardization. Front. Physiol. 2021, 12, 681632. [Google Scholar] [CrossRef] [PubMed]
- Sferruzzi-Perri, A.N.; Camm, E.J. The Programming Power of the Placenta. Front. Physiol. 2016, 7, 33. [Google Scholar]
- Nambadan, S.B.; Stanley, M.; Zhang, Y.; Athanasopoulos, V.; Jiang, S.H. A protocol to evaluate immunoglobulin deposits in mouse glomeruli. STAR Protoc. 2022, 3, 101375. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, K.; Lopez-Tello, J.; Vriens, J.; Sferruzzi-Perri, A.N. Double-label immunohistochemistry to assess labyrinth structure of the mouse placenta with stereology. Placenta 2020, 94, 44–47. [Google Scholar] [CrossRef]
- Romero-Calvo, I.; Ocón, B.; Martínez-Moya, P.; Suárez, M.D.; Zarzuelo, A.; Martínez-Augustin, O.; de Medina, F.S. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Anal. Biochem. 2010, 401, 318–320. [Google Scholar] [CrossRef]
- Salazar-Petres, E.; Carvalho, D.P.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. Placental structure, function and mitochondrial phenotype relate to fetal size in each fetal sex in mice. Biol. Reprod. 2022, ioac056. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]




| Protein Name | Direction of the Change in Pregnancy | Presence in the Urine | Top Three Organs—Enrichment (Expression Levels Based on FPKM) | GO Function Biological Process |
|---|---|---|---|---|
| AMBP | ↑ | NP | Liver (465.23) Heart (3.41) Bone marrow (1.87) | Receptor-mediated endocytosis and cell adhesion |
| CFB | ↑ | NP | Liver (110.86) Kidney (38.95) Intestine (18.33) | Immune system process and proteolysis |
| IGFBP4 | ↑ | NP | Liver (231.39) Kidney (151.64) Spleen (33.41) | Regulation of cell growth and inflammatory response |
| LYZ2 | ↓ | NP | Lung (1308.75) Intestine (337.09) Spleen (283.58) | Inflammatory response and metabolic process |
| THBS1 | ↓ | NP | Bone Marrow (13.92) Lung (5.57) Cortex (4.49) | Activation of MAPK activity and response to hypoxia |
| GLYCAM1 | ↑ | Pregnant | Spleen (0.28) Kidney (0.20) Intestine (0.18) | Regulation of immune response |
| ITIH4 | ↑ | Pregnant | Liver (114.27) Lung (3.43) Heart (0.33) | Platelet degranulation and acute-phase response |
| SERPINA1B | ↑ | Pregnant | Liver (844.81) Heart (6.34) Kidney (5.08) | Platelet degranulation and ER to Golgi vesicle-mediated transport |
| SERPINA6 | ↑ | Pregnant | Liver (332.31) Heart (0.28) Bone marrow (0.20) | Glucocorticoid biosynthetic and metabolic processes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Tello, J.; Jimenez-Martinez, M.A.; Salazar-Petres, E.; Patel, R.; George, A.L.; Kay, R.G.; Sferruzzi-Perri, A.N. Identification of Structural and Molecular Signatures Mediating Adaptive Changes in the Mouse Kidney in Response to Pregnancy. Int. J. Mol. Sci. 2022, 23, 6287. https://doi.org/10.3390/ijms23116287
Lopez-Tello J, Jimenez-Martinez MA, Salazar-Petres E, Patel R, George AL, Kay RG, Sferruzzi-Perri AN. Identification of Structural and Molecular Signatures Mediating Adaptive Changes in the Mouse Kidney in Response to Pregnancy. International Journal of Molecular Sciences. 2022; 23(11):6287. https://doi.org/10.3390/ijms23116287
Chicago/Turabian StyleLopez-Tello, Jorge, Maria Angeles Jimenez-Martinez, Esteban Salazar-Petres, Ritik Patel, Amy L. George, Richard G. Kay, and Amanda N. Sferruzzi-Perri. 2022. "Identification of Structural and Molecular Signatures Mediating Adaptive Changes in the Mouse Kidney in Response to Pregnancy" International Journal of Molecular Sciences 23, no. 11: 6287. https://doi.org/10.3390/ijms23116287







