1. Introduction
The transition to a terrestrial lifestyle confronted plants with a fundamentally different situation where they had to develop self-supporting structures to compensate for the mechanic load, which was no longer carried by buoyancy. This transition left fundamental traces in the organisation and composition of the cytoskeleton (for a review, see [
1]). One of the most striking changes was the progressive loss of dynein motors [
2], while the specific class of the minus-end-directed class-XIV kinesins expanded [
3], acquiring novel functions. For instance, the kinesins with a calponin-domain homologue link the mechanically rigid microtubules with the flexible actin filaments, which helps the pre-mitotic nucleus to locate the cell centre before initiating mitosis [
4]. The kinesins KatA and ATK5 shorten the spindle during anaphase, thus adopting functions exerted by dyneins in animal cells [
5,
6].
In our previous work [
7], we studied a rice homologue of KatA and ATK5, which turned out to occur at two sites in the cell and was, therefore, named the Dual Localisation Kinesin (DLK). While decorating cortical microtubules during the interphase and associating with different mitotic microtubule arrays during mitosis, it also moved into the nucleus in response to cold. Motility assays in vitro showed that this unconventional kinesin can drive the mutual sliding of microtubules and moves towards the minus-end of microtubules (i.e., in a direction typical for dynein motors) with a velocity comparable to other class-XIV kinesins. The accumulation of this kinesin in the nucleus could also be promoted in the absence of cold, when the cells were treated with Leptomycin B, a blocker of the nuclear export.
When a kinesin motor accumulates in the nucleus in response to a signal (cold) and is actively exported, there must be a functional reason, because the cytoskeleton is supposed to be tightly excluded from the karyoplasm during interphase. However, this textbook dogma has been progressively perforated during the last two decades. The seminal discovery of chromokinesin, a KIF4 kinesin constitutively localised in the mammalian nucleus [
8], was later followed by findings where a KIF7 motor acts as a transcriptional regulator in the hedgehog signalling pathway [
9]. Furthermore, in plants, a shuttling kinesin, OsBC12, was found to act as a transcription factor for the
ent-kaurene oxidase OsKO2, such that the respective mutant is a gibberellin deficient dwarf [
10].
This nuclear import of kinesins might be functionally linked to the nuclear import of tubulin that can be observed in plant cells in response to cold stress [
11]. After the end of the cold period, tubulin rapidly leaves the nucleus again to build up a new cortical array of microtubules. Although tubulins lack canonical Nuclear Localisation Sequences (NLS), they are endowed with specific Nuclear Export Sequences (NES) that mediate the interaction with the Exportin receptor complex. These NES exist in plants as well and are functional as shown by constructs, where these sequences were placed in front of a GFP reporter [
12]. While this export might be important to remove tubulin heterodimers that had been trapped during the formation of daughter nuclei in the telophase, the export inhibitor Leptomycin B can induce conspicuous accumulation of tubulin in the non-cycling stationary phase, which means that tubulin is also imported to a certain extent during the interphase, but is normally removed rapidly, such that the steady-state level of nuclear tubulin is low. Whether the intranuclear accumulation of tubulin in response to cold is caused by the higher sensitivity of this export as compared to the import or by the increased level of non-assembled tubulin from the eliminated microtubules remains to be elucidated.
When a kinesin motor, as well as its substrate, tubulin, partition to the nucleus in response to cold, there must be a functional relevance of this co-transport. The current work was motivated by the attempt to gain insight into this potential function. The analysis of the loss-of-function for DLK in rice and the patterns observed for the accumulation of the OsDLK transcript indicate that this kinesin, under normal conditions, acts in concert with cortical microtubule arrays required for cell elongation. The cold-induced nuclear import seems to function with a nuclear localisation signal, a putative leucin zipper, and the recognition of a specific DNA binding motif identified by a DPI-ELISA screen with N-terminal domain of recombinant OsDLK. The presence of this motif in the promoter of Cold-Box Factor 4, a transcription factor crucial for cold adaptation, along with the finding that the overexpression of OsDLK in tobacco specifically modulates the expression of this Cold Box Factor, point to a function of this unusual kinesin motor in the transduction of and adaptation to cold stress.
3. Discussion
The current study tried to gain insight into the biological function of the unusual kinesin motor Dual Localisation Kinesin (DLK), by analysing the phenotype for rice loss-of-function mutants, the regulatory pattern in rice, and the effect of overexpression in tobacco BY−2 cells. Since the localisation of this protein depends on temperature, one needs to consider its role under normal temperature separately from its function under cold stress. In fact, our data are compatible with a model where DLK under normal conditions sustains cell elongation by fulfilling a function associated with the cortical microtubule. However, in response to cold stress, this kinesin enters the nucleus, and binds specifically to target motifs in the DNA. As a result, the induction of transcripts for Cold-Box Factor 4 in response to cold is modulated, indicating a function of this unusual kinesin motor in the transduction of and adaptation to cold stress. These findings stimulate the following questions to be discussed: 1. What is the potential link between DLK and cell elongation? 2. What is the molecular base for the transcriptional activation? 3. How is the nuclear import of DLK connected to cold signalling and cold acclimation?
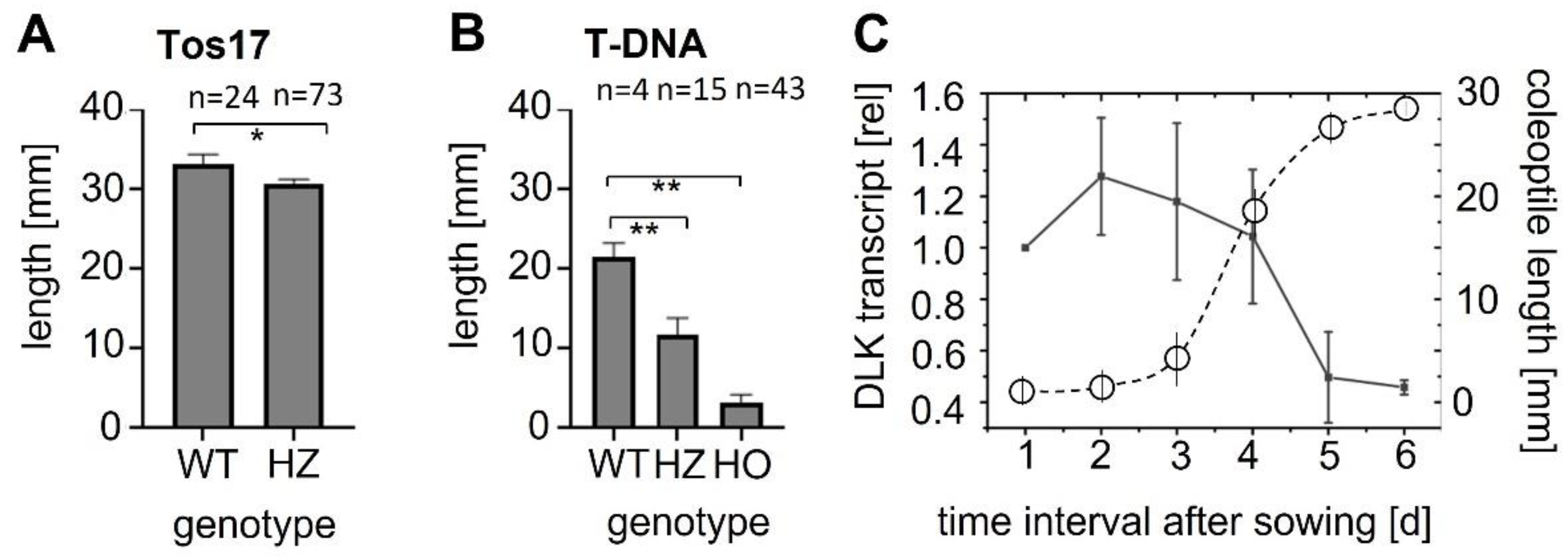
OsDLK and elongation. Under normal temperatures, OsDLK prevails at the cortical microtubules, which holds true, both for heterologous ([
7] tobacco BY−2, cotyledons of
Arabidopsis thaliana), as well as homologous, systems (
Figure 3: Rice coleoptile epidermis). This localisation indicates a function for cell elongation. This is supported by the finding that loss-of-function mutants (T-DNA, Tos−17 retrotransposon) show impaired elongation in the heterozygous state (
Figure 1A,B). In the homozygous state, they are very short (T-DNA,
Figure 1B) or even non-viable (Tos−17 retrotransposon,
Figure 1A). The fact that OsDLK is necessary for coleoptile elongation is further corroborated by the temporal pattern of OsDLK transcripts in etiolated coleoptiles (that grow exclusively via cell elongation). Here, the steady-state transcript levels are high up to day 4 after sowing and then decrease substantially during the phase of logarithmic expansion (
Figure 1C). As to be expected, the expression of OsDLK is strong in rapidly expanding parts of the leaves (second sheath, third leaf), while it is low in the roots, where growth proceeds concomitantly with mitosis (
Figure 2A). Conversely, the transient induction of OsDLK transcripts by the natural auxin indole-acetic acid is consistent with a role for OsDLK in elongation (
Figure 2B).
These considerations lead to the question by which mechanism can OsDLK sustain cell elongation. Coleoptiles grow exclusively by cell elongation without proliferation, and this expansion is constrained and directed by the mechanic properties of the outer epidermal cell wall [
21]. Elongation is sustained by a transverse orientation of newly deposited cellulose microfibrils that reinforce the otherwise preferentially transverse stress from the expanding protoplast [
22]. When microtubules re-orient into more longitudinal arrays in response to phototropic or gravitropic, this is followed by a decrease in the elongation rate and in one flank of the coleoptile, culminating in tropistic bending [
23]. In
Arabidopsis thaliana, the class-XIV kinesin ATK5 (the closest homologue of rice DLK) is not only found in spindles and phragmoplasts, but also decorates cortical microtubules [
6]. This protein has been shown to exert a bundling activity in the spindle. So, it is straightforward to assume that ATK5 (as well as its rice homologue DLK) will do so in the cortical array, thus stabilising a transverse orientation of microtubules and, in this way, sustaining coleoptile elongation.
OsDLK and transcriptional regulation. We find that the overexpression of OsDLK specifically modulates the expression of
NtCf9, the tobacco homologue of Cold-Box Factor 4 (
Figure 4B), while the responses of other cold-related transcripts did not show a significant difference compared to non-transformed tobacco cells (
Figure 4A). Recombinantly expressed OsDLK bound to a specific DNA motif, 11 bp in length (
Figure 5B–D), which has been known to be a target for transcription factor OPAQUE 2, a transcriptional key regulator that activates the accumulation of seed storage proteins by binding to this motif by virtue of its leucin zipper [
24,
25]. Interestingly, there exist several functional analogies between OPAQUE 2 and OsDLK.
OPAQUE2 regulates zein genes and has been shown to move into the nucleus after ubiquitination through the E3 ubiquitin ligase RFWD2 [
26]. The sucrose-responsive kinase SnRK1 phosphorylates RFWD2 and initiates its degradation, which is a mechanism to regulate the accumulation of zein depending on sucrose. SnRK1, in turn, is negatively regulated by the phosphatases ABI1 and PP2CA [
27]. At this point, there is an interesting bifurcation: When these phosphatases are activated, this is also a release of the kinase Open Stomata 1 from the membrane, such that it can activate the master switch ICE1, governing the cold-induced transcriptional cascade in the nucleus (for a review, see [
28]). The activation of ABI1 and PPC2A by cold stress would, thus, activate SnRK1, such that RFWD2 is phosphorylated and removed and OPAQUE2 would not move into the nucleus. Under the same conditions, OsDLK would move into the nucleus. Thus, both proteins not only share motifs in the promoters of their targets, but also move into the nucleus depending on signals, and they share upstream elements of signalling. However, their behaviour in response to cold stress is expected to be a mirror image.
That a kinesin function is physically linked to a function in transcriptional regulation appears exotic at first, but OsDLK is not the only case for such a link: For instance, the KIF7 motor regulates hedgehog signalling in mammalian cells [
9], and the human class-XIV motor KIFC1 was shown recently to be required for DNA replication during the S-phase [
29]. Likewise, the human motor KIF4 is localised in the nucleus throughout the interphase and participates in chromatin remodelling [
30]. In plants, so far, there is only one further case known where a kinesin exerts a function in DNA regulation, namely the rice kinesin OsBC12 activating expression of the gibberellin synthesis gene
ent-kaurene oxidase [
10].
Are nuclear kinesins rudiments of evolution? Microtubules and DNA are strictly sequestered during the interphase, separated by the nuclear envelope that tightly controls which molecules are allowed to pass through nuclear pores (for a classical review, see [
31]). The directionality is maintained by a gradient of the small GTPase Ran, which is found as Ran-GTP in the karyoplasm but as Ran-GDP in the cytoplasm. These forms of Ran are maintained by accessory proteins, such as RanGAP, which, in plants, is located at the outer nuclear envelope [
32] Ran-GTP can induce microtubule nucleation and initiate spindle formation. However, this is prevented during the interphase because tubulin is strictly separated from Ran-GTP. When the nuclear envelope breaks down at the onset of the metaphase, Ran-GTP can initiate microtubules in the former karyoplasm. In fact, plant RanGAP associates with tubulin in mitotic cells, but not in cells that are not cycling [
32]. Tubulin is void of a canonical NLS, but it does harbour functional NES motifs [
12] indicating that the exclusion of tubulin from the interphasic karyoplasm is essential. The strict sequestering of chromatin from tubulin is not a primordial phenomenon, but evolved from an ancestral situation, where tubulin was in the nucleus and only later “explored” the surrounding cytoplasm (for a review, see [
33]). Numerous transitional stages, where the spindle can develop without the breakdown of the nuclear envelope, can be considered evolutionary witnesses of this shift out of the nucleus. In response to cold stress, tubulin can return swiftly to this evolutionarily ancient state and accumulate in the nucleus, from where, upon rewarming, it rapidly returns to the cytoplasm, organising new microtubular structures [
11]. Since tubulin dimers are approximately twice as large as the size exclusion limit of nuclear pores, they would be expected to require an NLS in doing so. Since, so far, no NLS could be identified in any tubulin, it is more likely that tubulin enters the nucleus as part of a complex. Whether DLK is a component of this complex is not known but represents an attractive hypothesis that should be tested in future experiments.
Is DLK a negative regulator of cellular cold acclimation? Low temperature is among the major environmental factors shaping the geographical distribution of plants and can deploy a plethora of damage-related and adaptive responses, depending on the species, the harshness of the condition, and the time course of the stress (classical and comprehensive reviews that are still worth reading are those by Lyons and Burke [
34,
35]). Not surprisingly, the plant responses to this environmental complexity are complex and specific as well. Many plants can deploy efficient cellular adaptation to even harsh cold stress if this stress is preceded by prolonged but mild chilling, a phenomenon known as cold acclimation or cold hardening. The inducing effect of this chilling pre-treatment can be phenocopied by a transient elimination of microtubules (winter wheat [
36] and grapevine cells [
37]). Cold hardening is accompanied by the high expression of Cold-Box Factor 4 (grapevine plants [
38] and grapevine cells [
37]), and the overexpression of Cold-Box Factor 4 can induce freezing tolerance in grapevine without the need for pre-chilling [
39]. Since the perception of cold occurs at the plasma membrane, while the activation of transcriptional regulators proceeds in the nucleus, signals must be conveyed from the membrane to the nucleus. In fact, MAPK cascades are deployed in response to cold stress and these cascades modulate the stability of transcriptional regulators in a complex, partially antagonistic manner. In parallel, other signals, such as 14−3−3 proteins [
40] or the kinase Open Stomata 1 [
41] can, in response to cold stress, travel to the nucleus and modulate the activity of these transcriptional regulators. While this signalling appears redundant at first, it can be functionally dissected depending on time—some of these signals (such as Open Stomata 1) are rapid and might be elements of signal transduction, while others (such as inhibitory activities of MAP kinases) occur later and might be elements of signal habituation (for a review, see [
28]). The multitude of events linked to membrane–nucleus crosstalk leads to the question of how the nuclear import of DLK integrates with this complex network and what the functional relevance of this phenomenon might be.
A slow and steady drop in temperature, as typically occurs in autumn, allows for cellular adaptation, for instance, by producing proteins that protect membranes and organelles from ice damage as would otherwise occur later in the winter. The accumulation of these cold-responsive (COR) proteins is under the control of the CBF-transcriptional cascade (for a classical review, see [
42]), and CBF4 as a slower, but a sustained member of this cascade might be the pivotal driver of long-term acclimation. However, the response to a rapid cold snap must be different because there is no time to bring cold acclimation to a successful end. In contrast, the plant will only have a chance to survive if it succeeds in mobilising resources from older and more dispensable organs towards meristematic tissues, from where new organs can be re-generated once the cold-stress episode has faded [
43]. This scenario would require cold-induced programmed cell death (PCD), a process that has, indeed, been demonstrated for tobacco BY−2 cells [
44]. To initiate CBF4-dependent cold acclimation under these conditions would be counterproductive. Thus, it is expected that, under conditions of harsh and acute cold stress, the induction of CBF4 that would otherwise ensue from the response to the accumulation of ICE1, has to be suppressed. DLK, as a negative regulator that, in response to such harsh cold stress, is released from the disassembled microtubules (and thus, also will expose its DNA binding leucin zipper in the N-terminal domain), might act as a switch that quells cold acclimation under conditions where cells undergo cold-induced PCD. An implication of this working hypothesis is that the overexpression of CBF4 should mitigate cold-induced mortality. In fact, this is what we see in ongoing experiments.
4. Materials and Methods
Database search for cDNA clones and knock-out mutants. The genomic sequence of OsDLK (accession number Os07g01490) was screened for the availability of knock-out mutants in the RiceGE database (
http://signal.salk.edu/cgi-bin/RiceGE, accessed on 18 October 2013) leading to the identification of the T-DNA insertion line PFG_3A-07110.R (in the background of the
japonica cultivar ‘Dongjin’), and the Tos-17-insertion line ND4501_0_508_1A (in the background of the
japonica cultivar ‘Nipponbare’, NIAS, Tsukuba, JapanDongjinPFG_3A-07110.ROryza sativa L. japonica cv. Dongjin, T-DNA insertion line; Postech, South Korea). Seed material for the T-DNA insertion lines was kindly provided by the National Research Laboratory of Plant Functional Genomics, Division of Molecular and Life Sciences, Pohang University of Science and Technology (POSTECH), Pohang 790−784, Korea, while material for the Tos−17 insertion line was from the National Institute of Agricultural Science (NIAS), Tsukuba, Japan.
Cultivation of rice. For genotyping, the sterilised seeds were grown on floating meshes in photo-biological darkness (using boxes wrapped in black cloth) at 25 °C for 4 days, as described by Nick et al. [
45]. The etiolated coleoptiles were excised and immediately frozen in liquid nitrogen and stored at −80 °C until isolation. For the segregation analysis of coleoptile elongation, plants were raised in the dark, to compare the genotyped homozygotes or heterozygotes from the T-DNA or the Tos−17 insertion lines with the respective wildtype background, as well as with the segregating wildtype seedlings. Seedlings were digitalised on a scanner, and then the coleoptile length from the node to the tip was measured using the periphery tool of ImageJ (
https://imagej.nih.gov/ij/, accessed on 13 April 2015). For the analysis of tissue regulation, seedlings were grown in Magenta boxes (Sigma-Aldrich, St. Louis, MO, USA) on 0.4% [
w/
v] phytoagar (0.6%
w/
v, Duchefa, Haarlem, The Netherlands) under sterile conditions as described in Tang et al. [
46], either at 25 °C in darkness or under continuous white light.
Genotyping of rice mutants. Etiolated coleoptiles were shock-frozen in liquid nitrogen, and ground to a powder in a TissueLyser (Qiagen, Hilden, Germany). Genomic DNA was isolated from 100−200 mg of powdered material based on the standard cetyl trimethyl ammonium bromide (CTAB) protocol using chloroform-isopropanol for partitioning [
47]. Diagnostic sequences for the genotype of the Tos−17 or T-DNA insertion mutants were amplified by two complementary polymerase chain reactions [
48]. The first pair probed the flanking sequences of the putative insertion site, i.e., motifs that would be found in both wildtype and transformant (however, in the transformant, the amplicon would be larger), while the second pair probed a target site located in the respective insert (T-DNA or Tos−17, respectively), while the second site targeted one flanking region of the putative insertion. This reaction would only be amplified in the case of a transformation event. In the segregated wildtype (SeWT) of the T-DNA insertion line, the primer pair KinF1/R1 produced a fragment of 330 bp, while the transgenic allele did not lead to an amplicon. In the case of amplification with the same upstream primer (KinF1) and the T-DNA-specific downstream primer TR, a fragment of 592 bp was amplified for the transgenic allele, while the wildtype allele did not produce a product. Those plants that showed bands with both primers KinF1/R1 and KinF1/TR (flanking T-DNA insertion site) could, thus, be defined as heterozygotes (HZ), while in homozygotes (HO), only the band at 592 bp should be detected. The Tos−17 line was genotyped in a similar manner. In heterozygotes, both primers KinF2/R2 and KinF2/Tos17R would produce bands at 350 bp and 546 bp, respectively, while KinF2/R2 should work for SeWT plants only and KinF2/Tos17R for HO plants only. The sequences of primers for different mutants are listed in
Supplementary Table S1, independently. All PCR reactions used Taq DNA polymerase (NEB) and the following conditions: Initial denaturation for 5 min at 95 °C, followed by 42 cycles of denaturation for 30 s at 95 °C; annealing for 30 s at 60 °C; synthesis for 30 s at 72 °C. The PCR products were separated on 1% [
w/
v] agarose gels.
Transient transformation of rice seedlings by biolistics. Seedlings of rice (
Oryza sativa L.
japonica cultivar
Nihonmasari) were raised in darkness at 25 °C for 4 days. The rice coleoptiles were transiently transformed with the recombinant plasmid pK7FWG2-OsDLK in which OsDLK was C-terminally fused with GFP, via gold particle bombardment as described by Holweg et al. [
49]. During biolistic transformation, the coleoptiles were arranged in the middle of PetriSlides (Millipore, Schwalbach, Germany) with 0.4% phytoagar and fixed with a wire grid. For each plasmid solution, three Petri dishes were prepared. The Petri dishes were then placed in the particle gun and were bombarded three times at a pressure of 2.5 bar in the vacuum chamber at −0.8 bar. Following bombardment, the transformed rice coleoptiles were returned to the dark at 25 °C for 24 h and transferred into an ice bath for the chilling assay.
Microscopy and image analysis. Following biolistic transformation and incubation for 24 h in the dark for the expression of the transgene, individual cells of the coleoptiles were followed over time during cold stress using an AxioImager Z.1 microscope (Zeiss, Jena, Germany) equipped with an ApoTome microscope slider for optical sectioning and a cooled digital CCD camera (AxioCam MRm). GFP fluorescence was recorded through the filter set at 38 HE (Zeiss; excitation at 470 nm, beamsplitter at 495 nm and emission at 525 nm). Acquired images were operated via the AxioVision (Rel. 4.8.2) software. The coleoptiles were excised from the seeds, split longitudinally, and then placed with their epidermal side positioned towards the objective and covered with coverslips. After collecting the z-stack, the slide with a coleoptile slice was returned to the ice bath for further incubation. The whole process was conducted in a dark room.
RNA isolation. The plant material was harvested into liquid nitrogen and kept at −80 °C until RNA extraction. The samples were ground to a powder using the TissueLyser (Qiagen, Hilden, Germany), and RNA was extracted from aliquots of 100 mg of these powdered samples using the innuPREP plant RNA kit (Analytik Jena, Germany), combined with RNase-free DNAse I (Qiagen) to avoid contamination with genomic DNA, according to the manufacturer’s instructions. The purity and integrity of the extracted RNA were determined both spectrophotometrically (Nano-Drop 2000) and by gel electrophoresis (on 1% [w/v] agarose gels).
Measuring steady-state levels of OsDLK transcripts in rice seedlings. The expression patterns of OsDLK were assessed in the O. sativa ssp. japonica cv. Nipponbare wildtype. To address the developmental time course of steady-state levels for the OsDLK transcript in rice, RNA was extracted at daily intervals from seedlings raised for up to 6 days at 25 °C under continuous white light (120 µmol·m
−2·s
−1 of photosynthetically available radiation; Neo tube TLD 36 W/25, Philips, Hamburg). For specimens that still had not initiated germination, the mature embryo was dissected, and for specimens that had germinated, the caryopsis was separated from the seedling. At least three entire individuals were pooled to extract RNA for each time point and experimental replication. To investigate the tissue specificity of OsDLK expression, the rice seedlings were grown under the same conditions for 10 days. Then, different organs, including the entire first leaf (having terminated growth at this stage), the blade and sheath of the second leaf (still elongating), the entire third leaf (in the early phase of elongation), the seminal root (having terminated growth at this stage), and the crown root (beginning to elongate), were separated carefully. Tissues for at least 12 individual seedlings were pooled for each data point and replication. For the auxin dose–response curve of DLK expression, segments of 10 mm length were excised under green safelight from etiolated coleoptiles grown for 4 days at 25 °C as described by [
50]. The young leaf inside each coleoptile was removed with tweezers. The segments were first incubated in water for 1 h to deplete them from endogenous auxin. Subsequently, they were transferred to different concentrations of indole-3-acetic acid (IAA) and incubated in a shaker for either 1 h or 6 h. At least ten segments per data point and replication were pooled to extract RNA extraction and quantitative analysis. All data on transcript levels represent mean values and standard errors from at least three independent experimental series.
Steady-state transcript levels were measured by a quantitative Real-time polymerase chain reaction (RT-PCR) using GoTaq Polymerase (NEB) with an initial denaturation for 3 min at 95 °C, followed by 39 cycles of denaturation for 15 s at 95 °C, annealing for 40 s at 60 °C, and synthesis for 30 s at 72 °C, using a Bio-Rad CFX detection System (Bio-Rad, München, Germany) according to the manufacturer instructions. Signals were visualised using the iQ SYBR Green Supermix (Bio-Rad). The primer pair qDLK fw/re was used to detect the OsDLK transcripts level, and the primer pairs Ubiquitin-10 fw/re and GAPDH fw/re were used for the endogenous control gene (
Supplementary Table S2).
Cold response gene expression in transgenic BY−2 cells. The cold response of gene expression was mapped in tobacco BY−2 (
Nicotiana tabacum L. cv.
Bright Yellow 2) cells that were either non-transformed (wildtype) or that overexpressed a GFP fusion of OsDLK [
7]. Both cell lines were cultivated at 25 °C until the end of their cycling phase (day 3 after subcultivation). To administer cold stress, the flasks with cells were transferred into an ice bath on an orbital shaker at 100 rpm for up to additional four days. In addition to the foreign OsDLK transcripts, several cold-responsive genes of tobacco itself were followed over time. The details for the respective oligonucleotide primers are given in
Supplementary Table S3. The ribosomal gene L25 and elongation factor gene EF-1α were used as housekeeping genes for normalisation. Quantitative RT-PCR was conducted using GoTaq Polymerase (NEB) with an initial denaturation for 3 min at 95 °C, followed by 39 cycles of denaturation for 15 s at 95 °C, annealing for 40 s at 58 °C, and synthesis for 30 s at 72 °C.
Recombinant protein expression and Western Blot. To screen protein–DNA interactions, OsDLKT, a partial fragment containing the tail part harbouring the motor domain of OsDLK, was cloned into the binary plasmid pET-DEST42 via GATEWAY
® cloning as described by Klotz and Nick [
51]. The partial construct OsDLKT was amplified using the forward primer:
5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTCCACGCGCGCCACTCGCC-3′
and the reverse primer:
5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATTCTCTCCGTCCAAAATTTGTT-3′.
The recombinant plasmid pET-DEST42-OsDLKT was transferred into the E. coli strain BL21-Codon Plus (DE3)-RIL. Protein expression was induced by 200 µM of isopropyl-β-D-thiogalactopyranoside (IPTG) in culture flasks containing LB medium supplemented with ampicillin, at 37 °C for 4 h. Cells were collected by centrifugation at 4500× g at 4 °C for 20 min (Hermle Universal centrifuge, Wehingen, Germany), and the sediments were washed with DPI-ELISA buffer (4 mM HEPES pH 7.5, 100 mM KCl, 8% glycerol), supplemented with 1 mM phenyl-methyl-sulphonyl-fluoride (PMSF) and a proteinase inhibitor cocktail (Roche, Germany). The cells were lysed by sonication (UP100H, Hielscher, Germany) with a frequency of 6 cycles of 15 s with 15 s interruption, at 80% power. The crude extracts for proteins of interest were collected through centrifugation at 4500× g at 4 °C for 20 min. As a negative control, cells transformed by the empty vector pET-DEST42 were included as well. The crude extract samples were mixed with the sample buffer. After heating at 95 °C for 5 min, the mixture was split equally into two SDS-polyacrylamide gels and run at 25 mA for 90 min. One of the gels was stained for recording the loading, while the lanes of the other gel were electroblotted onto a polyvinylidene fluoride (PVDF) membrane (Pall Gelman Laboratory, Dreieich, Germany). The expression of the protein was detected with the mouse monoclonal antibody anti-penta His (Qiagen, Hilden; Germany) in a 2000-fold dilution of Tris-Buffered Saline with 0.1% Tween®-20 (TBST). The primary antibody was visualised using anti-mouse IgG conjugated with alkaline phosphatase in a dilution of 1:50,000 of TBST. The signal was developed using the Alkaline Phosphatase Color Development Kit (Thermo Scientific, Langenselbold, Germany).
Identification of DNA binding motifs. To identify candidates for DNA-binding targets of OsDLKT, a DNA–protein-interaction (DPI)-ELISA strategy was used [
20] based on an optimised double-stranded DNA (dsDNA) probe library that allows the high-throughput identification of hexa-nucleotide DNA-binding motifs. The presence of putative
cis elements in potential target promoters was predicted via Plant Care (
http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 6 April 2017).