miR2119, a Novel Transcriptional Regulator, Plays a Positive Role in Woody Plant Drought Tolerance by Mediating the Degradation of the CkBI-1 Gene Associated with Apoptosis
Abstract
1. Introduction
2. Results
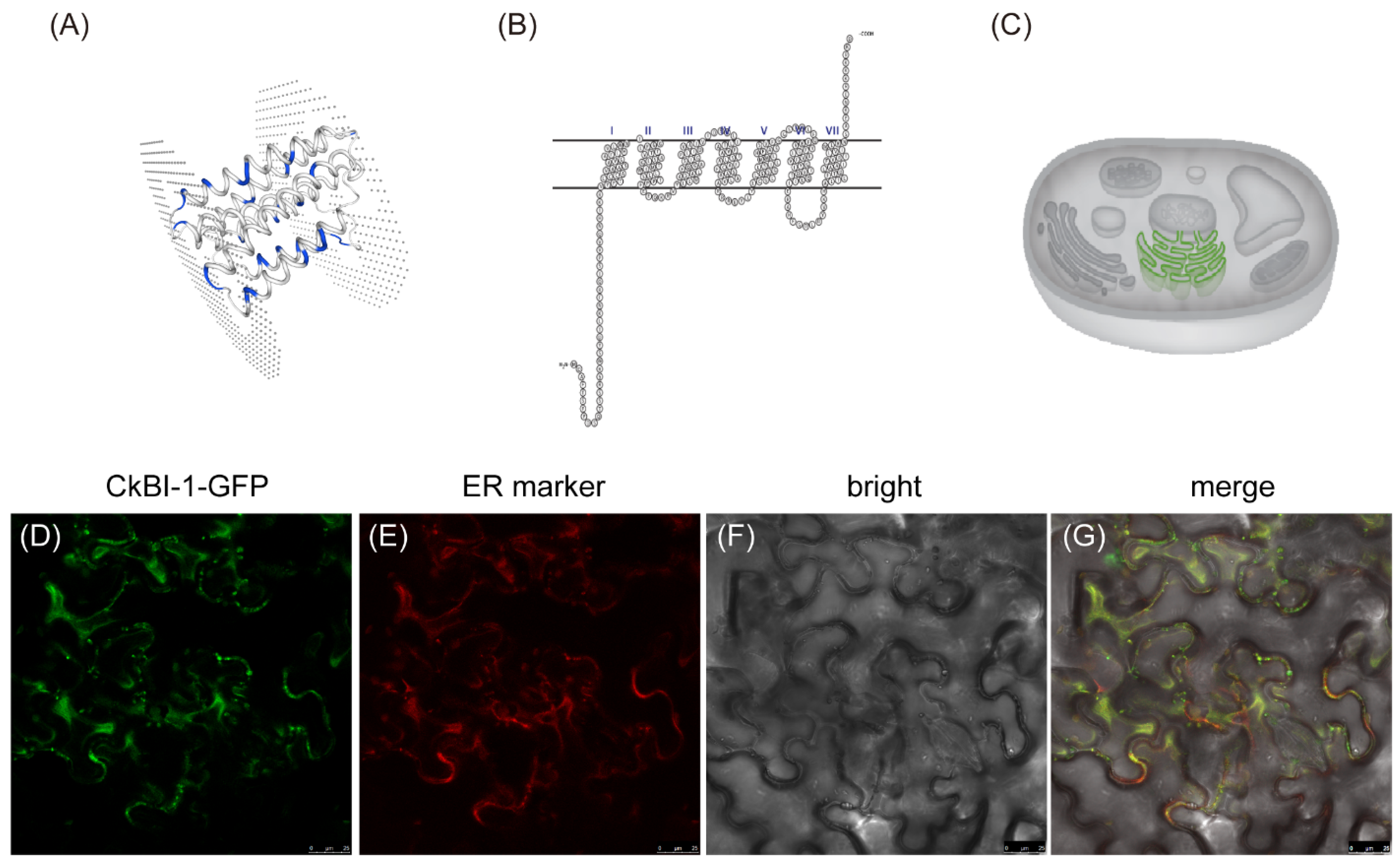
2.1. Protein Structure Prediction and Subcellular Localization of CkBI-1
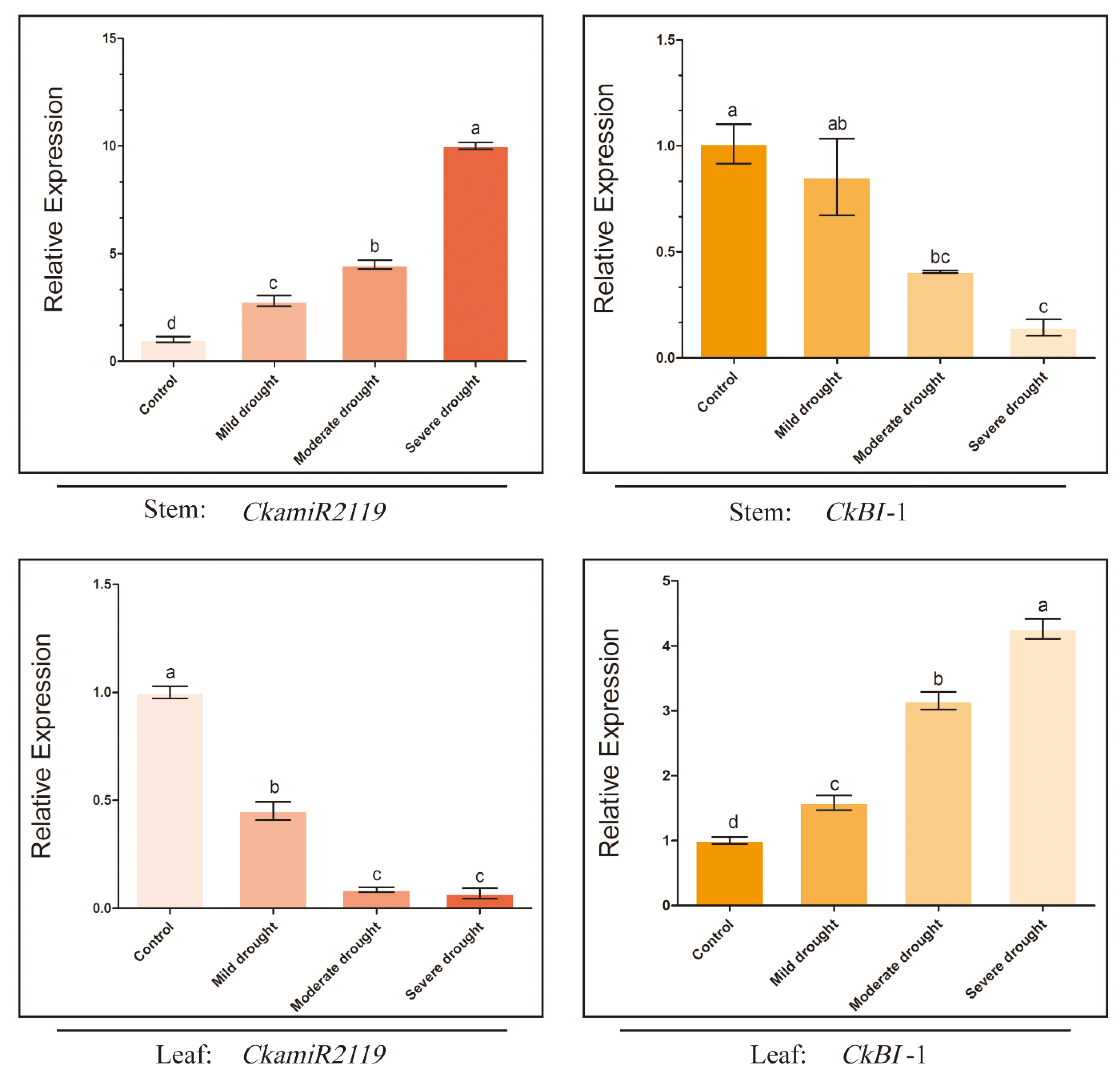
2.2. Different Expression Patterns of CkmiR2119 and CkBI-1 in Stems and Leaves of C. korshinskii
2.3. CkmiR2119 Can Cleave CkBI-1 and Reduce Its Expression Level
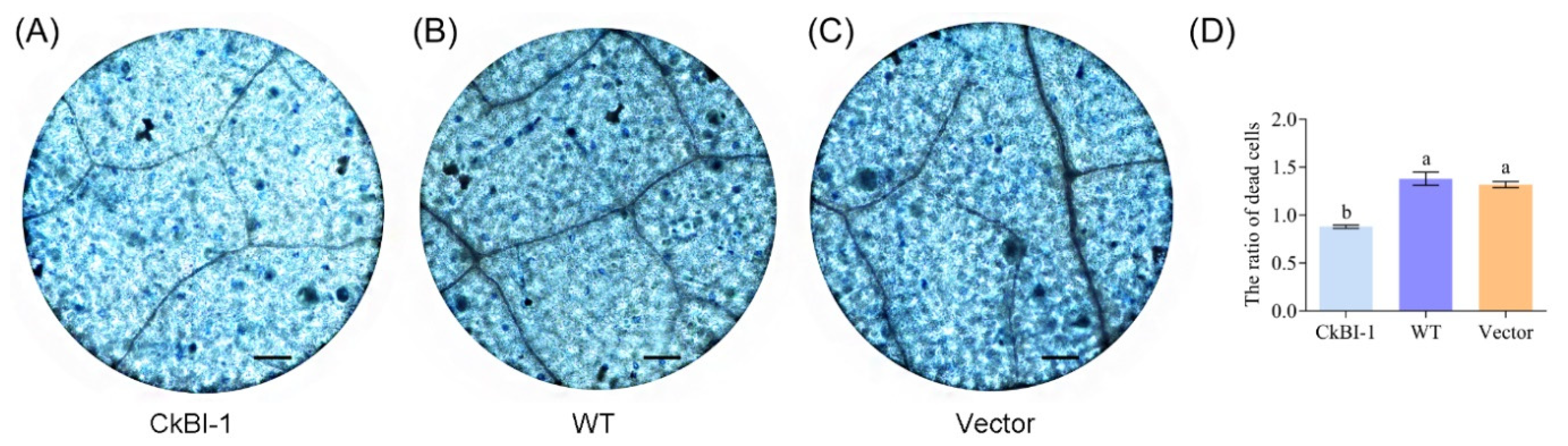
2.4. CkBI-1-OE Line Has the Effect of Inhibiting Cell Death
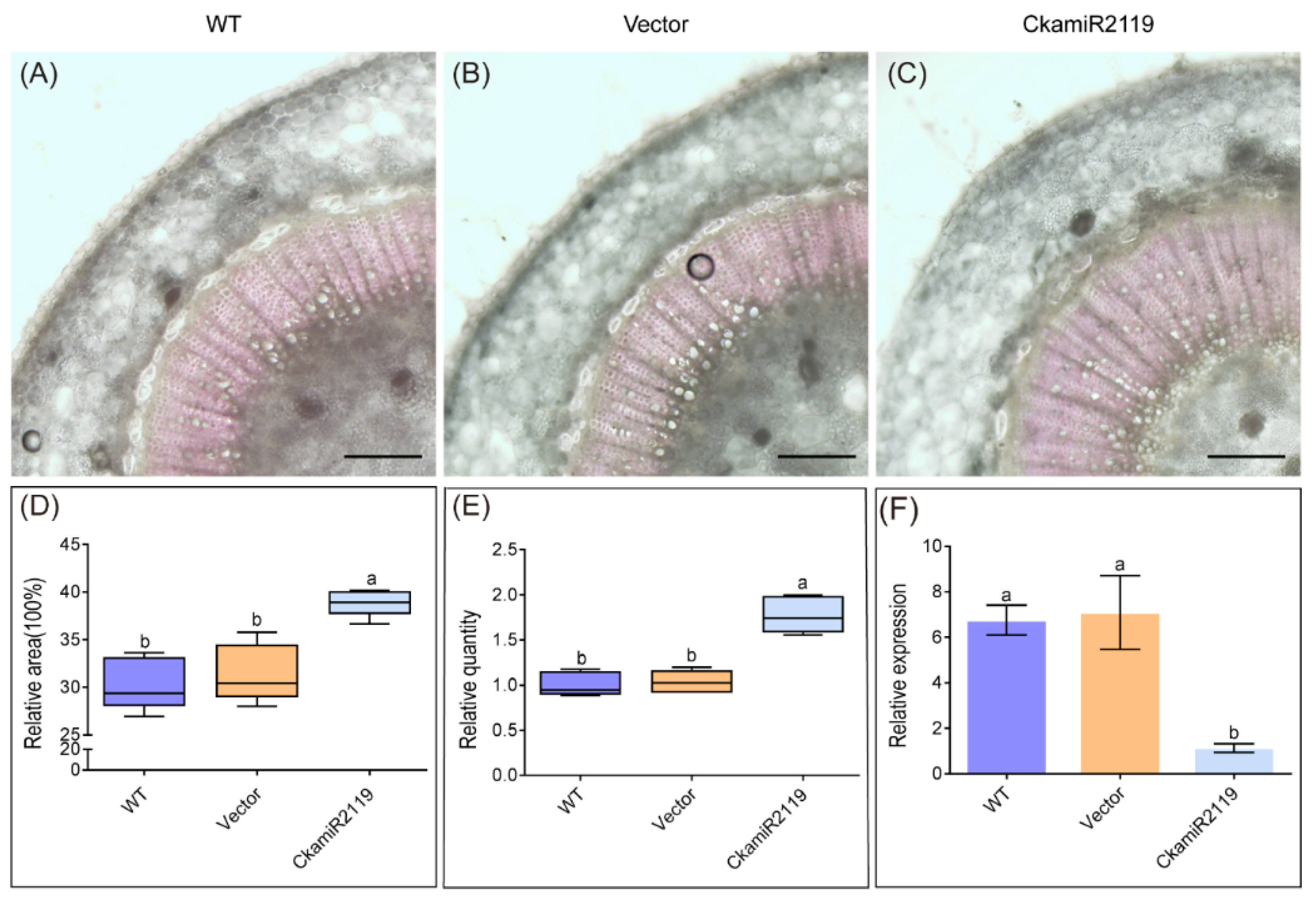
2.5. CkamiR2119-OE Line Has More Developed Stem Xylems
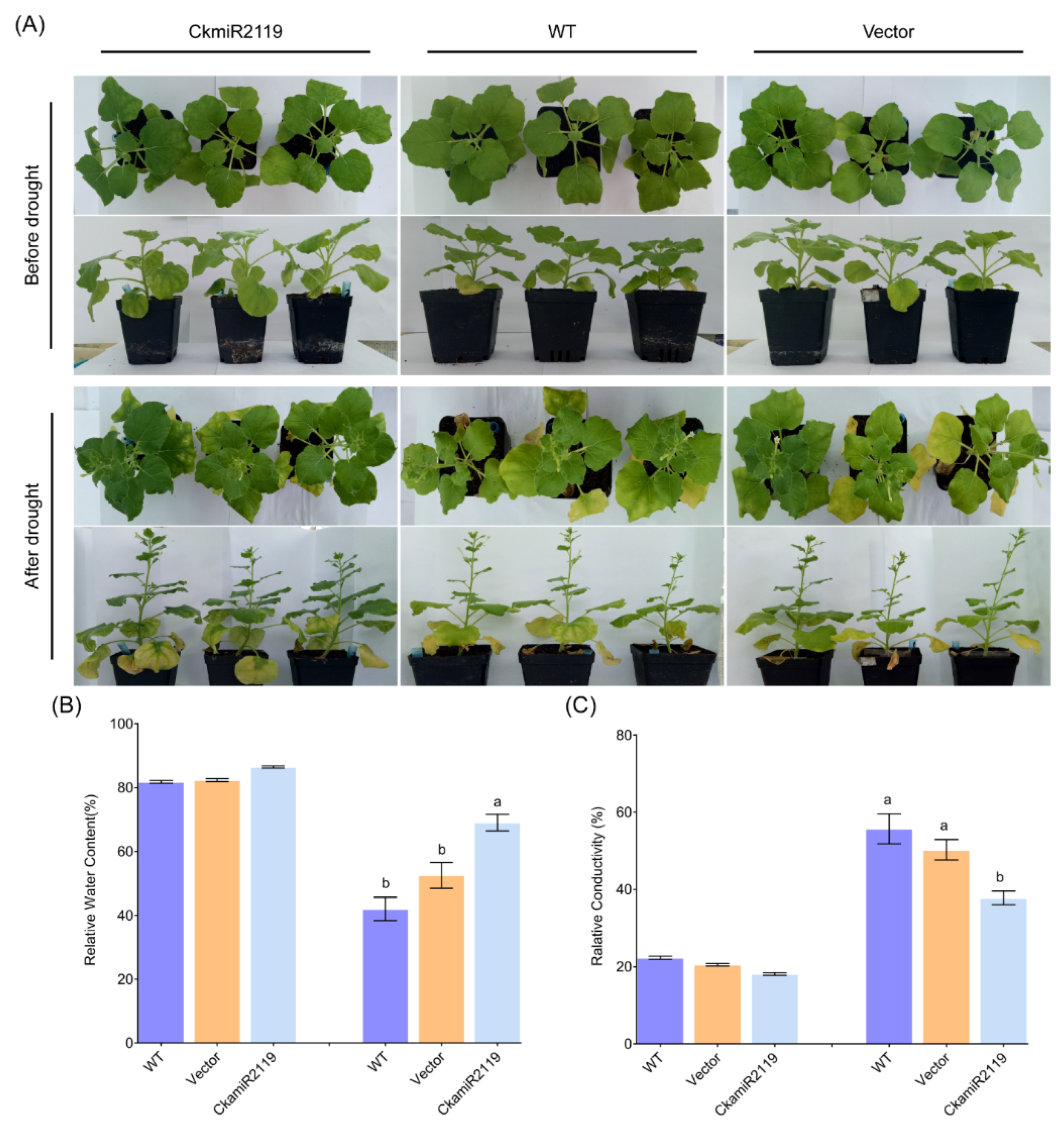
2.6. Drought Tolerance Analysis of CkmiR2119 Transgenic Plants under Drought Stress Treatment
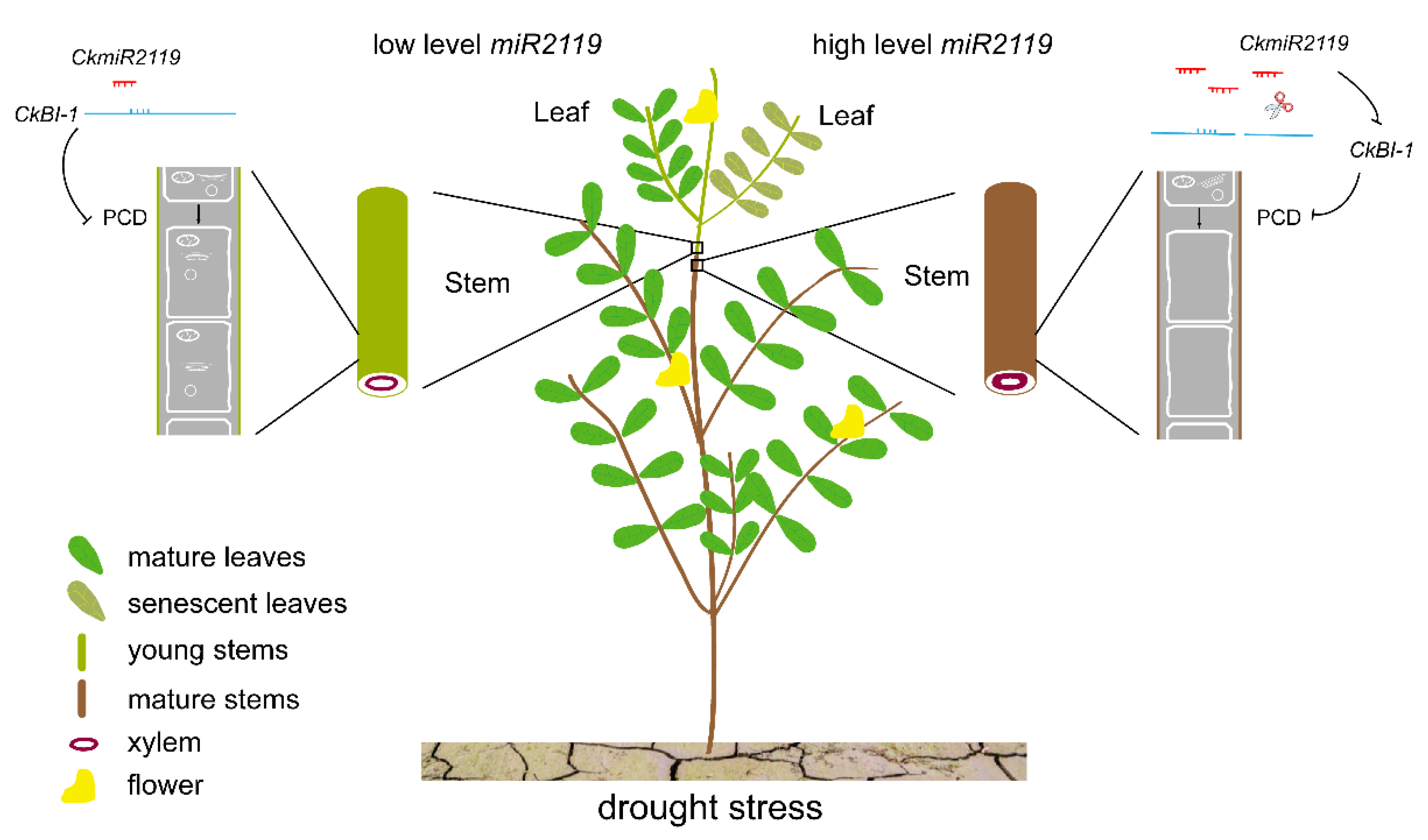
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Protein Structure Prediction
4.3. Relevant Genes Cloning and Plasmids Construction and Subcellular Localization
4.4. Verification of CkBI-1 Cleaves Sites
4.5. Tobacco Transient Expression Experiment and Identification of Stable Expression Transgenic CkmiR2119 and CkBI-1 Tobacco Plants
4.6. Quantitative Real-Time PCR (qRT-PCR) Assay
4.7. Physiological Analysis of the Transgenic and WT Plants under Drought
4.8. Safranin Staining
4.9. Trypan Blue Staining
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sternberg, T. Regional drought has a global impact. Nature 2011, 472, 169. [Google Scholar] [CrossRef]
- Xu, E.; Fan, G.; Niu, S.; Zhao, Z.; Deng, M.; Dong, Y. Transcriptome-wide profiling and expression analysis of diploid and autotetraploid Paulownia tomentosa × Paulownia fortunei under drought stress. PLoS ONE 2014, 9, e113313. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global synthesis of drought effects on cereal, legume, tuber and root crops production: A review. Agric. Water Manag. 2016, 179, 18–33. [Google Scholar] [CrossRef]
- Brendan, C.; Brodribb, T.J.; Brodersen, C.R.; Duursma, R.A.; Rosana, L.; Medlyn, B.E. Triggers of tree mortality under drought. Nature 2018, 558, 531–539. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought resistance in rice from conventional to molecular breeding: A review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim. Biophys. Acta 2008, 1779, 743–748. [Google Scholar] [CrossRef]
- Sun, G. MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 2012, 80, 17–36. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, S.; Yang, T.; Zhang, S.; Chen, Y.Q.; Liu, B. Small RNAs in regulating temperature stress response in plants. J. Integr. Plant Biol. 2017, 59, 774–791. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Zhao, B.; Liang, R.; Ge, L.; Li, W.; Xiao, H.; Lin, H.; Ruan, K.; Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, W.; Li, X.; Sun, D.; Xu, K.; Feng, C.; Kue Foka, I.C.; Ketehouli, T.; Gao, H.; Wang, N.; et al. Integration of sRNA, degradome, transcriptome analysis and functional investigation reveals gma-miR398c negatively regulates drought tolerance via GmCSDs and GmCCS in transgenic Arabidopsis and soybean. BMC Plant Biol. 2020, 20, 190. [Google Scholar] [CrossRef]
- Fei, X.; Li, J.; Kong, L.; Hu, H.; Tian, J.; Liu, Y.; Wei, A. miRNAs and their target genes regulate the antioxidant system of Zanthoxylum bungeanum under drought stress. Plant Physiol. Biochem. 2020, 150, 196–203. [Google Scholar] [CrossRef]
- Ning, P.; Zhou, Y.; Gao, L.; Sun, Y.; Zhou, W.; Liu, F.; Yao, Z.; Xie, L.; Wang, J.; Gong, C. Unraveling the microRNA of Caragana korshinskii along a precipitation gradient on the Loess Plateau, China, using high-throughput sequencing. PLoS ONE 2017, 12, e0172017. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De la Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009, 70, 385–401. [Google Scholar] [CrossRef]
- De la Rosa, C.; Covarrubias, A.A.; Reyes, J.L. A dicistronic precursor encoding miR398 and the legume-specific miR2119 coregulates CSD1 and ADH1 mRNAs in response to water deficit. Plant Cell Environ. 2019, 42, 133–144. [Google Scholar] [CrossRef]
- Yu, F.; Xie, Q. Non-26S Proteasome Endomembrane Trafficking Pathways in ABA Signaling. Trends Plant Sci. 2017, 22, 976–985. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Drakakaki, G. Plant TGN in the stress response: A compartmentalized overview. Curr. Opin. Plant Biol. 2018, 46, 122–129. [Google Scholar] [CrossRef]
- Watanabe, N.; Lam, E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 2008, 283, 3200–3210. [Google Scholar] [CrossRef]
- Isbat, M.; Zeba, N.; Kim, S.R.; Hong, C.B. A BAX inhibitor-1 gene in Capsicum annuum is induced under various abiotic stresses and endows multi-tolerance in transgenic tobacco. J. Plant Physiol. 2009, 166, 1685–1693. [Google Scholar] [CrossRef]
- Jian, S.; Zhao, C.; Fang, S.; Yu, K. Soil water content and water balance simulation of Caragana korshinskii Kom. in the semiarid Chinese Loess Plateau. J. Hydrol. Hydromech. 2014, 62, 89–96. [Google Scholar] [CrossRef]
- Yang, Q.; Yin, J.; Li, G.; Qi, L.; Yang, F.; Wang, R.; Li, G. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 2014, 41, 2325–2334. [Google Scholar] [CrossRef]
- Tao, Q.; Yuanchun, Y.U.; Zhang, W.; Gao, H.; Chen, R.; Bai, L. Pelleting and germination of Caragana korshinskii and Caragana microphylia seeds. J. Anhui Agric. Univ. 2015, 42, 60–64. [Google Scholar] [CrossRef]
- Fu, B.; Wang, S.; Liu, Y.; Liu, J.; Liang, W.; Miao, C. Hydrogeomorphic ecosystem responses to natural and anthropogenic changes in the Loess Plateau of China. Annu. Rev. Earth Planet. Sci. 2016, 45, 223–243. [Google Scholar] [CrossRef]
- Li, Y.J.; Zhao, Z.; Sun, D.X.; Han, G. Hydrological physiological characteristics of Caragana korshinskii under water stress. J. Northwest For. Univ. 2008, 3, 1–4. [Google Scholar]
- Zhou, M. The Adaption of Epidermal Micromorphology and Physiological Mechanism of Caragana Fabr. to Drought Stress; The College of Life Science of Lanzhou University: Lanzhou, China, 2016. [Google Scholar]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef]
- Chen, L.; Luan, Y.; Zhai, J. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 2015, 34, 2013–2025. [Google Scholar] [CrossRef]
- Watanabe, N.; Lam, E. Bax Inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int. J. Mol. Sci. 2009, 10, 3149–3167. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Reed, J.C. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell 1998, 1, 337–346. [Google Scholar] [CrossRef]
- Kawai-Yamada, M.; Jin, L.; Yoshinaga, K.; Hirata, A.; Uchimiya, H. Mammalian bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. USA 2001, 98, 12295–12300. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, N.; Lamb, G.N.; Cessna, S.G.; Brisson, L.F. Modulation of Bax Inhibitor-1 and cytosolic Ca2+ by cytokinins in Nicotiana tabacum cells. Biochimie 2007, 89, 961–971. [Google Scholar] [CrossRef]
- Tyree, M.T. The Cohesion-Tension theory of sap ascent: Current controversies. J. Exp. Bot. 2002, 48, 1753–1765. [Google Scholar] [CrossRef]
- Greenberg, J.T. Programmed cell death: A way of life for plants. Proc. Natl. Acad. Sci. USA 1996, 93, 12094–12097. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Koizumi, K.; Motomatsu, K.; Motose, H.; Sugiyama, M. Molecular mechanisms of vascular pattern formation. Prog. Biotechnol. 2001, 18, 53–61. [Google Scholar]
- Obara, K.; Kuriyama, H.; Fukuda, H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 2001, 125, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Gladish, D.K. Hypoxic stress triggers a programmed cell death pathway to induce vascular cavity formation in Pisum sativum roots. Physiol. Plant. 2012, 146, 413–426. [Google Scholar] [CrossRef]
- Liu, F.; Xie, L.; Yao, Z.; Zhou, Y.; Gong, C. Caragana korshinskii phenylalanine ammonialyase is up-regulated in the phenylpropanoid biosynthesis pathway in response to drought stress. Biotechnol. Biotechnol. Equip. 2019, 33, 842–854. [Google Scholar] [CrossRef]
- Sablok, G.; Pérez-Quintero, A.L.; Hassan, M.; Tatarinova, T.V.; López, C. Artificial microRNAs (amiRNAs) engineering—On how microRNA-based silencing methods have affected current plant silencing research. Biochem. Biophys. Res. Commun. 2011, 406, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Zhang, P.; Li, J.; Zhang, T.; Xie, L.; Gong, C. miR2119, a Novel Transcriptional Regulator, Plays a Positive Role in Woody Plant Drought Tolerance by Mediating the Degradation of the CkBI-1 Gene Associated with Apoptosis. Int. J. Mol. Sci. 2022, 23, 6306. https://doi.org/10.3390/ijms23116306
Liu F, Zhang P, Li J, Zhang T, Xie L, Gong C. miR2119, a Novel Transcriptional Regulator, Plays a Positive Role in Woody Plant Drought Tolerance by Mediating the Degradation of the CkBI-1 Gene Associated with Apoptosis. International Journal of Molecular Sciences. 2022; 23(11):6306. https://doi.org/10.3390/ijms23116306
Chicago/Turabian StyleLiu, Furong, Puzhi Zhang, Jiayang Li, Tianxin Zhang, Lifang Xie, and Chunmei Gong. 2022. "miR2119, a Novel Transcriptional Regulator, Plays a Positive Role in Woody Plant Drought Tolerance by Mediating the Degradation of the CkBI-1 Gene Associated with Apoptosis" International Journal of Molecular Sciences 23, no. 11: 6306. https://doi.org/10.3390/ijms23116306
APA StyleLiu, F., Zhang, P., Li, J., Zhang, T., Xie, L., & Gong, C. (2022). miR2119, a Novel Transcriptional Regulator, Plays a Positive Role in Woody Plant Drought Tolerance by Mediating the Degradation of the CkBI-1 Gene Associated with Apoptosis. International Journal of Molecular Sciences, 23(11), 6306. https://doi.org/10.3390/ijms23116306







