A NAC Transcription Factor from ‘Sea Rice 86′ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings
Abstract
:1. Introduction
2. Results
2.1. Isolation and Sequence Analysis of OsNACL35
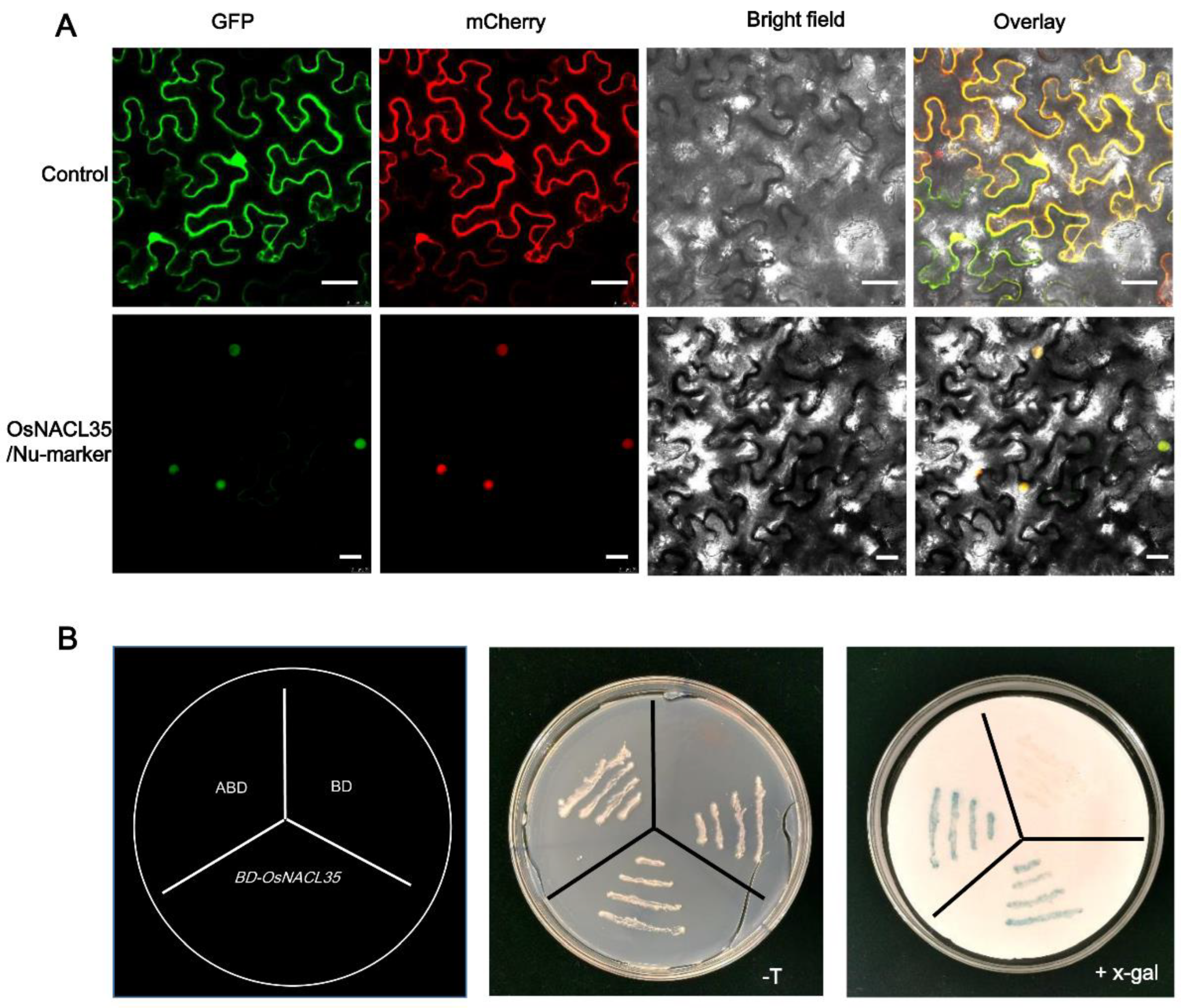
2.2. OsNACL35 Is Localized in the Nucleus and Displays Transactivation Activity in Yeast
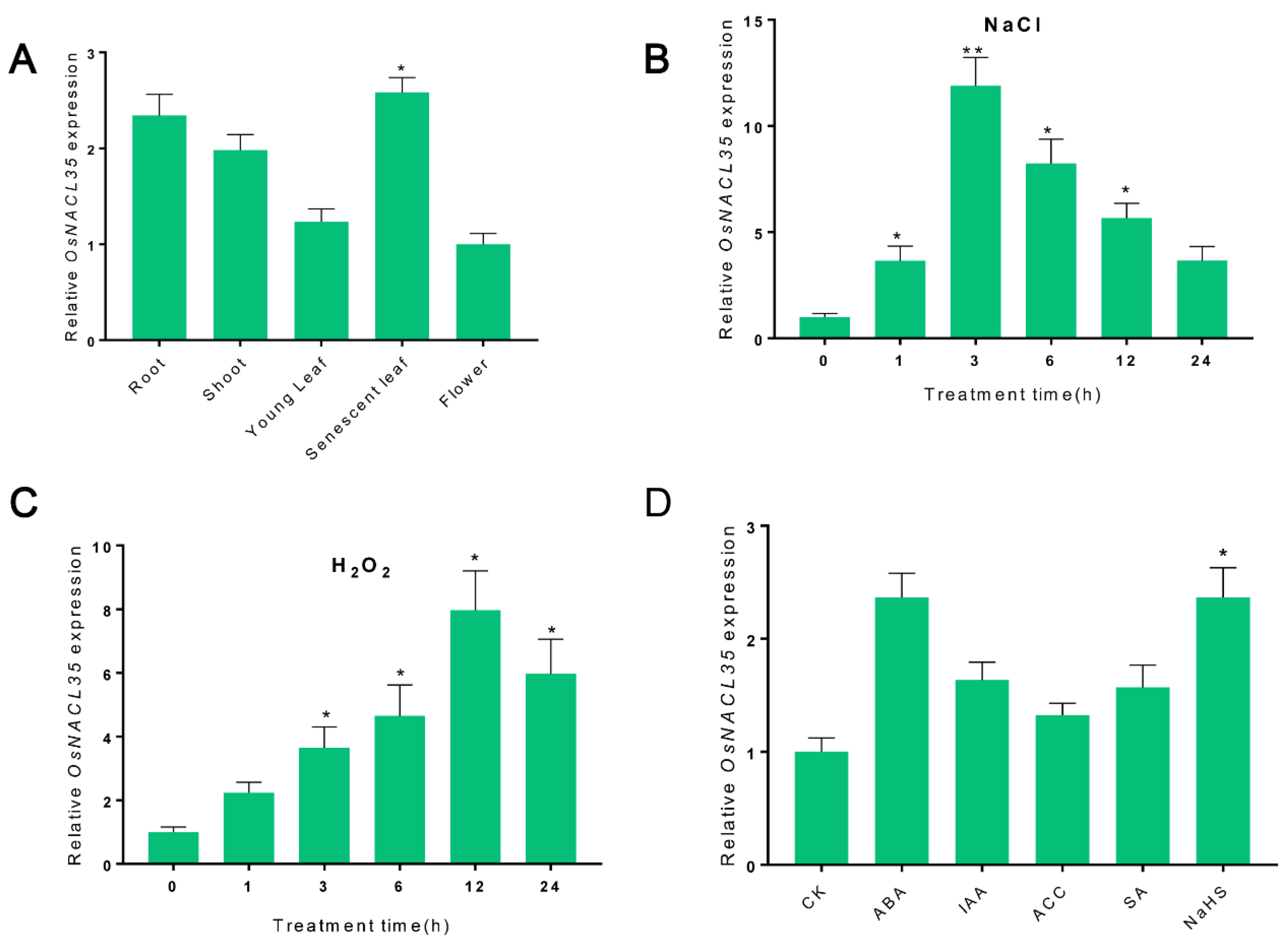
2.3. Expression Pattern of OsNACL35 in SR86
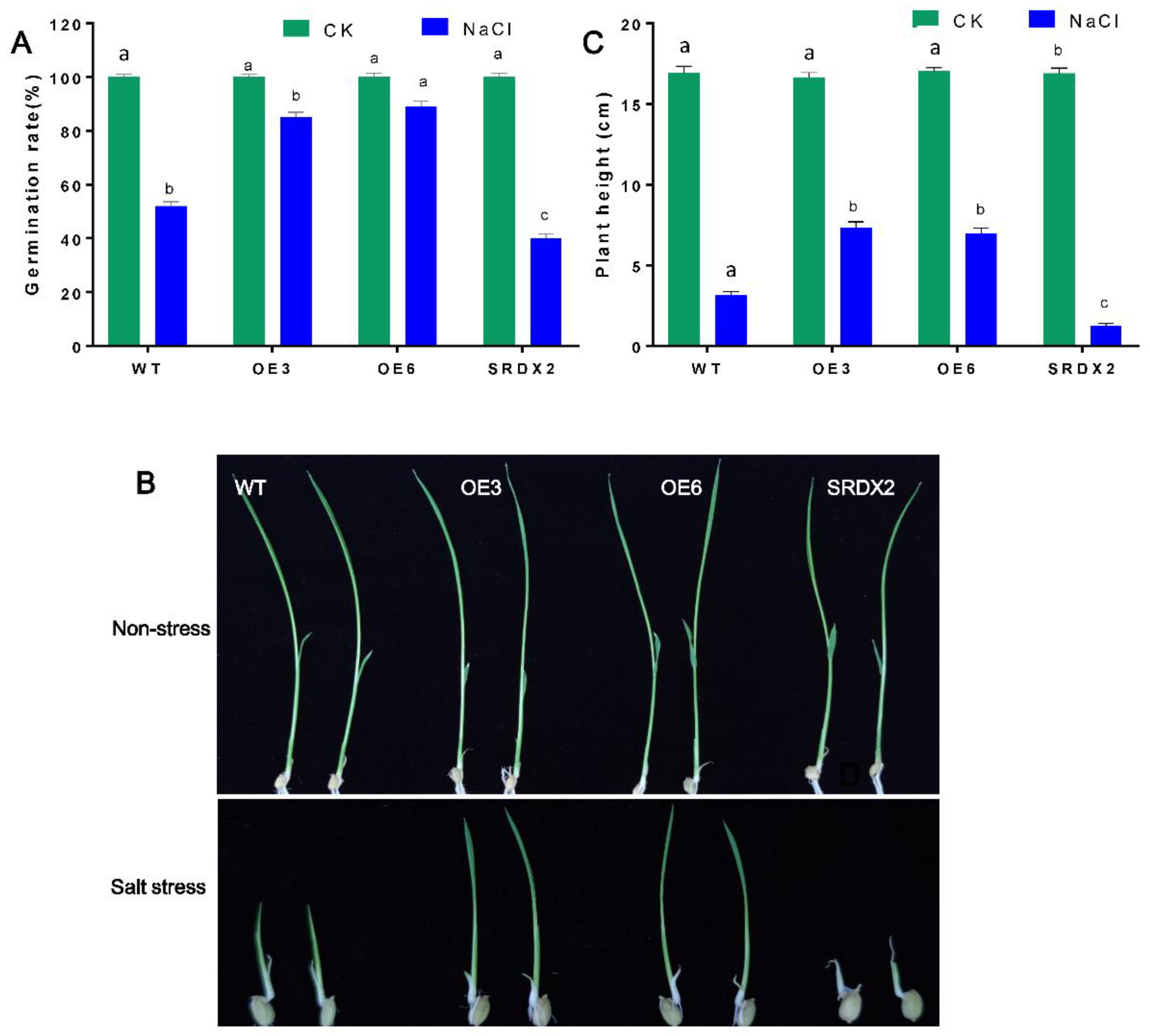
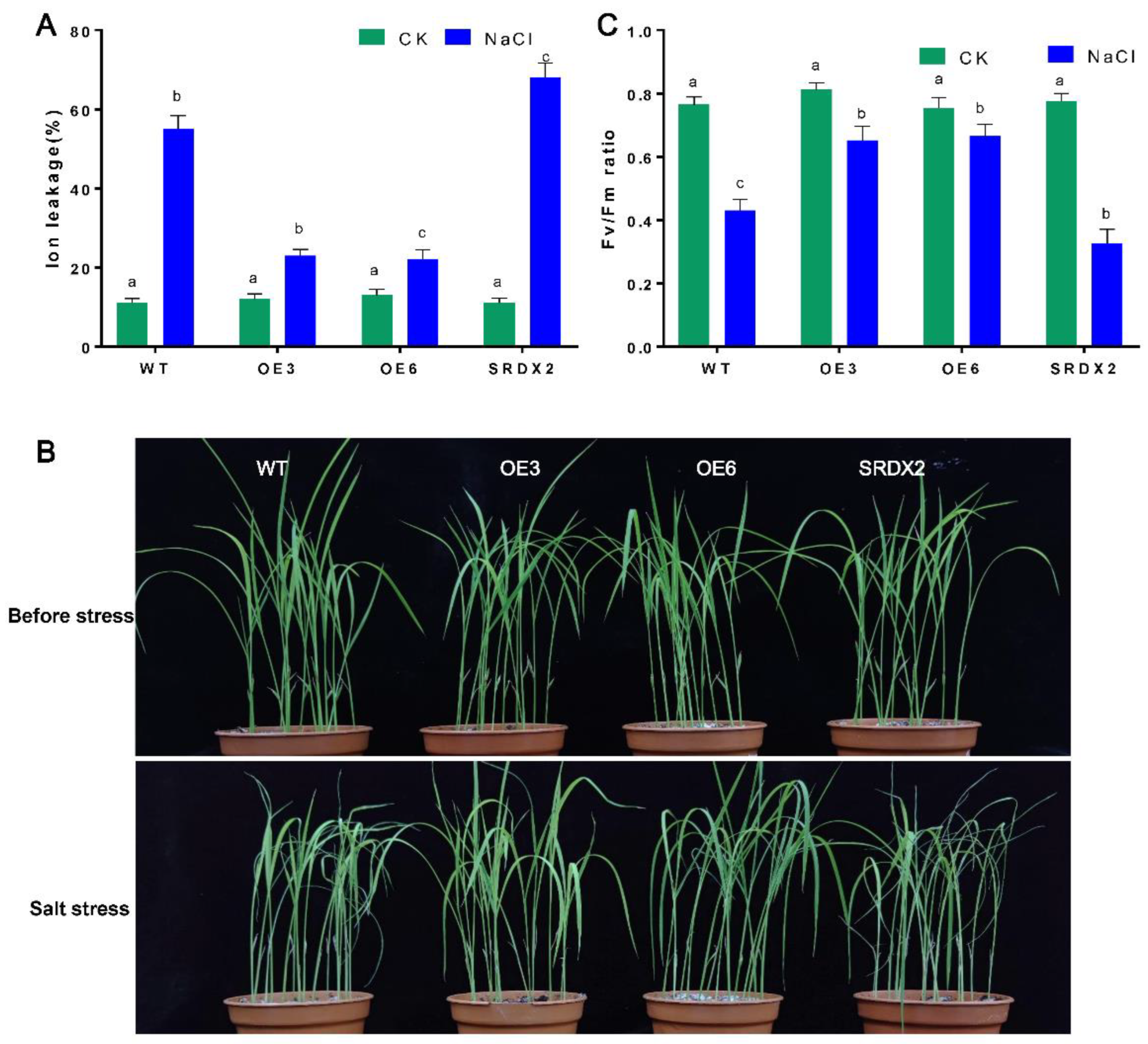
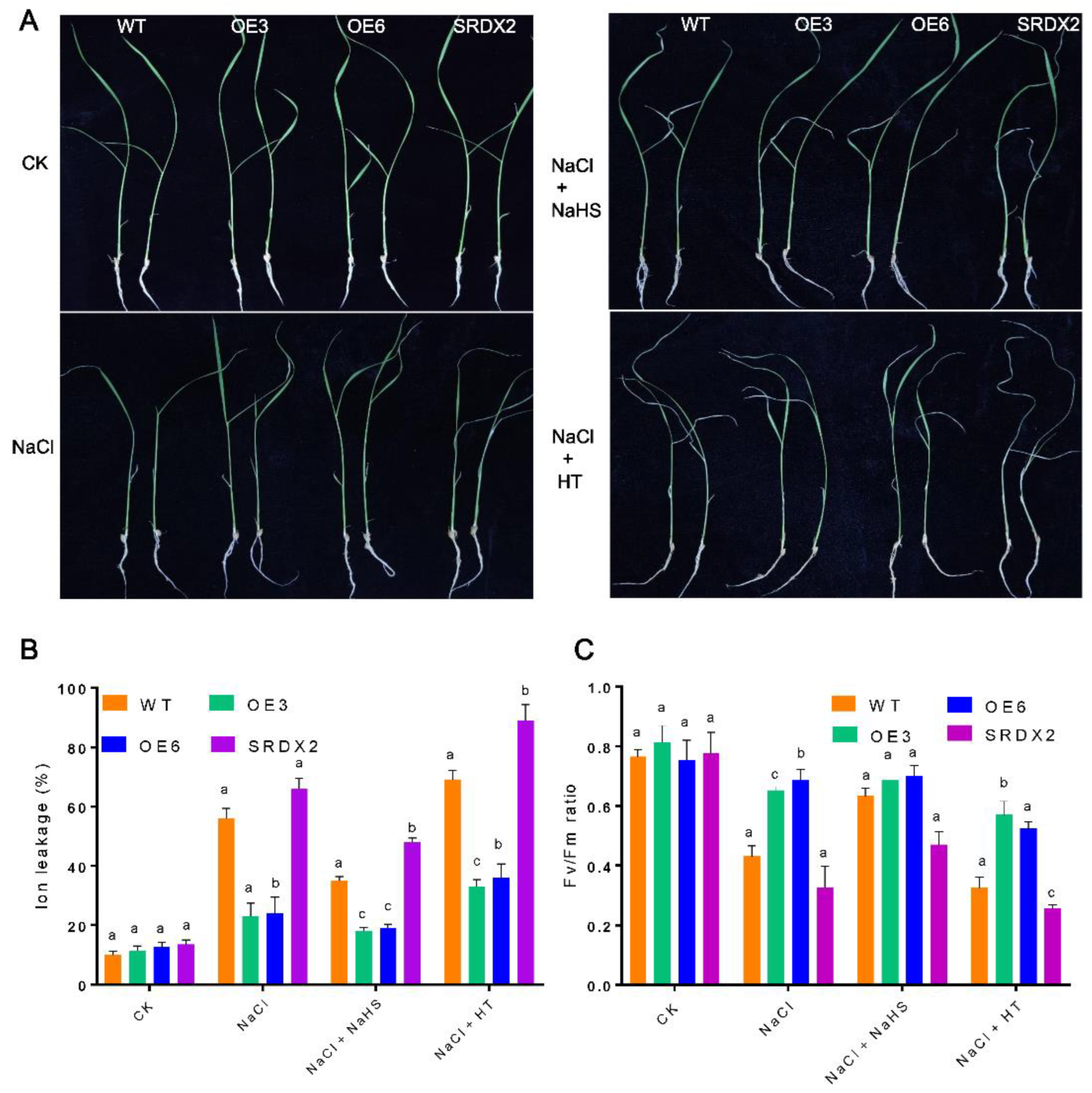
2.4. Overexpression of OsNACL35 Confers Tolerance to Salt Stress
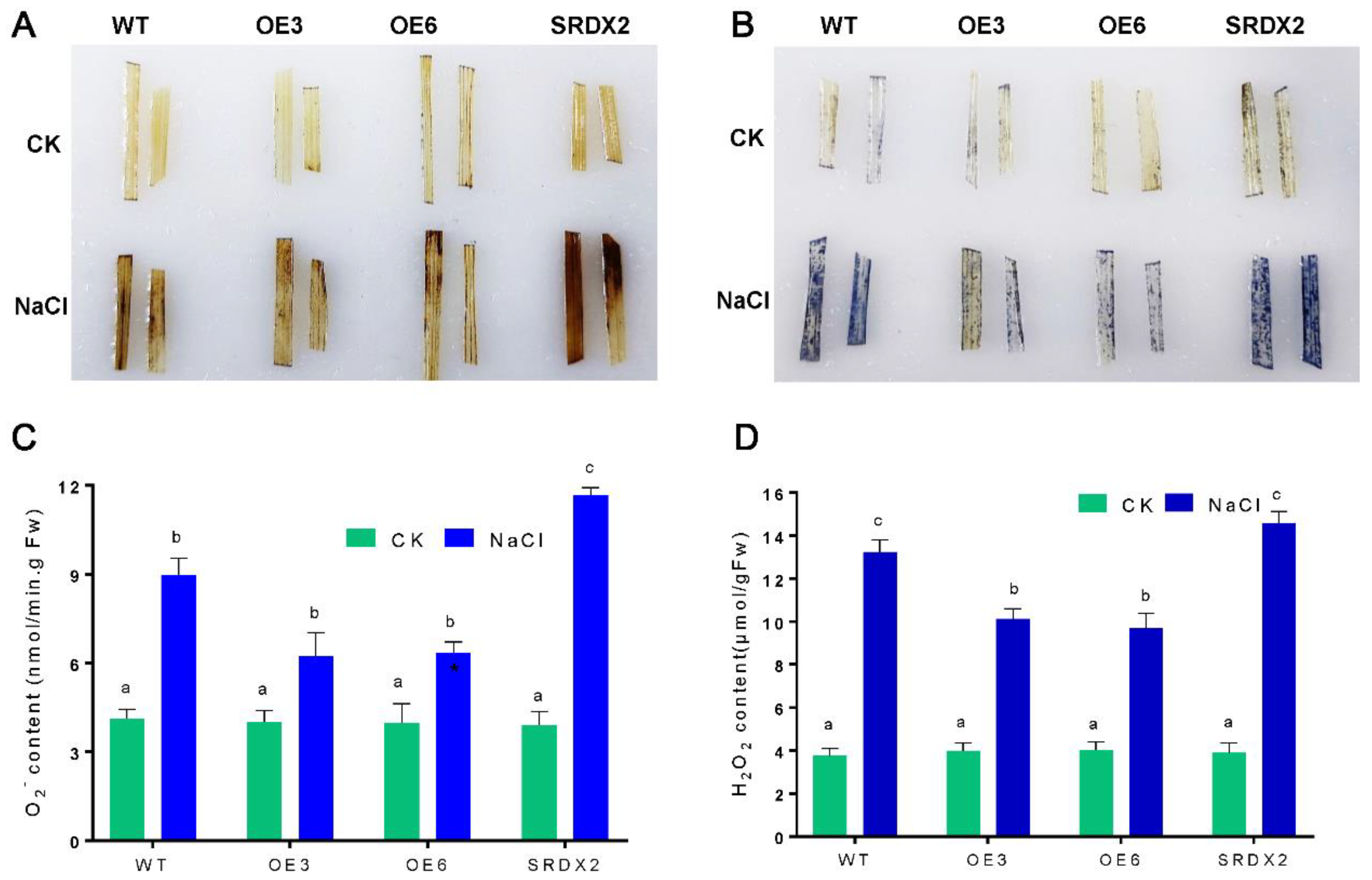
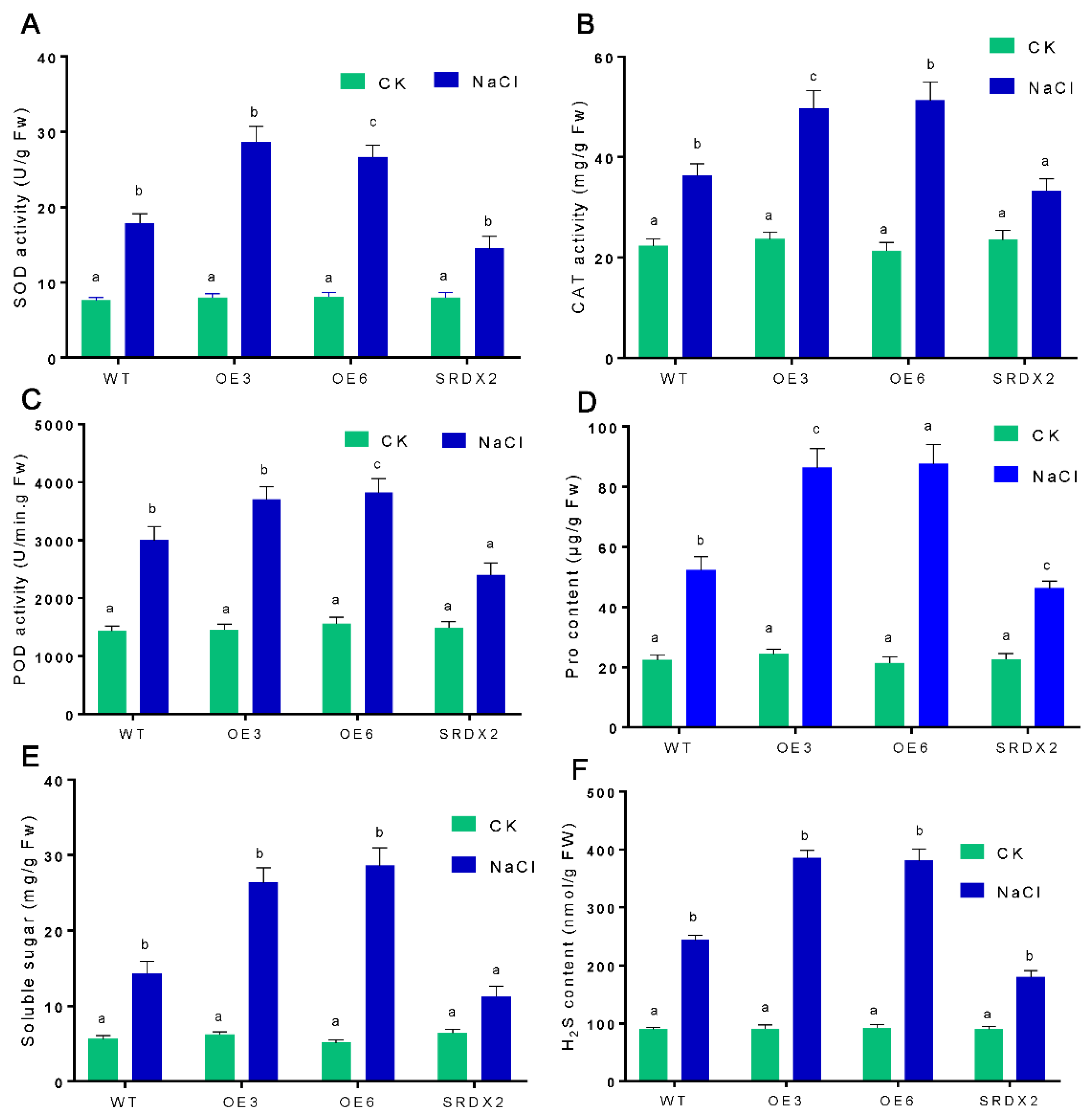
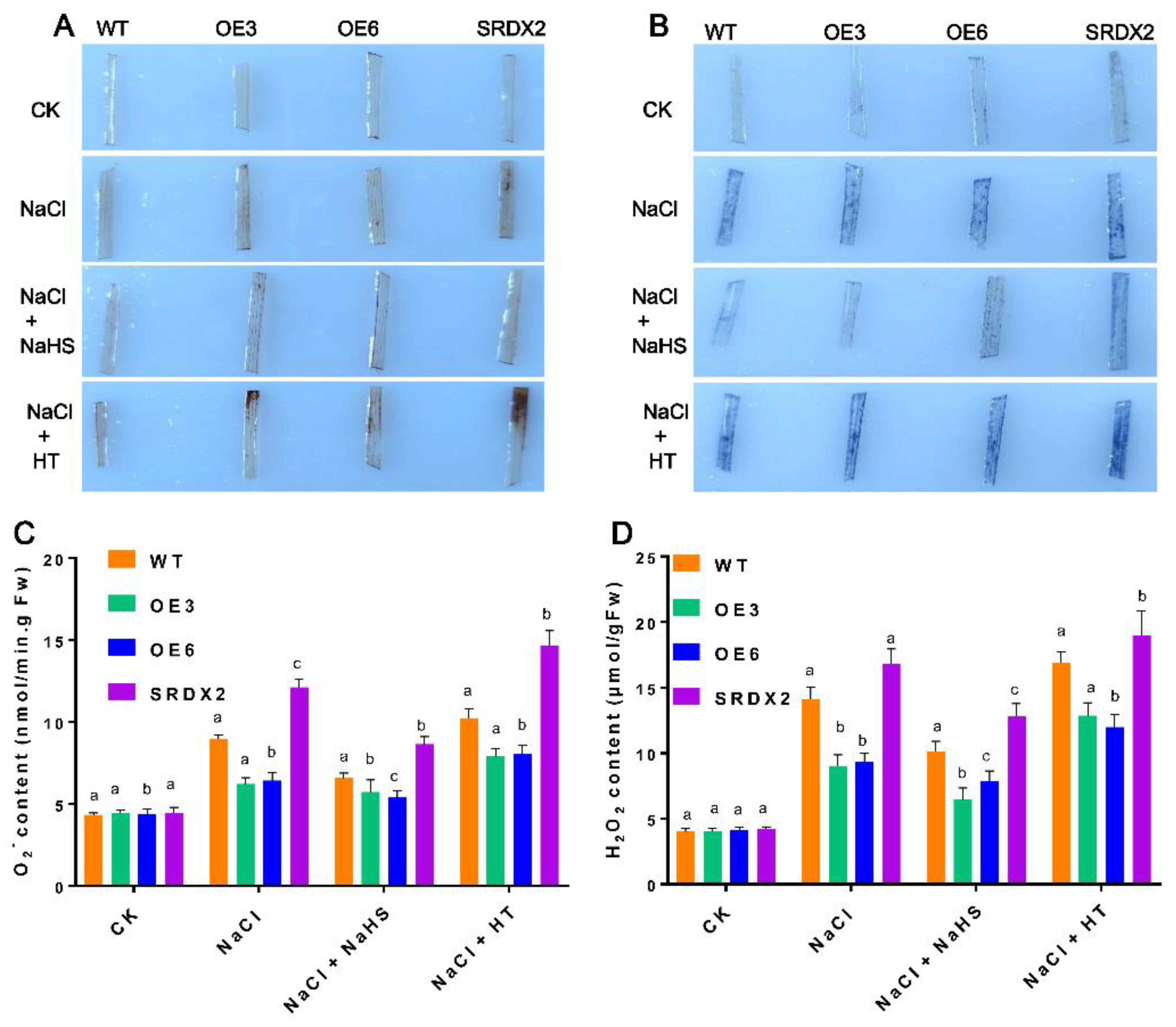
2.5. Overexpression of OsNACL35 Promotes Scavenging of Reactive Oxygen Species and Accumulation of Osmotic Substance and H2S
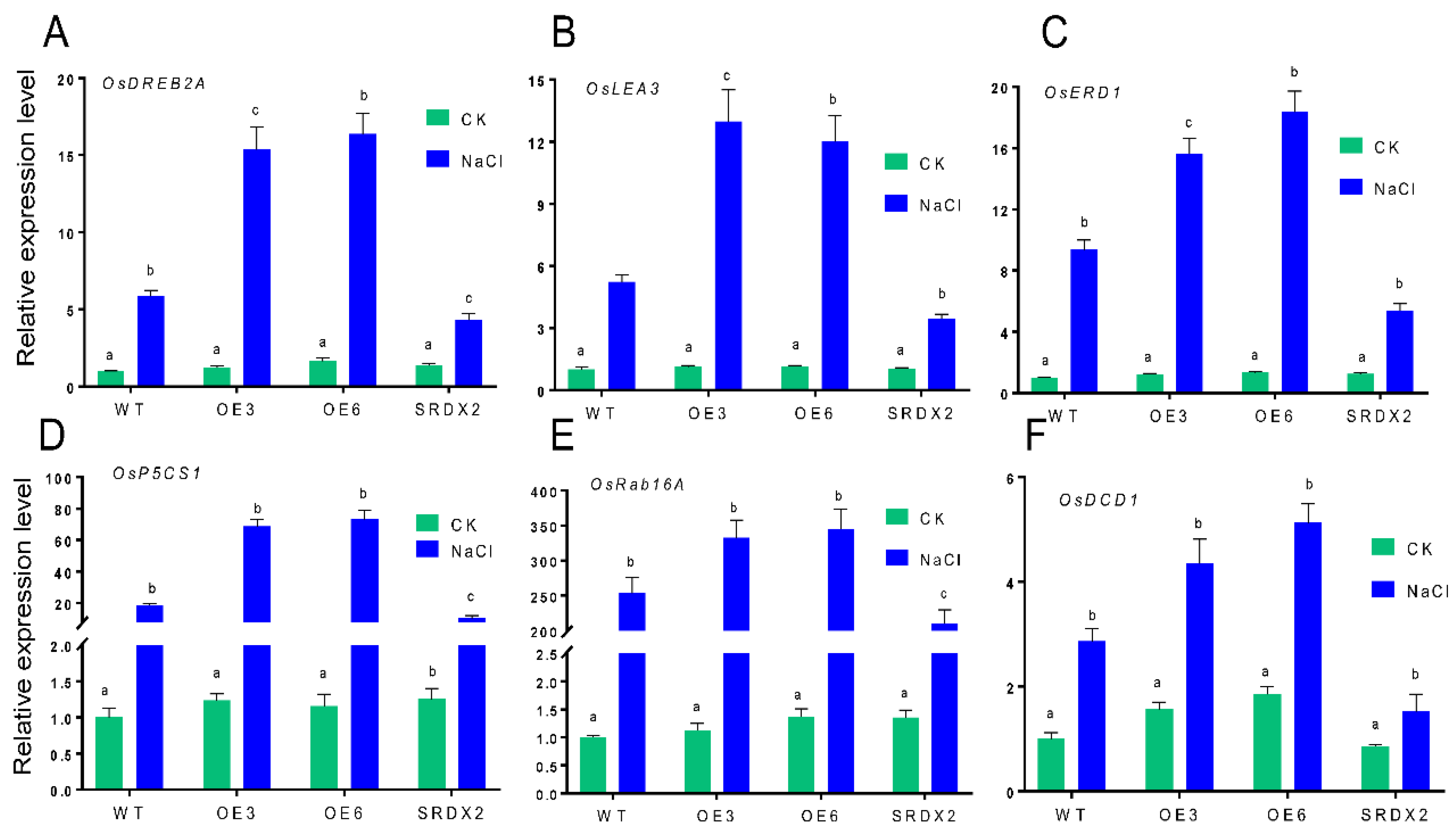
2.6. Overexpression of OsNACL35 Alters the Expression of Various Stress-Related Genes
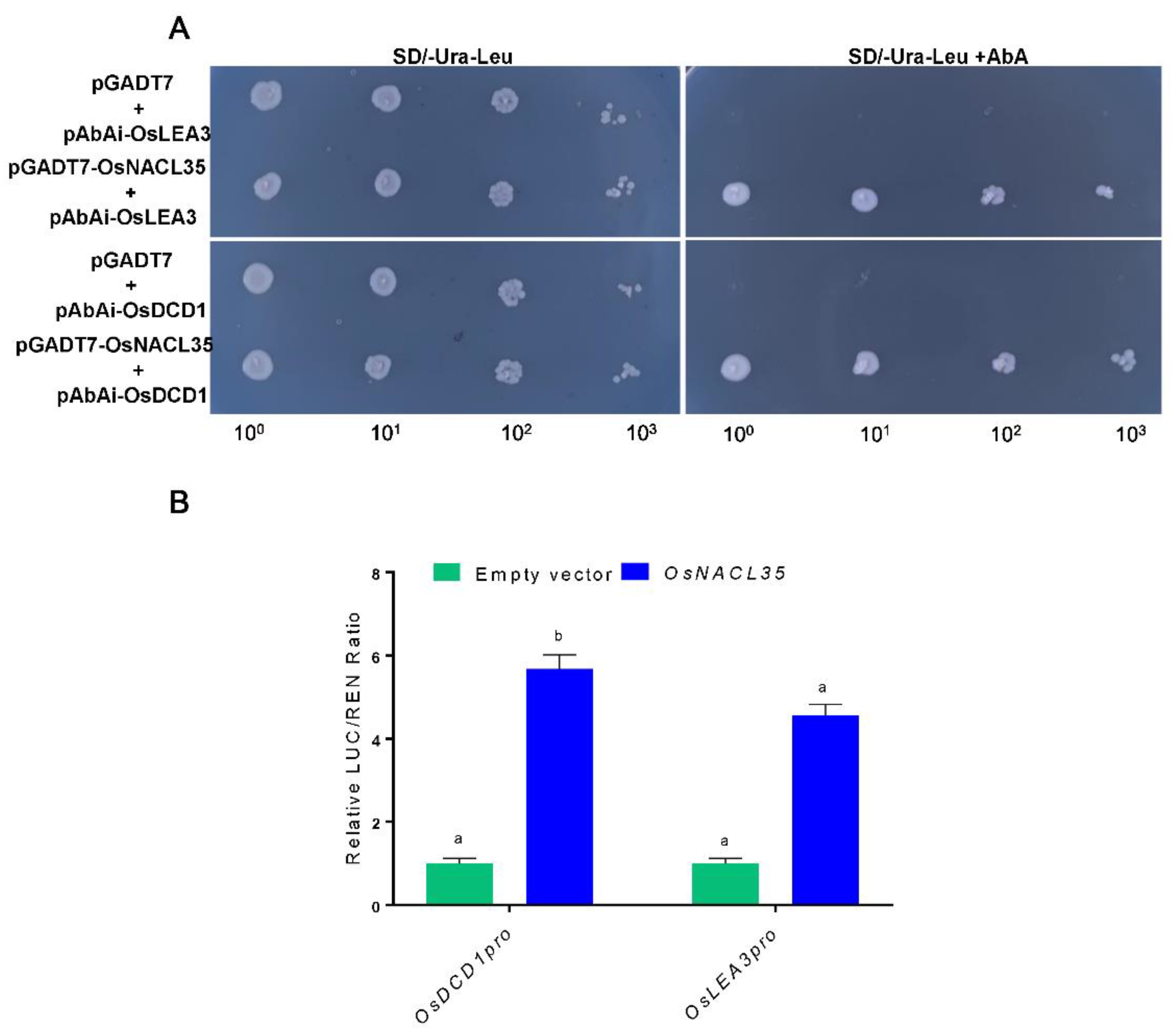
2.7. OsNACL35 Directly Regulates the Expression of OsDCD1 and OsLEA3
2.8. H2S Acts a Positive Molecule to Promote Salt Stress Tolerance in Rice Seedlings
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. RNA Isolation and qRT-PCR
4.3. Generation of Transgenic Lines
4.4. Subcellular Localization
4.5. Transactivation Activity Assays
4.6. Yeast One-Hybrid Assays
4.7. Physiological and Biochemical Measurements
4.8. Histochemical Staining
4.9. Measurement of Endogenous H2S Content
4.10. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular Mechanisms of Salinity Tolerance in Rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant Salt-Tolerance Mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Mao, B.; Yuan, D.; Chu, C.; Duan, M. Salt Tolerance in Rice: Physiological Responses and Molecular Mechanisms. Crop J. 2022, 10, 13–25. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. OsNAC45 Is Involved in ABA Response and Salt Tolerance in Rice. Rice 2020, 13, 79. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-Type MYB Gene, OsMYB2, Is Involved in Salt, Cold, and Dehydration Tolerance in Rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Yang, X.; Song, B.; Cui, J.; Wang, L.; Wang, S.; Luo, L.; Gao, L.; Mo, B.; Yu, Y.; Liu, L. Comparative Ribosome Profiling Reveals Distinct Translational Landscapes of Salt-Sensitive and -Tolerant Rice. BMC Genom. 2021, 22, 612. [Google Scholar] [CrossRef]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene Expression Profiles during the Initial Phase of Salt Stress in Rice. Plant Cell 2001, 13, 889–905. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, D.; Han, S.H.; Kim, S.H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.C. Multilayered Regulation of Membrane-Bound ONAC054 Is Essential for Abscisic Acid-Induced Leaf Senescence in Rice. Plant Cell 2020, 32, 630–649. [Google Scholar] [CrossRef]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 Regulatory Module Promotes Chlorophyll Degradation via Aba Biosynthesis in Arabidopsis Leavesw Open. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Balazadeh, S.; Mueller-Roeber, B. Tomato Fruit Ripening Factor NOR Controls Leaf Senescence. J. Exp. Bot. 2019, 70, 2727–2740. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Li, W.; Liang, X.; Ren, C.; Liu, W.; Yang, G.; Zhao, M.; Yang, T.; Li, X.; Han, D. Molecular Cloning and Characterization of MbMYB108, a Malus Baccata MYB Transcription Factor Gene, with Functions in Tolerance to Cold and Drought Stress in Transgenic Arabidopsis Thaliana. Int. J. Mol. Sci. 2022, 23, 4846. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, X.; Ma, H.; Fan, H.; Lin, F.; Chen, J.; Chai, T.; Wang, H. PcWRKY11, an II-d WRKY Transcription Factor from Polygonum Cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis Thaliana. Int. J. Mol. Sci. 2022, 23, 4357. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, J.; Guo, S.; Yuan, X.; Zhao, S.; Tian, H.; Dai, S.; Kong, X.; Ding, Z. AtHB7/12 Regulate Root Growth in Response to Aluminum Stress. Int. J. Mol. Sci. 2020, 21, 4080. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wu, X.; Gao, S.; Zhang, S.; Wang, W.; Fang, Z.; Liu, S.; Wang, X.; Zhao, C.; Tang, Y. Systematic Analysis and Identification of Drought-Responsive Genes of the CAMTA Gene Family in Wheat (Triticum Aestivum L.). Int. J. Mol. Sci. 2022, 23, 4542. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-Wide Analysis of NAC Transcription Factor Family in Rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC Transcription Factors: Structurally Distinct, Functionally Diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Mao, C.; Ding, J.; Zhang, B.; Xi, D.; Ming, F. OsNAC2 Positively Affects Salt-Induced Cell Death and Binds to the OsAP37 and OsCOX11 Promoters. Plant J. 2018, 94, 454–468. [Google Scholar] [CrossRef] [Green Version]
- Mao, C.; He, J.; Liu, L.; Deng, Q.; Yao, X.; Liu, C.; Qiao, Y.; Li, P.; Ming, F. OsNAC2 Integrates Auxin and Cytokinin Pathways to Modulate Rice Root Development. Plant Biotechnol. J. 2020, 18, 429–442. [Google Scholar] [CrossRef] [Green Version]
- Xi, D.; Chen, X.; Wang, Y.; Zhong, R.; He, J.; Shen, J.; Ming, F. Arabidopsis ANAC092 Regulates Auxin-Mediated Root Development by Binding to the ARF8 and PIN4 Promoters. J. Integr. Plant Biol. 2019, 61, 1015–1031. [Google Scholar] [CrossRef] [Green Version]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice Nac Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sablowski, R.W.; Meyerowitz, E.M. A Homolog of NO APICAL MERISTEM Is an Immediate Target of the Floral Homeotic Genes APETALA3/PISTILLATA. Cell 1998, 92, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, W.; Xian, Z.; Hu, N.; Lin, D.; Ren, H.; Chen, J.; Su, D.; Li, Z. Overexpression of SLGRAS40 in Tomato Enhances Tolerance to Abiotic Stresses and Influences Auxin and Gibberellin Signaling. Front. Plant Sci. 2017, 8, 1659. [Google Scholar] [CrossRef]
- van Beek, C.R.; Guzha, T.; Kopana, N.; van der Westhuizen, C.S.; Panda, S.K.; van der Vyver, C. The SlNAC2 Transcription Factor from Tomato Confers Tolerance to Drought Stress in Transgenic Tobacco Plants. Physiol. Mol. Biol. Plants 2021, 27, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiong, L. Genetic Engineering and Breeding of Drought-Resistant Crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Nishiyama, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Potential Utilization of NAC Transcription Factors to Enhance Abiotic Stress Tolerance in Plants by Biotechnological Approach. GM Crops 2010, 1, 32–39. [Google Scholar] [CrossRef]
- Wilson, L.G.; Bressan, R.A.; Filner, P. Light-Dependent Emission of Hydrogen Sulfide from Plants. Plant Physiol. 1978, 61, 184–189. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Liu, Z.; Long, Y.; Liang, Y.; Jin, Z.; Zhang, L.; Liu, D.; Li, H.; Zhai, J.; Pei, Y. The Ca2+/Calmodulin2-Binding Transcription Factor TGA3 Elevates LCD Expression and H2S Production to Bolster Cr6+ Tolerance in Arabidopsis. Plant J. 2017, 91, 1038–1050. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhou, M.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Shen, J.; Ge, Z.; Zhang, Z.; Shen, W.; et al. Hydrogen Sulfide, a Signaling Molecule in Plant Stress Responses. J. Integr. Plant Biol. 2021, 63, 146–160. [Google Scholar] [CrossRef]
- Wang, R. Physiological Implications of Hydrogen Sulfide: A Whiff Exploration That Blossomed. Physiol. Rev. 2012, 92, 791–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wang, W.-H.; Wu, F.-H.; He, E.-M.; Liu, X.; Shangguan, Z.-P.; Zheng, H.-L. Hydrogen Sulfide Enhances Salt Tolerance through Nitric Oxide-Mediated Maintenance of Ion Homeostasis in Barley Seedling Roots. Sci. Rep. 2015, 5, 12516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, K.; Yamada, N.; Yoshida, R.; Ihara, H.; Sawa, T.; Akaike, T.; Iwai, S. 8-Mercapto-Cyclic GMP Mediates Hydrogen Sulfide-Induced Stomatal Closure in Arabidopsis. Plant Cell Physiol. 2015, 56, 1481–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Hu, Y.; Fan, T.; Li, J. Hydrogen Sulfide Modulates Actin-Dependent Auxin Transport via Regulating ABPs Results in Changing of Root Development in Arabidopsis. Sci. Rep. 2015, 5, 8251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papanatsiou, M.; Scuffi, D.; Blatt, M.R.; García-Mata, C. Hydrogen Sulfide Regulates Inward-Rectifying K+ Channels in Conjunction with Stomatal Closure. Plant Physiol. 2015, 168, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, B.; Zhang, W.; Chao, J.; Zhang, T.; Zhao, T.; Noctor, G.; Liu, Y.; Han, Y. Functional Analysis of the Role of Hydrogen Sulfide in the Regulation of Dark-Induced Leaf Senescence in Arabidopsis. Sci. Rep. 2017, 7, 2615. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chen, S.; Wang, X.; Shi, C.; Liu, H.; Yang, J.; Shi, W.; Guo, J.; Jia, H. Hydrogen Sulfide Disturbs Actin Polymerization via S -Sulfhydration Resulting in Stunted Root Hair Growth. Plant Physiol. 2018, 178, 936–949. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.-L.; Tian, Y.; Li, L.; Yu, M.; Hou, R.-P.; Ren, X.-M. H2S Alleviates Salinity Stress in Cucumber by Maintaining the Na+/K+ Balance and Regulating H2S Metabolism and Oxidative Stress Response. Front. Plant Sci. 2019, 678. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.D.; Zhang, X.Y.; Yao, G.F.; Rong, Y.L.; Ding, C.; Tang, J.; Yang, F.; Huang, Z.Q.; Xu, Z.M.; Chen, X.Y.; et al. A Nuclear-Localized Cysteine Desulfhydrase Plays a Role in Fruit Ripening in Tomato. Hortic. Res. 2020, 7, 211. [Google Scholar] [CrossRef]
- Guo, H.; Xiao, T.; Zhou, H.; Xie, Y.; Shen, W. Hydrogen Sulfide: A Versatile Regulator of Environmental Stress in Plants. Acta Physiol. Plant. 2016, 38, 16. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Jia, H.; Li, F.; Ma, Y.; Liesche, J.; Liao, M.; Ding, X.; Liu, C.; Chen, Y.; et al. Persulfidation-Induced Structural Change in SnRK2.6 Establishes Intramolecular Interaction between Phosphorylation and Persulfidation. Mol. Plant 2021, 14, 1814–1830. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen Sulfide Positively Regulates Abscisic Acid Signaling through Persulfidation of SnRK2.6 in Guard Cells. Mol. Plant 2020, 13, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen Sulfide-Linked Persulfidation of ABI4 Controls ABA Responses through the Transactivation of MAPKKK18 in Arabidopsis. Mol. Plant 2021, 14, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.-H.; Zhao, J.-F.; Song, Y.; Zhang, C.-J.; Guo, Y. Identification of an Apoplastic Protein Involved in the Initial Phase of Salt Stress Response in Rice Root by Two-Dimensional Electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostofa, M.G.; Saegusa, D.; Fujita, M.; Tran, L.-S.P. Hydrogen Sulfide Regulates Salt Tolerance in Rice by Maintaining Na+/K+ Balance, Mineral Homeostasis and Oxidative Metabolism Under Excessive Salt Stress. Front. Plant Sci. 2015, 6, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Murad, M.; Khan, A.L.; Muneer, S. Silicon in Horticultural Crops: Cross-Talk, Signaling, and Tolerance Mechanism under Salinity Stress. Plants 2020, 9, 460. [Google Scholar] [CrossRef] [Green Version]
- Jia, T.; Wang, J.; Chang, W.; Fan, X.; Sui, X.; Song, F. Proteomics Analysis of E. Angustifolia Seedlings Inoculated with Arbuscular Mycorrhizal Fungi under Salt Stress. Int. J. Mol. Sci. 2019, 20, 788. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.-C.; Chen, K.-C.; Cheng, T.-S.; Lee, C.; Lin, S.-H.; Tung, C.-W. Chlorophyll Fluorescence Analysis in Diverse Rice Varieties Reveals the Positive Correlation between the Seedlings Salt Tolerance and Photosynthetic Efficiency. BMC Plant Biol. 2019, 19, 403. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Tran, L.-S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of a NAC-Type Transcription Factor OsNAC6 Involved in Abiotic and Biotic Stress-Responsive Gene Expression in Rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The Abiotic Stress-Responsive NAC-Type Transcription Factor OsNAC5 Regulates Stress-Inducible Genes and Stress Tolerance in Rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-Type Transcription Factor OsNAC2 Regulates ABA-Dependent Genes and Abiotic Stress Tolerance in Rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) Transcription Factor Enhances Drought Resistance and Salt Tolerance in Rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H.; et al. A NAC Transcription Factor OsNAC3 Positively Regulates ABA Response and Salt Tolerance in Rice. BMC Plant Biol. 2021, 21, 546. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Z.; Chen, Z.; Cao, B.; Xu, K. Comparative Transcriptome Analysis of Two Contrasting Chinese Cabbage (Brassica Rapa L.) Genotypes Reveals That Ion Homeostasis Is a Crucial Biological Pathway Involved in the Rapid Adaptive Response to Salt Stress. Front. Plant Sci. 2021, 12, 1093. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice Transcription Factor OsMADS25 Modulates Root Growth and Confers Salinity Tolerance via the ABA–Mediated Regulatory Pathway and ROS Scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Huang, Z.; Li, M.; Hou, Z. Growth, Ionic Homeostasis, and Physiological Responses of Cotton under Different Salt and Alkali Stresses. Sci. Rep. 2020, 10, 21844. [Google Scholar] [CrossRef]
- Rossatto, T.; do Amaral, M.N.; Benitez, L.C.; Vighi, I.L.; Braga, E.J.B.; de Magalhães Júnior, A.M.; Maia, M.A.C.; da Silva Pinto, L. Gene Expression and Activity of Antioxidant Enzymes in Rice Plants, Cv. BRS AG, under Saline Stress. Physiol. Mol. Biol. Plants 2017, 23, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Jadamba, C.; Kang, K.; Paek, N.-C.; Lee, S.I.; Yoo, S.-C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef] [Green Version]
- Mancardi, D.; Penna, C.; Merlino, A.; Del Soldato, P.; Wink, D.A.; Pagliaro, P. Physiological and Pharmacological Features of the Novel Gasotransmitter: Hydrogen Sulfide. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 864–872. [Google Scholar] [CrossRef] [Green Version]
- Christou, A.; Manganaris, G.A.; Papadopoulos, I.; Fotopoulos, V. Hydrogen Sulfide Induces Systemic Tolerance to Salinity and Non-Ionic Osmotic Stress in Strawberry Plants through Modification of Reactive Species Biosynthesis and Transcriptional Regulation of Multiple Defence Pathways. J. Exp. Bot. 2013, 64, 1953–1966. [Google Scholar] [CrossRef]
- Thakur, M.; Anand, A. Hydrogen sulfide: An Emerging Signaling Molecule Regulating Drought Stress Response in Plants. Physiol. Plant 2021, 172, 1227–1243. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Deng, Y.; Liu, Z.; Liao, W. Hydrogen Sulfide in Plants: Crosstalk with Other Signal Molecules in Response to Abiotic Stresses. Int. J. Mol. Sci. 2021, 22, 12068. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Chen, Y.; Zhai, F.; Zhang, J.; Zhang, F.; Yuan, X.; Xie, Y. Hydrogen Sulfide Promotes Rice Drought Tolerance via Reestablishing Redox Homeostasis and Activation of ABA Biosynthesis and Signaling. Plant Physiol. Biochem. 2020, 155, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.Q.; Zhang, J.H.; Sun, L.M.; Zhu, L.F.; Abliz, B.; Hu, W.J.; Zhong, C.; Bai, Z.G.; Sajid, H.; Cao, X.C.; et al. Hydrogen Sulfide Alleviates Aluminum Toxicity via Decreasing Apoplast and Symplast Al Contents in Rice. Front. Plant Sci. 2018, 9, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostofa, M.G.; Rahman, A.; Ansary, M.M.U.; Watanabe, A.; Fujita, M.; Tran, L.-S.P. Hydrogen Sulfide Modulates Cadmium-Induced Physiological and Biochemical Responses to Alleviate Cadmium Toxicity in Rice. Sci. Rep. 2015, 5, 14078. [Google Scholar] [CrossRef] [Green Version]
- Lv, W.; Yang, L.; Xu, C.; Shi, Z.; Shao, J.; Xian, M.; Chen, J. Cadmium Disrupts the Balance between Hydrogen Peroxide and Superoxide Radical by Regulating Endogenous Hydrogen Sulfide in the Root Tip of Brassica Rapa. Front. Plant Sci. 2017, 8, 232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Cai, W.; Ji, T.-T.; Ye, L.; Lu, Y.-T.; Yuan, T.-T. WRKY13 Enhances Cadmium Tolerance by Promoting D-CYSTEINE DESULFHYDRASE and Hydrogen Sulfide Production. Plant Physiol. 2020, 183, 345–357. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-Based Modification of Cysteine Desulfhydrase and the NADPH Oxidase RBOHD Controls Guard Cell Abscisic Acid Signaling. Plant Cell 2020, 32, 1000–1017. [Google Scholar] [CrossRef]
- Jia, H.; Chen, S.; Liu, D.; Liesche, J.; Shi, C.; Wang, J.; Ren, M.; Wang, X.; Yang, J.; Shi, W.; et al. Ethylene-Induced Hydrogen Sulfide Negatively Regulates Ethylene Biosynthesis by Persulfidation of ACO in Tomato Under Osmotic Stress. Front. Plant Sci. 2018, 9, 1517. [Google Scholar] [CrossRef] [Green Version]
- Wei, M.-Y.; Liu, J.-Y.; Li, H.; Hu, W.-J.; Shen, Z.-J.; Qiao, F.; Zhu, C.-Q.; Chen, J.; Liu, X.; Zheng, H.-L. Proteomic Analysis Reveals the Protective Role of Exogenous Hydrogen Sulfide against Salt Stress in Rice Seedlings. Nitric Oxide 2021, 111, 14–30. [Google Scholar] [CrossRef]
- Lv, Y.; Guo, Z.; Li, X.; Ye, H.; Li, X.; Xiong, L. New Insights into the Genetic Basis of Natural Chilling and Cold Shock Tolerance in Rice by Genome-Wide Association Analysis. Plant. Cell Environ. 2016, 39, 556–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexieva, V.; Sergiev, I.; Mapelli, S.; Karanov, E. The Effect of Drought and Ultraviolet Radiation on Growth and Stress Markers in Pea and Wheat. Plant. Cell Environ. 2001, 24, 1337–1344. [Google Scholar] [CrossRef]
- Miao, B.-H.; Han, X.-G.; Zhang, W.-H. The Ameliorative Effect of Silicon on Soybean Seedlings Grown in Potassium-Deficient Medium. Ann. Bot. 2010, 105, 967–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Wang, Z.; Gong, G.; Zhu, Y.; Ye, Q.; Lu, S.; Liu, X. Hydrogen Sulfide Alleviates Manganese Stress in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 5046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, L.-Y.; Hu, K.-D.; He, Y.-D.; Wang, S.-H.; Luo, J.-P. Hydrogen Sulfide Promotes Wheat Seed Germination and Alleviates Oxidative Damage against Copper Stress. J. Integr. Plant Biol. 2008, 50, 1518–1529. [Google Scholar] [CrossRef] [PubMed]

| Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| OsNACL35 | ATTGCTGTCCAATTTCAGTC | GATCATCAATAGTACATCATGAC |
| OsRab16A | CACACCACAGCAAGAGCTAAGTG | TGGTGCTCC ATCCTGCTTAAG |
| OsLEA3 | CGGCAG CGTCCTCCAAC | CGGTCATCCCCAGCGTG |
| OsDREB2A | GCTGCACATCAGCACCTTCA | TCCTGC ACCTCAGGGACTAC |
| OsDCD1 | GTGGATCAAGCGAGACGACA | CGCTCCCACCAATCTCTCAA |
| OsERD1 | TCAAAGGGAAGACGAAGCATGG | GGGACGGAATACAACCATCTCA |
| OsP5CS1 | GCTGACATGGATATGGCAAAAC | GTAAGGTCTCCATTGCATTGCA |
| OsPOD | AACGCAACCACCAAGCCG | CCTCGATCATGCCCATCTTGA |
| OsCATA | CCCCAAGGTCTCCCCTGA | AACGACTCATCACACTGGGAGAG |
| OsAPX8 | ATCATCGCCAGCGGATGA | GCAGCGACGAAGGGCTC |
| OsRab16A | CACACCACAGCAAGAGCTAAGTG | TGGTGCTCC ATCCTGCTTAAG |
| OsLEA3pro | GAGTGAACAGCCGAATTCCTC | ACTTAGGATTCTCAAATTCC |
| OsDCD1pro | GATTTACATGTCAGCACCTTCA | TGACTACACCTCAGGGACTAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Song, K.; Guo, M.; Wu, H.; Ji, X.; Hou, L.; Liu, X.; Lu, S. A NAC Transcription Factor from ‘Sea Rice 86′ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings. Int. J. Mol. Sci. 2022, 23, 6435. https://doi.org/10.3390/ijms23126435
Sun Y, Song K, Guo M, Wu H, Ji X, Hou L, Liu X, Lu S. A NAC Transcription Factor from ‘Sea Rice 86′ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings. International Journal of Molecular Sciences. 2022; 23(12):6435. https://doi.org/10.3390/ijms23126435
Chicago/Turabian StyleSun, Yan, Kaiqiang Song, Miaomiao Guo, Hao Wu, Xuan Ji, Lixia Hou, Xin Liu, and Songchong Lu. 2022. "A NAC Transcription Factor from ‘Sea Rice 86′ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings" International Journal of Molecular Sciences 23, no. 12: 6435. https://doi.org/10.3390/ijms23126435
APA StyleSun, Y., Song, K., Guo, M., Wu, H., Ji, X., Hou, L., Liu, X., & Lu, S. (2022). A NAC Transcription Factor from ‘Sea Rice 86′ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings. International Journal of Molecular Sciences, 23(12), 6435. https://doi.org/10.3390/ijms23126435






