Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.)
Abstract
:1. Introduction
2. Results
2.1. Identification of MsNF-Y Family Genes
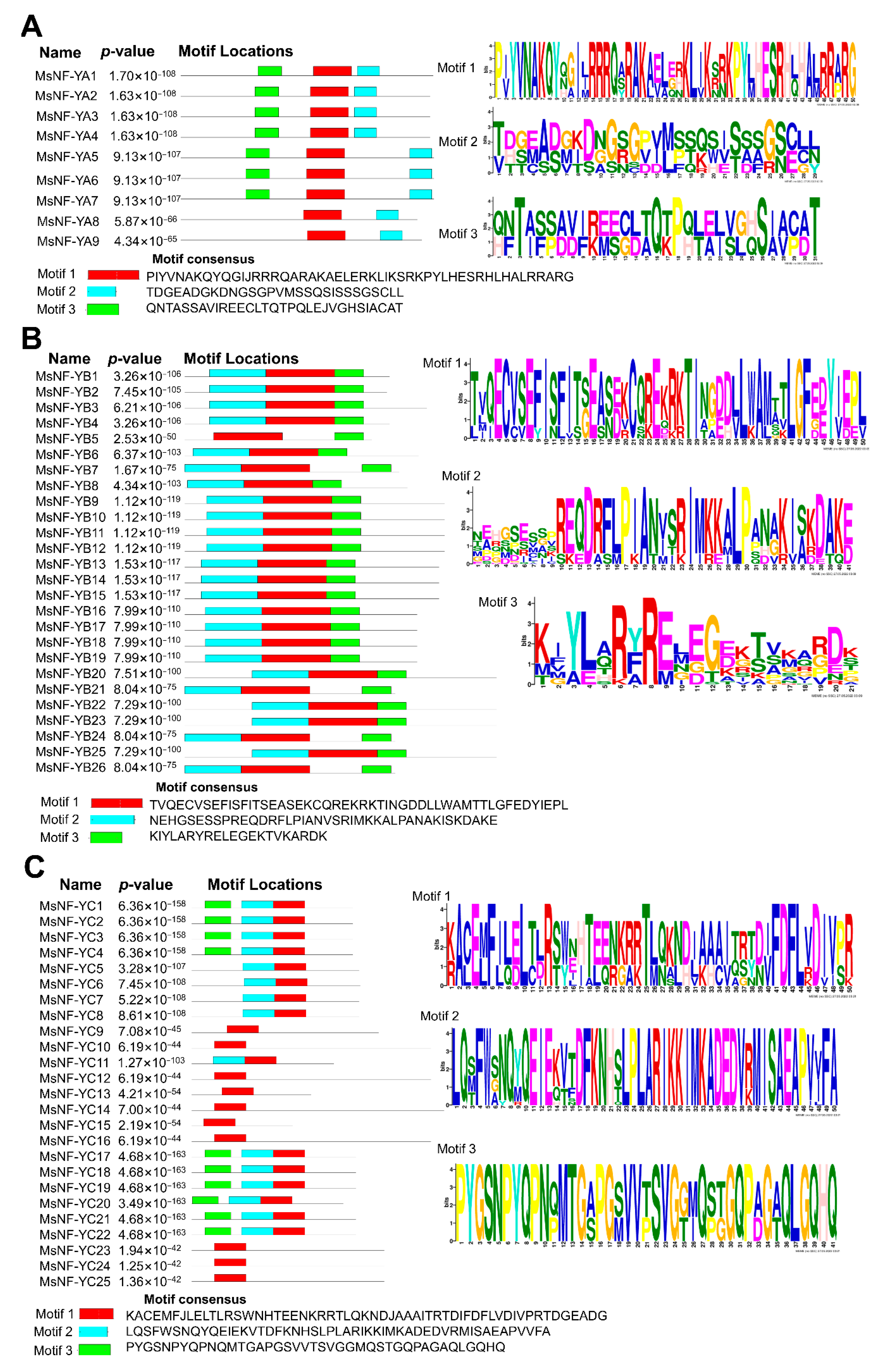
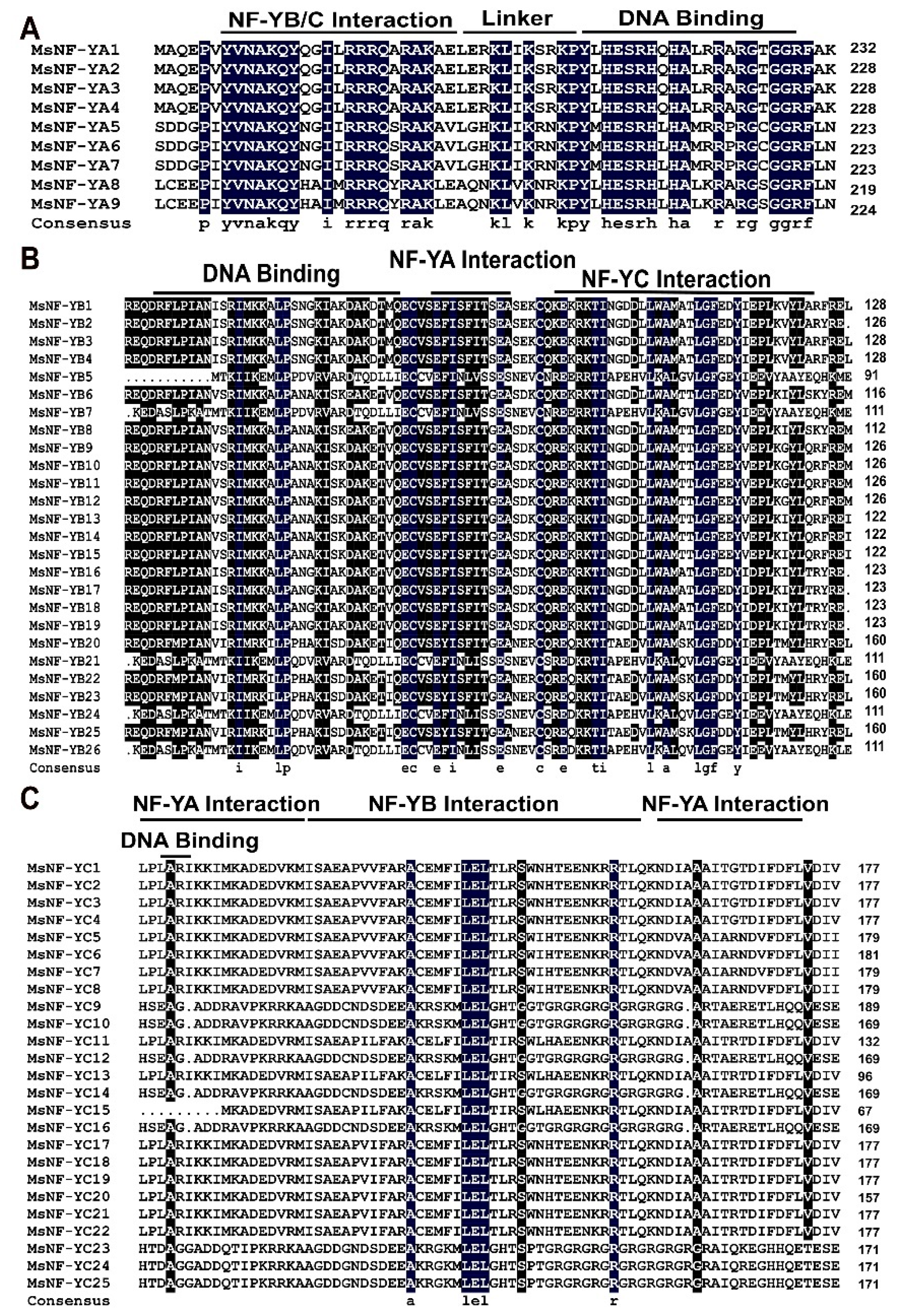
2.2. Analysis of the Motifs and Conserved Domains of MsNF-Y Family Members
2.3. Phylogenetic Relationships and Gene Structure of the MsNF-Y Family Members
2.4. Genome Distribution and Gene Duplication Events of the MsNF-Ys in the Alfalfa Genome
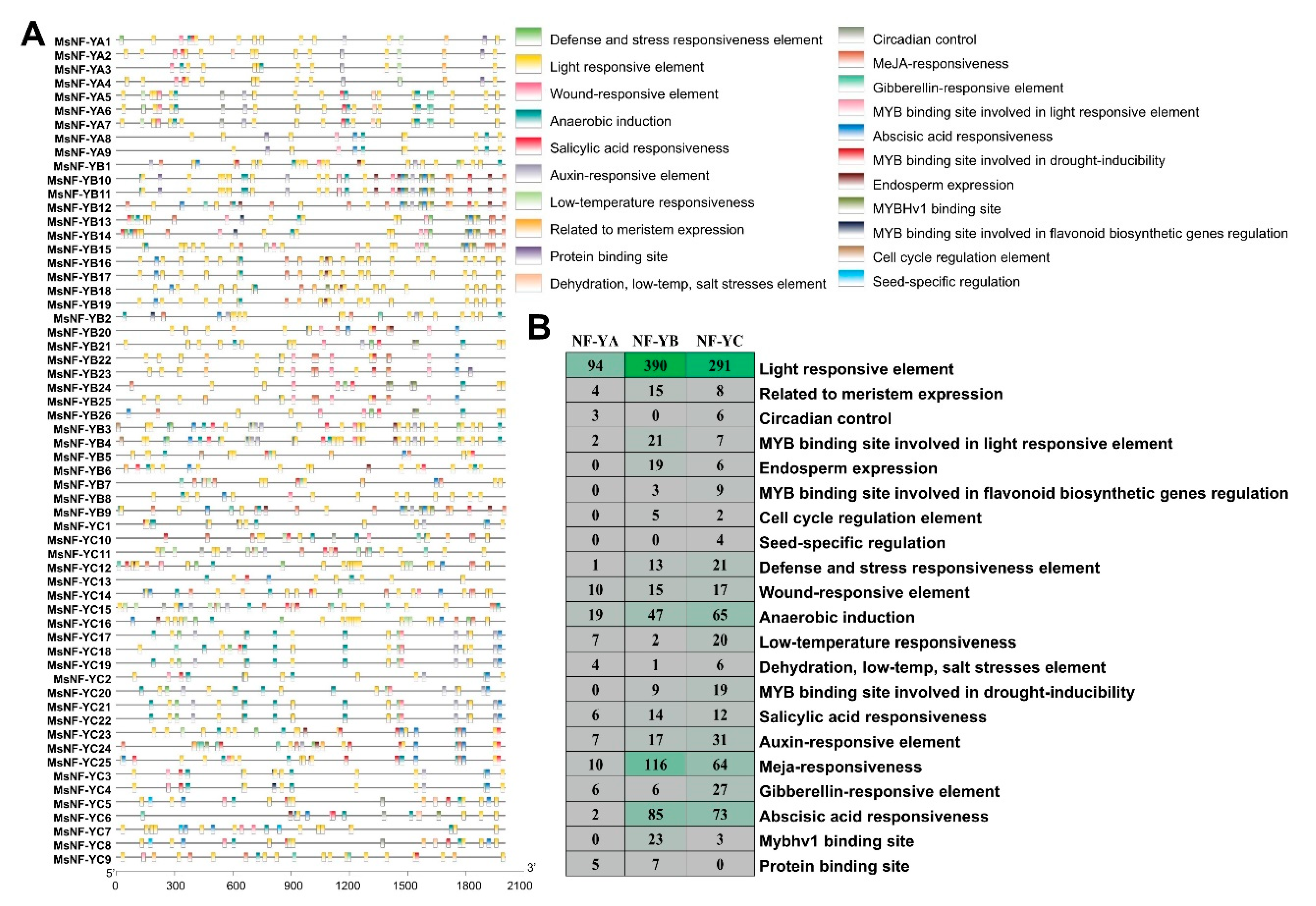
2.5. cis-Element Analysis in the Promoter Regions of MsNF-Y Genes
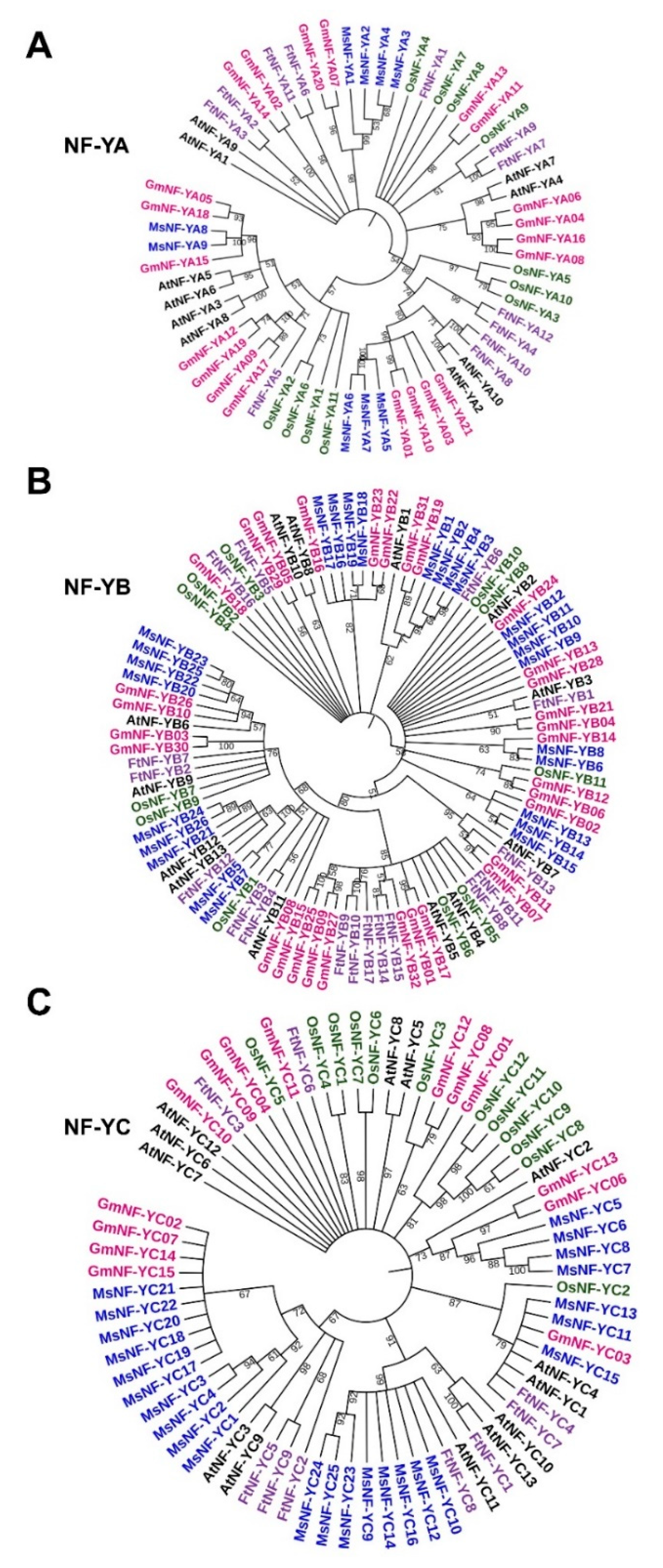
2.6. NF-Y Gene Evolutionary Analysis of Different Plant Species
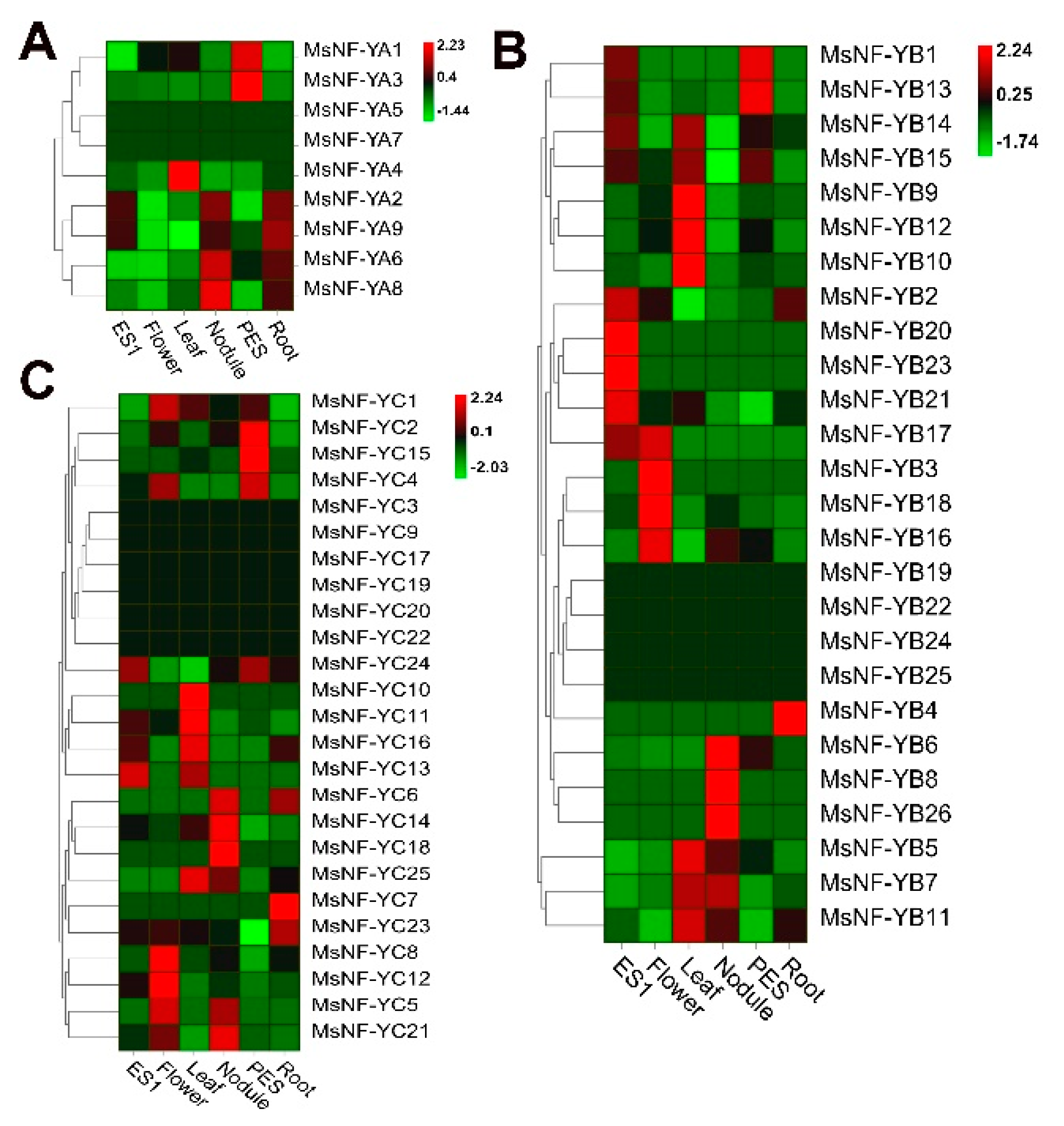
2.7. Expression Analysis of MsNF-Y Genes in Different Tissues from Alfalfa
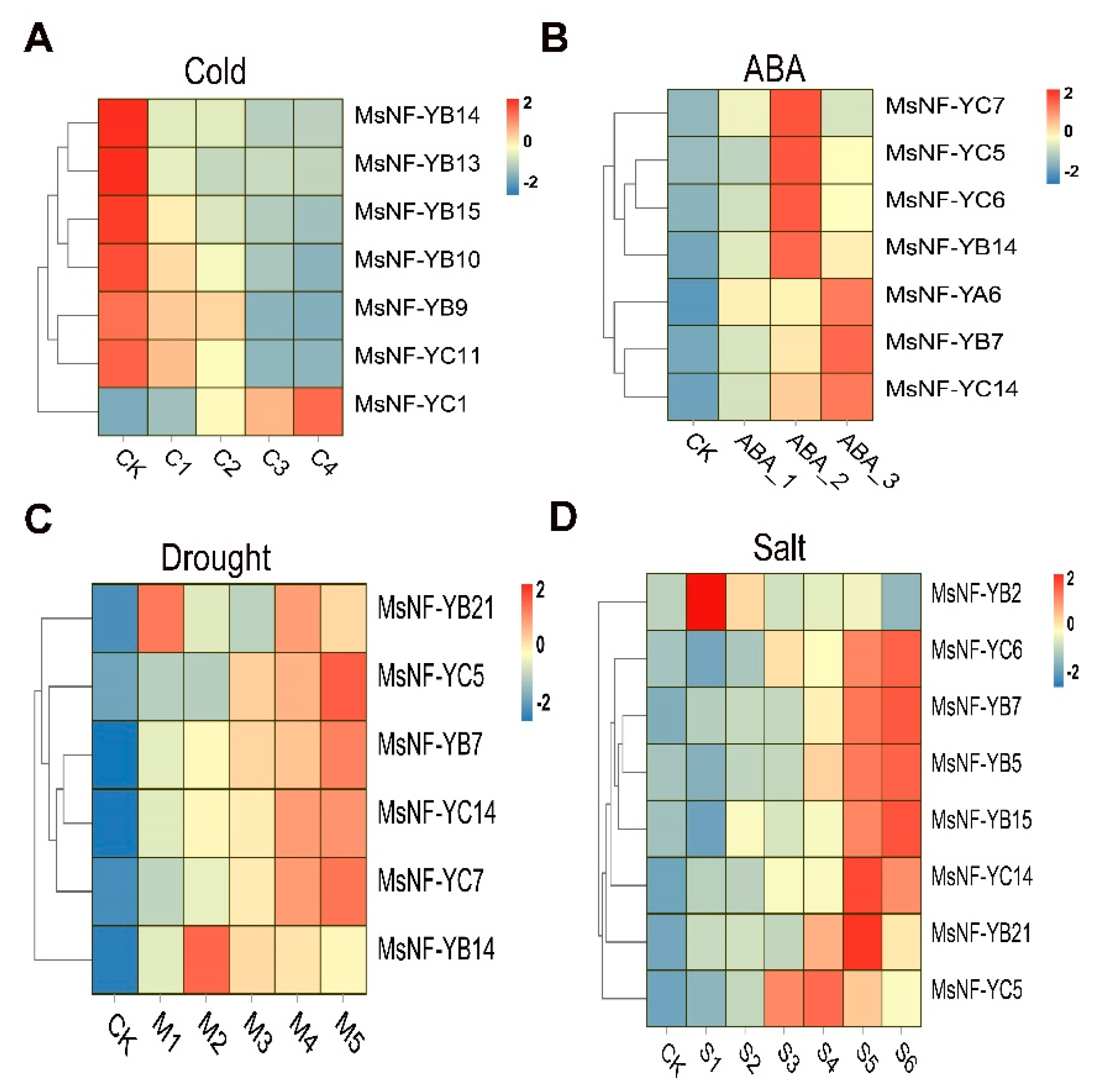
2.8. Expression Analysis of MsNF-Y Genes in Alfalfa under Different Abiotic Stresses
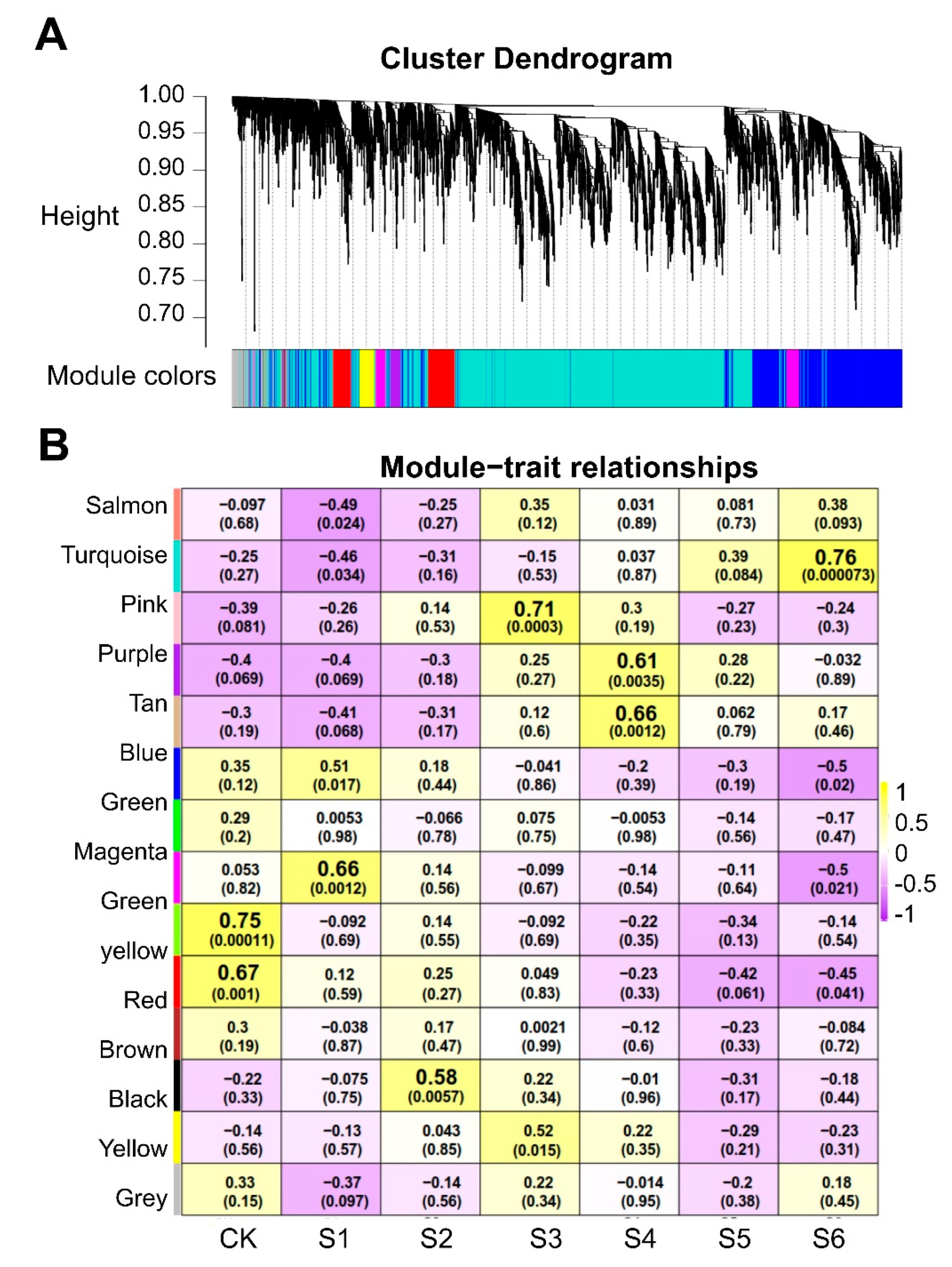
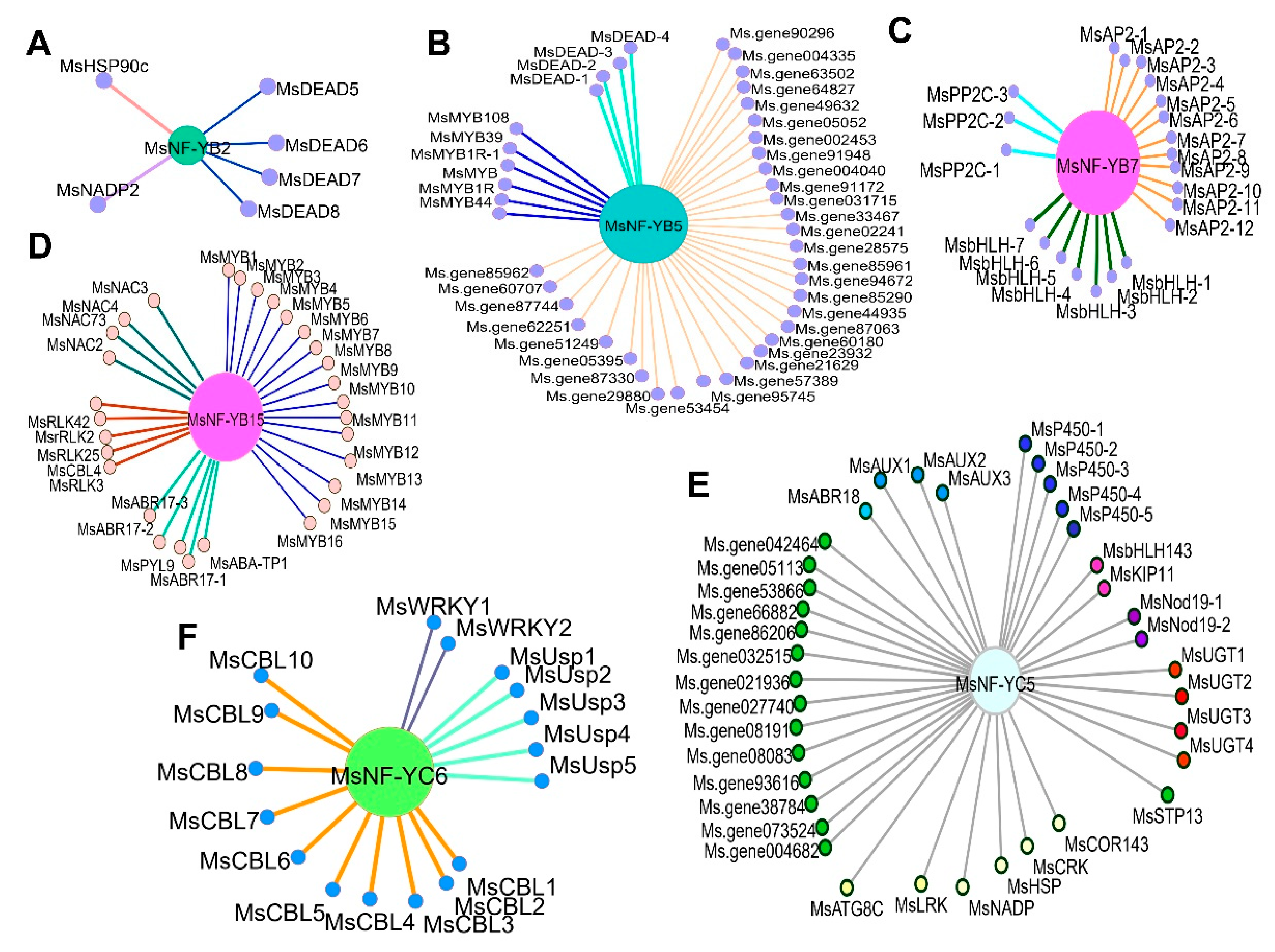
2.9. Important Candidate MsNF-Y Genes in Salt Stress
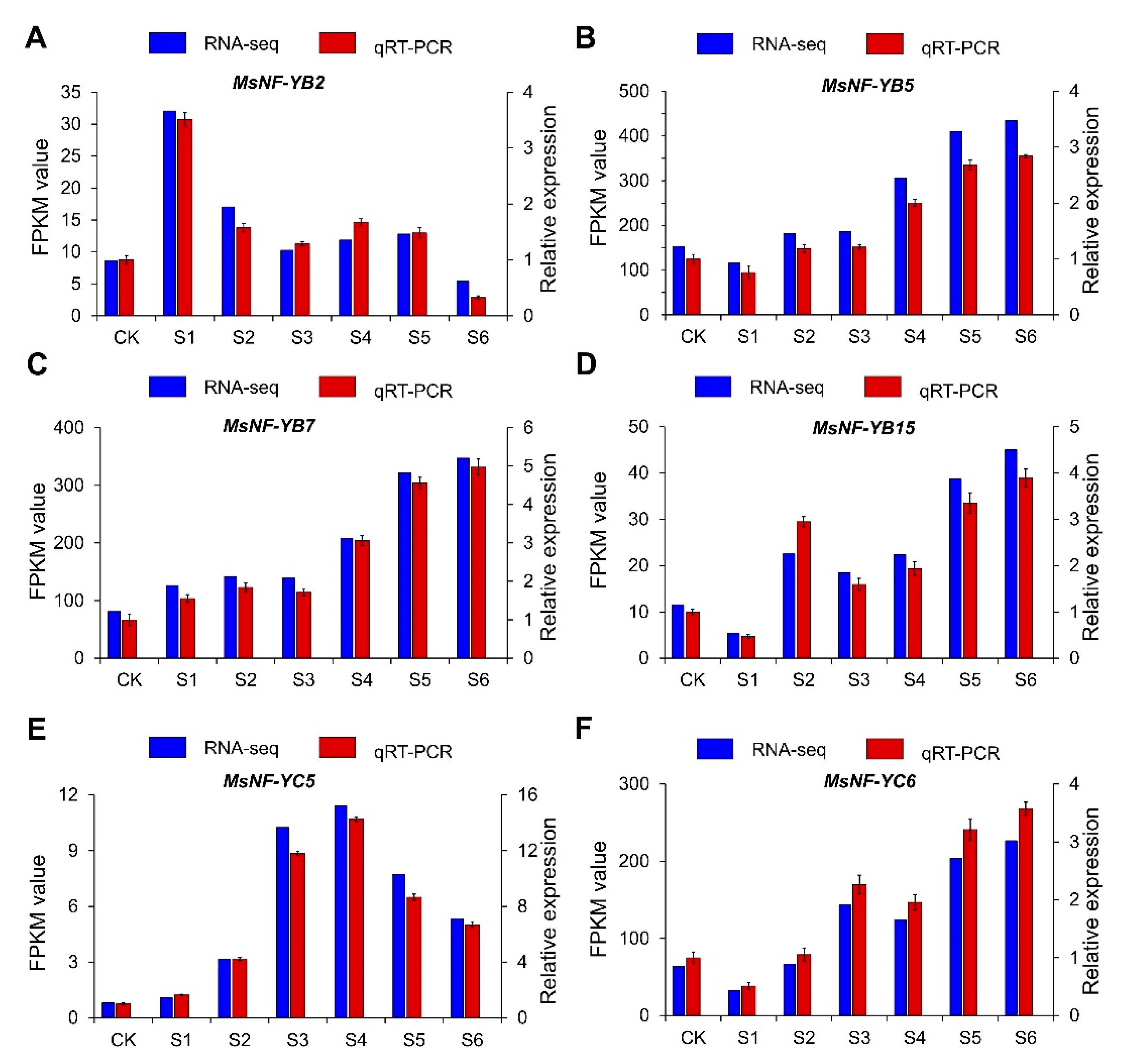
2.10. RT-qPCR Analysis of MsNF-Y Genes under Salt Conditions
3. Discussion
4. Materials and Methods
4.1. Identification of NF-Y Genes in the Alfalfa Genome
4.2. Motif Analysis and Sequence Alignments
4.3. Phylogenetic Tree Construction, Genome Localization and Gene Duplication
4.4. cis-Element Analysis
4.5. Transcription Data Analysis
4.6. Weighted Gene Coexpression Network Construction
4.7. Expression Analysis by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; He, F.; Long, R.; Zhang, F.; Li, M.; Wang, Z.; Kang, J.; Yang, Q. A global alfalfa diversity panel reveals genomic selection signatures in Chinese varieties and genomic associations with root development. J. Integr. Plant Biol. 2021, 63, 1937–1951. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wei, C.; Zhang, Y.; Long, R.; Li, M.; Wang, Z.; Yang, Q.; Kang, J.; Chen, L. Genome-wide association analysis coupled with transcriptome analysis reveals candidate genes related to salt stress in alfalfa (Medicago sativa L.). Front. Plant Sci. 2022, 12, 826584. [Google Scholar] [CrossRef] [PubMed]
- Myers, Z.A.; Holt, B.F. NUCLEAR FACTOR-Y: Still Complex after All These Years? Curr. Opin. Plant Biol. 2018, 45, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Chaves-Sanjuan, A.; Nardini, M. Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Kahle, J.; Baake, M.; Doenecke, D.; Albig, W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin β and importin 13. Mol. Cell Biol. 2005, 25, 5339–5354. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Hu, Y.; Liu, X.; Li, Y.; Hou, X. Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 2015, 27, 3099–3111. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.X.; Deng, H.; Li, T.; Sharma, S.; Yun, Q.; Li, Q.; Zhiguo, E.; Chen, C. OsbZIP76 interacts with OsNF-YBs and regulates endosperm cellularization in rice (Oryza sativa). J. Integr. Plant Biol. 2020, 62, 1983–1996. [Google Scholar] [CrossRef]
- Das, S.; Parida, S.K.; Agarwal, P.; Tyagi, A.K. Transcription factor OsNF-YB9 regulates reproductive growth and development in rice. Planta 2019, 250, 1849–1865. [Google Scholar] [CrossRef]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.; Zhang, Z.; Zhang, J.; Zhou, Y.; Chen, C. The rice LEC1-like transcription factor OsNF-YB9 interacts with SPK, an endosperm-specific sucrose synthase protein kinase, and functions in seed development. Plant J. 2021, 106, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R.M.; et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. New Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef] [PubMed]
- Braybrook, S.A.; Harada, J.J. LECs go crazy in embryo development. Trends Plant Sci. 2008, 13, 624–630. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [Green Version]
- Kwong, R.W.; Bui, A.Q.; Lee, B.H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Liu, C.; Zhao, J.; Ye, X.; Wu, Q.; Yao, T.; Peng, L.; Zou, L.; Zhao, G. Genome-wide analysis of the NF-Y gene family and their roles in relation to fruit development in Tartary buckwheat (Fagopyrum tataricum). Int. J. Biol. Macromol. 2021, 190, 487–498. [Google Scholar] [CrossRef]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post-transcriptionally to promote drought resistance. Plant Cell 2008, 8, 2238–2251. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.J.; Yu, T.F.; Li, X.H.; Cao, X.Y.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Zhang, J.H.; et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 2020, 20, 123. [Google Scholar] [CrossRef] [Green Version]
- Quach, T.N.; Nguyen, H.T.M.; Valliyodan, B.; Joshi, T.; Xu, D.; Nguyen, H.T. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genom. 2015, 290, 1095–1115. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Li, Z.; Ran, Q.; Li, P.; Peng, Z.; Zhang, J. ZmNF-YB16 overexpression improves drought resistance and yield by enhancing photosynthesis and the antioxidant capacity of maize plants. Front. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.F.; Liu, Y.; Fu, J.D.; Ma, J.; Fang, Z.W.; Chen, J.; Zheng, L.; Lu, Z.W.; Zhou, Y.B.; Chen, M.; et al. The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 2021, 19, 2589–2605. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.; Liang, Z. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.). BMC Genom. 2016, 17, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xu, W.; Chen, Z.; Han, B.; Haque, M.E.; Liu, A. Gene structure, expression pattern and interaction of Nuclear Factor-Y family in castor bean (Ricinus communis). Planta 2018, 247, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E., 4th; Tayrose, G.; Holt, B.F., 3rd. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Zhang, C.; Zou, H.; Wu, Z. Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 2016, 478, 752–758. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 2017, 7, 2045. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.; Hu, Z.; Jiang, Q.; Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef]
- Han, X.; Tang, S.; An, Y.; Zheng, D.C.; Xia, X.L.; Yin, W.L. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 2013, 64, 4589–4601. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Cui, S.; Li, X.; Liu, B.; Deng, H.; Liu, Y.; Hou, M.; Yang, X.; Mu, G.; Liu, L. Transcriptome and co-expression network analyses reveal differential gene expression and pathways in response to severe drought stress in peanut (Arachis hypogaea L.). Front. Genet. 2021, 12, 672884. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, 200–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Fu, F.; Bucciarelli, B.; Yang, S.S.; Samac, D.A.; Lamb, J.A.F.S.; Monteros, M.J.; Graham, M.A.; Gronwald, J.W.; Krom, N.; et al. The Medicago sativa gene index 1.2: A web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genom. 2015, 16, 502–518. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Deng, H.; Ma, W.; Qiang, Z.; Liu, Z. Genome-wide identification of the MADS-box transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa L.) and expression analysis under abiotic stress. BMC Genom. 2021, 22, 603. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 36. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]


| Name | Gene ID | CDs | Length (AA) | Chromosome Localization | Mw (kDa) | pI | Homologs in Arabidopsis |
|---|---|---|---|---|---|---|---|
| MsNF-YA1 | MS.gene84539 | 1002 | 333 | chr3.1:46159735:46162043 | 36.08 | 5.68 | AtNF-YA1/9 |
| MsNF-YA2 | MS.gene04098 | 990 | 329 | chr3.2:52938927:52941216 | 35.83 | 5.81 | AtNF-YA1/9 |
| MsNF-YA3 | MS.gene08079 | 990 | 329 | chr3.3:49398206:49400492 | 35.79 | 5.81 | AtNF-YA1/9 |
| MsNF-YA4 | MS.gene04396 | 990 | 329 | chr3.4:57338770:57341055 | 35.77 | 5.81 | AtNF-YA1/9 |
| MsNF-YA5 | MS.gene007115 | 1005 | 334 | chr7.1:6286954:6289818 | 36.28 | 9.48 | AtNF-YA10 |
| MsNF-YA6 | MS.gene23087 | 1005 | 334 | chr7.2:7507261:7510125 | 36.31 | 9.48 | AtNF-YA10 |
| MsNF-YA7 | MS.gene26493 | 1005 | 334 | chr7.3:7757499:7760361 | 36.28 | 9.48 | AtNF-YA10 |
| MsNF-YA8 | MS.gene011285 | 939 | 312 | chr8.2:75039696:75042162 | 33.23 | 7.70 | AtNF-YA6 |
| MsNF-YA9 | MS.gene53329 | 957 | 318 | chr8.4:73707595:73710074 | 35.03 | 8.48 | AtNF-YA6 |
| MsNF-YB1 | MS.gene004698 | 450 | 149 | chr1.1:54900382:54901482 | 16.34 | 5.32 | AtNF-YB1/8/10 |
| MsNF-YB2 | MS.gene32082 | 444 | 147 | chr1.2:55567170:55568359 | 16.09 | 5.54 | AtNF-YB8/10 |
| MsNF-YB3 | MS.gene036253 | 531 | 176 | chr1.3:52425710:52429054 | 19.47 | 5.56 | AtNF-YB1/8/10 |
| MsNF-YB4 | MS.gene40015 | 450 | 149 | chr1.4:59578396:59579496 | 16.34 | 5.32 | AtNF-YB1/8/10 |
| MsNF-YB5 | MS.gene045804 | 411 | 136 | chr2.1:36197735:36200699 | 15.28 | 4.61 | AtNF-YB12/13 |
| MsNF-YB6 | MS.gene048249 | 513 | 170 | chr2.3:62568258:62569485 | 18.58 | 5.48 | AtNF-YB2/3 |
| MsNF-YB7 | MS.gene83228 | 471 | 156 | chr2.4:34709719:34712823 | 17.42 | 4.63 | AtNF-YB12/13 |
| MsNF-YB8 | MS.gene02911 | 489 | 162 | chr2.4:62033643:62034131 | 17.67 | 5.78 | AtNF-YB3 |
| MsNF-YB9 | MS.gene94950 | 570 | 189 | chr3.1:44574260:44574829 | 20.12 | 6.21 | AtNF-YB2/3 |
| MsNF-YB10 | MS.gene05647 | 570 | 189 | chr3.2:51228991:51229560 | 20.12 | 6.21 | AtNF-YB2/3 |
| MsNF-YB11 | MS.gene04197 | 570 | 189 | chr3.3:48302965:48303534 | 20.06 | 6.21 | AtNF-YB2/3 |
| MsNF-YB12 | MS.gene04298 | 570 | 189 | chr3.4:55907374:55907943 | 20.12 | 6.21 | AtNF-YB2/3 |
| MsNF-YB13 | MS.gene37951 | 558 | 185 | chr5.1:76486331:76486888 | 20.21 | 5.87 | AtNF-YB2/3 |
| MsNF-YB14 | MS.gene81327 | 558 | 185 | chr5.2:80485723:80486280 | 20.32 | 6.23 | AtNF-YB2/3 |
| MsNF-YB15 | MS.gene020793 | 558 | 185 | chr5.3:77328038:77328595 | 20.20 | 5.75 | AtNF-YB2/3 |
| MsNF-YB16 | MS.gene22469 | 510 | 169 | chr7.1:8232375:8233608 | 18.62 | 8.49 | AtNF-YB1/8/10 |
| MsNF-YB17 | MS.gene20033 | 510 | 169 | chr7.2:11459416:11460648 | 18.62 | 8.49 | AtNF-YB1/8/10 |
| MsNF-YB18 | MS.gene007467 | 510 | 169 | chr7.2:11511734:11512962 | 18.62 | 8.49 | AtNF-YB1/8/10 |
| MsNF-YB19 | MS.gene09802 | 510 | 169 | chr7.3:12232958:12234190 | 18.62 | 8.49 | AtNF-YB1/8/10 |
| MsNF-YB20 | MS.gene012740 | 714 | 237 | chr8.1:730789:733261 | 26.28 | 6.26 | AtNF-YB6 |
| MsNF-YB21 | MS.gene044533 | 462 | 153 | chr8.1:14534380:14537789 | 17.15 | 4.72 | AtNF-YB12/13 |
| MsNF-YB22 | MS.gene95097 | 708 | 235 | chr8.2:735520:737984 | 26.14 | 6.26 | AtNF-YB6 |
| MsNF-YB23 | MS.gene042085 | 708 | 235 | chr8.3:558353:560819 | 26.14 | 6.26 | AtNF-YB6 |
| MsNF-YB24 | MS.gene038788 | 462 | 153 | chr8.3:14583864:14587257 | 17.15 | 4.72 | AtNF-YB12/13 |
| MsNF-YB25 | MS.gene43428 | 708 | 235 | chr8.4:784437:786904 | 26.14 | 6.26 | AtNF-YB6 |
| MsNF-YB26 | MS.gene033001 | 462 | 153 | chr8.4:17356790:17360381 | 17.15 | 4.72 | AtNF-YB12/13 |
| MsNF-YC1 | MS.gene96763 | 768 | 255 | chr1.1:61200434:61201201 | 28.41 | 5.62 | AtNF-YC1/2/3/9 |
| MsNF-YC2 | MS.gene055556 | 768 | 255 | chr1.2:60238503:60239270 | 28.40 | 5.62 | AtNF-YC1/2/3/9 |
| MsNF-YC3 | MS.gene44078 | 768 | 255 | chr1.3:58221159:58221926 | 28.40 | 5.62 | AtNF-YC1/2/3/9 |
| MsNF-YC4 | MS.gene005213 | 768 | 255 | chr1.4:66923142:66923909 | 28.40 | 5.62 | AtNF-YC1/2/3/9 |
| MsNF-YC5 | MS.gene073979 | 798 | 265 | chr2.1:30297070:30297867 | 29.76 | 5.79 | AtNF-YC1/2/3/4/9 |
| MsNF-YC6 | MS.gene039020 | 804 | 267 | chr2.2:24489042:24489845 | 30.13 | 5.97 | AtNF-YC1/2/3/4/9 |
| MsNF-YC7 | MS.gene77621 | 798 | 265 | chr2.3:30013127:30013924 | 29.85 | 5.97 | AtNF-YC1/2/3/4/9 |
| MsNF-YC8 | MS.gene92165 | 798 | 265 | chr2.4:27659056:27659853 | 29.82 | 5.79 | AtNF-YC1/2/3/4/9 |
| MsNF-YC9 | MS.gene49550 | 891 | 296 | chr3.1:8392449:8394316 | 32.70 | 5.66 | AtNF-YC11 |
| MsNF-YC10 | MS.gene039544 | 1140 | 379 | chr3.1:8402787:8408425 | 41.59 | 5.75 | AtNF-YC11 |
| MsNF-YC11 | MS.gene06223 | 678 | 225 | chr3.1:79056763:79057440 | 25.17 | 5.65 | AtNF-YC1/4 |
| MsNF-YC12 | MS.gene97173 | 1140 | 379 | chr3.2:6048908:6054534 | 41.60 | 5.75 | AtNF-YC11 |
| MsNF-YC13 | MS.gene055499 | 570 | 189 | chr3.2:82525648:82526217 | 20.97 | 5.71 | AtNF-YC1/4 |
| MsNF-YC14 | MS.gene88253 | 1203 | 400 | chr3.3:7831439:7839020 | 43.94 | 8.95 | AtNF-YC11 |
| MsNF-YC15 | MS.gene013147 | 483 | 160 | chr3.3:81706695:81707177 | 17.32 | 4.96 | AtNF-YC1/4 |
| MsNF-YC16 | MS.gene046309 | 1140 | 379 | chr3.4:8318736:8325208 | 41.60 | 5.75 | AtNF-YC11 |
| MsNF-YC17 | MS.gene050860 | 783 | 260 | chr7.1:2796742:2797524 | 29.04 | 6.03 | AtNF-YC1/2/3/4/9 |
| MsNF-YC18 | MS.gene29325 | 783 | 260 | chr7.2:3652860:3653642 | 29.04 | 6.03 | AtNF-YC1/2/3/4/9 |
| MsNF-YC19 | MS.gene67561 | 783 | 260 | chr7.3:3320464:3321246 | 29.04 | 6.03 | AtNF-YC1/2/3/4/9 |
| MsNF-YC20 | MS.gene050859 | 723 | 240 | chr7.3:3331958:3332680 | 26.95 | 6.04 | AtNF-YC1/2/3/4/9 |
| MsNF-YC21 | MS.gene072458 | 783 | 260 | chr7.3:3363915:3364697 | 29.04 | 6.03 | AtNF-YC1/2/3/4/9 |
| MsNF-YC22 | MS.gene072460 | 783 | 260 | chr7.3:3435457:3436239 | 29.04 | 6.03 | AtNF-YC1/2/3/4/9 |
| MsNF-YC23 | MS.gene063839 | 918 | 305 | chr8.1:62879212:62882462 | 33.26 | 5.08 | AtNF-YC11 |
| MsNF-YC24 | MS.gene88394 | 918 | 305 | chr8.3:56793662:56796860 | 33.37 | 5.13 | AtNF-YC11 |
| MsNF-YC25 | MS.gene78092 | 918 | 305 | chr8.4:58034595:58037795 | 33.39 | 5.02 | AtNF-YC11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, Y.; Suo, X.; Niu, Q.; Yin, S.; Chen, L. Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 2022, 23, 6426. https://doi.org/10.3390/ijms23126426
An Y, Suo X, Niu Q, Yin S, Chen L. Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.). International Journal of Molecular Sciences. 2022; 23(12):6426. https://doi.org/10.3390/ijms23126426
Chicago/Turabian StyleAn, Yixin, Xin Suo, Qichen Niu, Shuxia Yin, and Lin Chen. 2022. "Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.)" International Journal of Molecular Sciences 23, no. 12: 6426. https://doi.org/10.3390/ijms23126426
APA StyleAn, Y., Suo, X., Niu, Q., Yin, S., & Chen, L. (2022). Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.). International Journal of Molecular Sciences, 23(12), 6426. https://doi.org/10.3390/ijms23126426








