Integration of Adenylate Kinase 1 with Its Peptide Conformational Imprint
Abstract
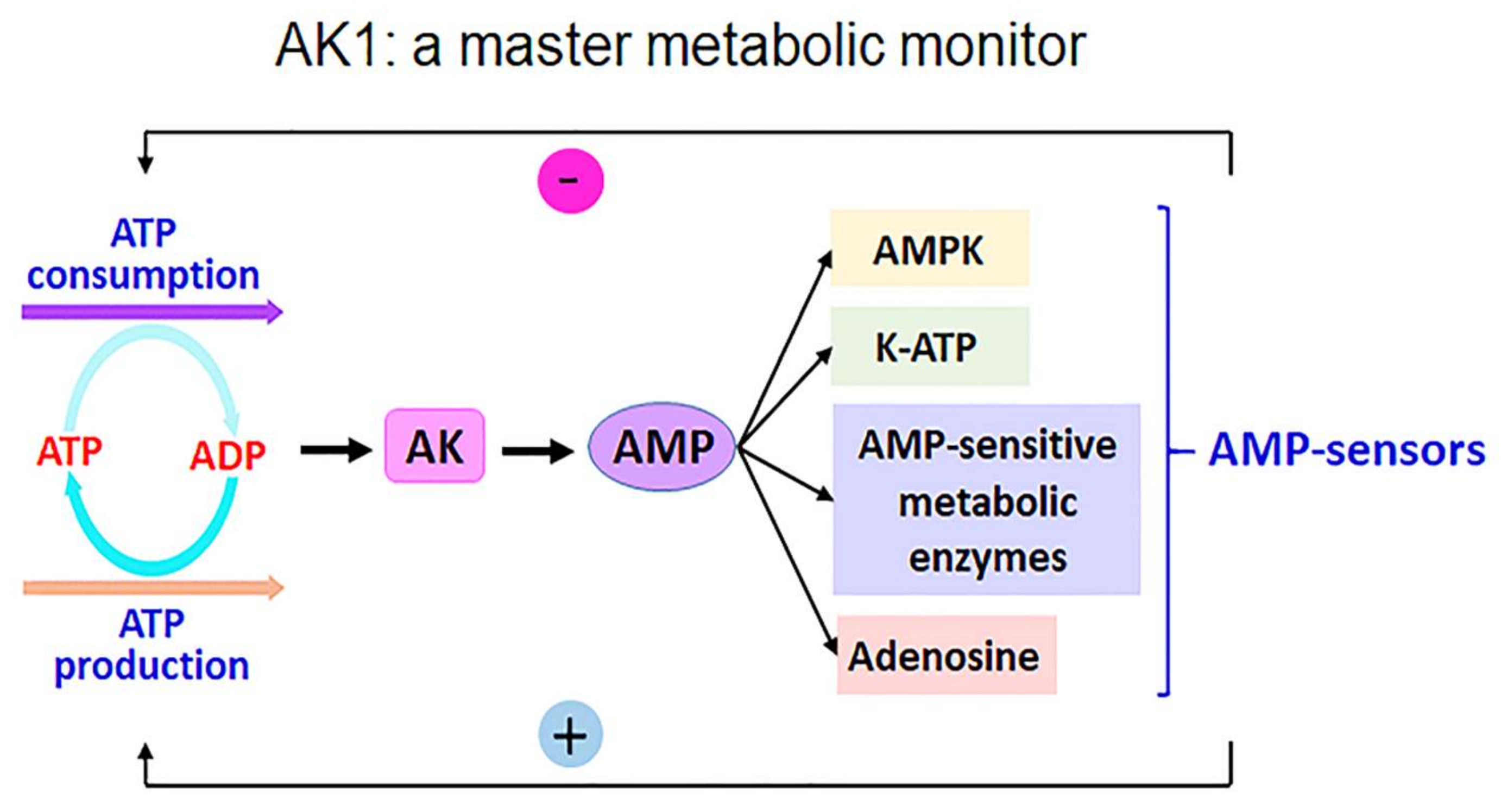
:1. Introduction
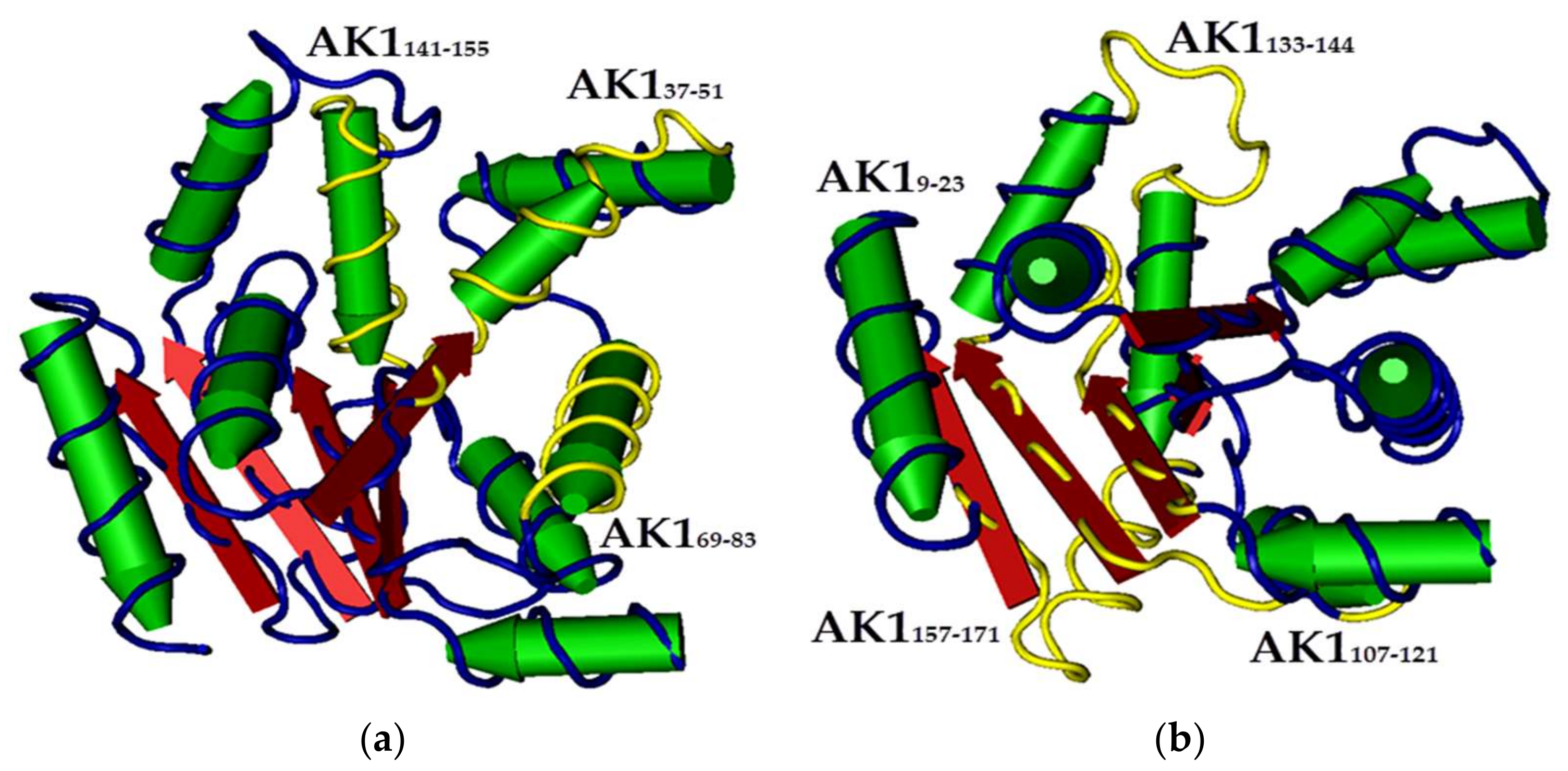
2. Results and Discussion
3. Materials and Methods
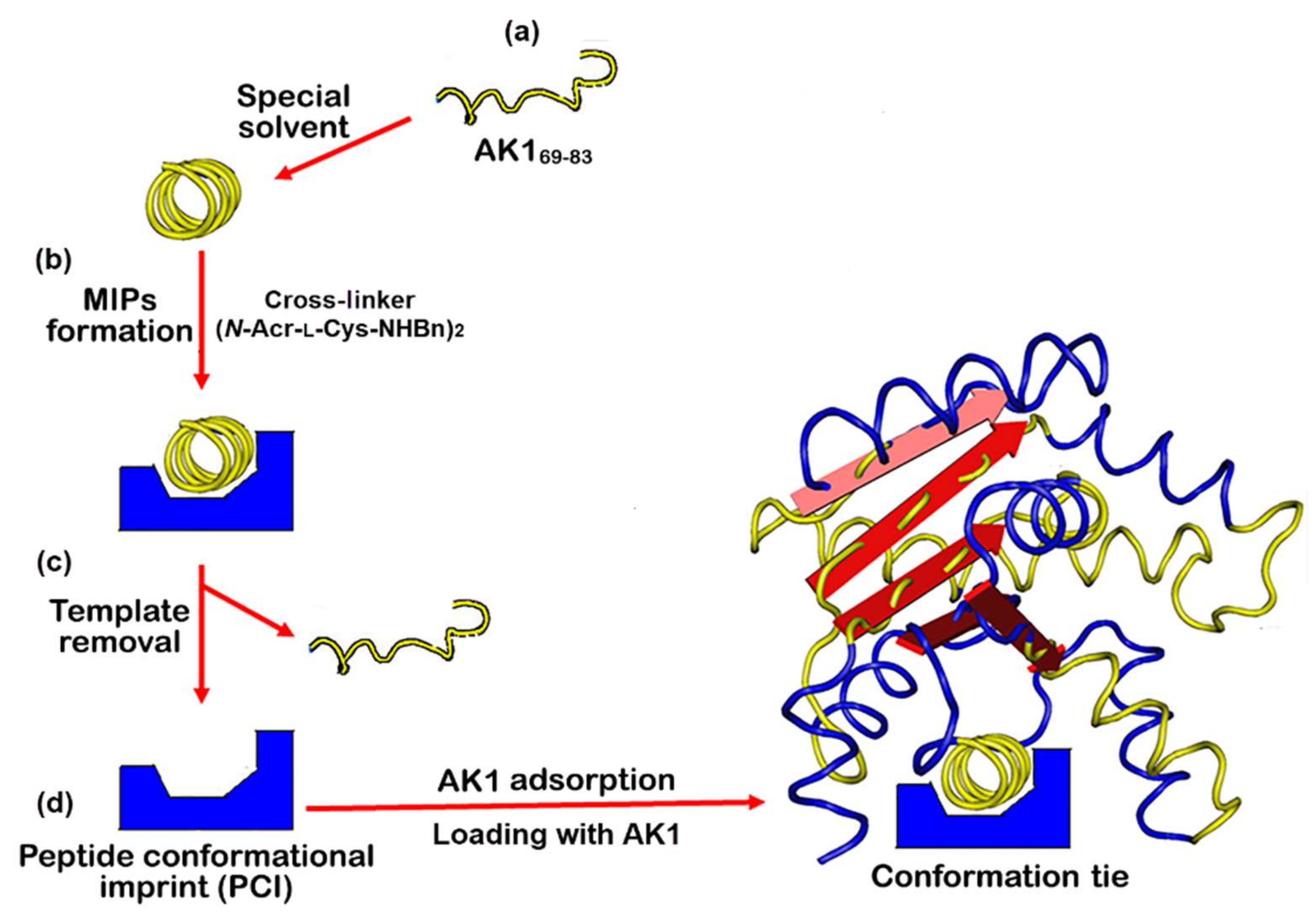
3.1. Synthesis of Templates
3.2. Preparation of Peptide Conformational Imprint Molecularly Imprinted Polymers on Quartz Crystal Microbalance Chips
3.3. The Quartz Crystal Microbalance System
3.4. Determination of Binding Affinities of Peptide Conformational Imprint Molecularly Imprinted Polymers on Quartz Crystal Microbalance Chips
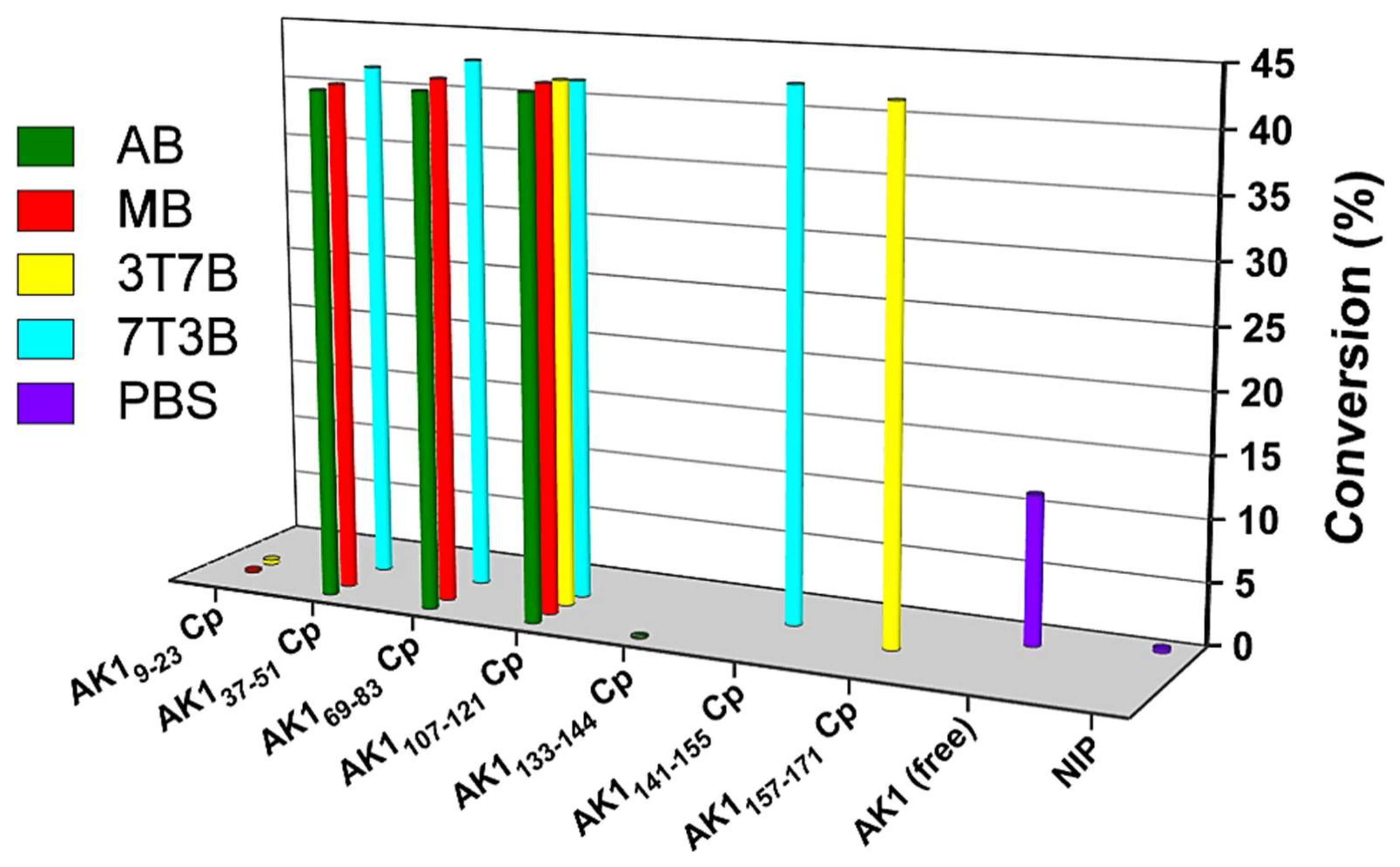
3.5. Catalytic Activity of Free AK1 and AK1-Integrated QCM Chips Was Measured Using the HPLC Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nanda, V.; Koder, R.L. Designing artificial enzymes by intuition and computation. Nat. Chem. 2010, 2, 15–24. [Google Scholar] [CrossRef]
- Cao, Y.; Li, X.; Ge, J. Enzyme Catalyst Engineering toward the Integration of Biocatalysis and Chemocatalysis. Trends Biotechnol. 2021, 39, 1173–1183. [Google Scholar] [CrossRef]
- Miller Neilan, R.; Majetic, G.; Gil-Silva, M.; Adke, A.P.; Carrasquillo, Y.; Kolber, B.J. Agent-based modeling of the central amygdala and pain using cell-type specific physiological parameters. PLoS Comput. Biol. 2021, 17, e1009097. [Google Scholar] [CrossRef] [PubMed]
- Wolfenden, R.; Snider, M.J. The depth of chemical time and the power of enzymes as catalysts. Acc. Chem. Res. 2001, 34, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Zorn, J.A.; Wells, J.A. Turning enzymes ON with small molecules. Nat. Chem. Biol. 2010, 6, 179–188. [Google Scholar] [CrossRef]
- Klinman, J.P. Enzyme dynamics: Control of active-site compression. Nat. Chem. 2010, 2, 907–909. [Google Scholar] [CrossRef] [PubMed]
- Piermattei, A.; Karthikeyan, S.; Sijbesma, R.P. Activating catalysts with mechanical force. Nat. Chem. 2009, 1, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Bassik, N.; Brafman, A.; Zarafshar, A.M.; Jamal, M.; Luvsanjav, D.; Selaru, F.M.; Gracias, D.H. Enzymatically triggered actuation of miniaturized tools. J. Am. Chem. Soc. 2010, 132, 16314–16317. [Google Scholar] [CrossRef] [PubMed]
- Adkar, B.V.; Jana, B.; Bagchi, B. Role of water in the enzymatic catalysis: Study of ATP + AMP → 2ADP conversion by adenylate kinase. J. Phys. Chem. A 2011, 115, 3691–3697. [Google Scholar] [CrossRef]
- Lanning, N.J.; Looyenga, B.D.; Kauffman, A.L.; Niemi, N.M.; Sudderth, J.; DeBerardinis, R.J.; MacKeigan, J.P. A mitochondrial RNAi screen defines cellular bioenergetic determinants and identifies an adenylate kinase as a key regulator of ATP levels. Cell Rep. 2014, 7, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Dzeja, P.; Terzic, A. Adenylate kinase and AMP signaling networks: Metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. [Google Scholar] [CrossRef]
- Rogne, P.; Rosselin, M.; Grundström, C.; Hedberg, C.; Sauer, U.H.; Wolf-Watz, M. Molecular mechanism of ATP versus GTP selectivity of adenylate kinase. Proc. Natl. Acad. Sci. USA 2018, 115, 3012–3017. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Gatti, D.L.; Bai, Y.; Manfredi, G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: Coupled mechanisms of energy metabolism regulation. Cell Metab. 2011, 13, 712–719. [Google Scholar] [CrossRef]
- Loukovaara, S.; Sandholm, J.; Aalto, K.; Liukkonen, J.; Jalkanen, S.; Yegutkin, G.G. Deregulation of ocular nucleotide homeostasis in patients with diabetic retinopathy. J. Mol. Med. 2017, 95, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Formentini, L.; Martínez-Reyes, I.; Cuezva, J.M. The mitochondrial bioenergetic capacity of carcinomas. IUBMB Life 2010, 62, 554–560. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Breuer, M.E.; Koopman, W.J.; Koene, S.; Nooteboom, M.; Rodenburg, R.J.; Willems, P.H.; Smeitink, J.A. The role of mitochondrial OXPHOS dysfunction in the development of neurologic diseases. Neurobiol. Dis. 2013, 51, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Pisliakov, A.V.; Cao, J.; Kamerlin, S.C.; Warshel, A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc. Natl. Acad. Sci. USA 2009, 106, 17359–17364. [Google Scholar] [CrossRef]
- Kamerlin, S.C.; Warshel, A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins 2010, 78, 1339–1375. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, H.J.; Sutton, J.M.; Jenkins, A.T. Study into the kinetic properties and surface attachment of a thermostable adenylate kinase. Biochem. Biophys. Rep. 2015, 1, 1–7. [Google Scholar] [CrossRef]
- Daily, M.D.; Phillips, G.N., Jr.; Cui, Q. Many local motions cooperate to produce the adenylate kinase conformational transition. J. Mol. Biol. 2010, 400, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Whitford, P.C.; Miyashita, O.; Levy, Y.; Onuchic, J.N. Conformational transitions of adenylate kinase: Switching by cracking. J. Mol. Biol. 2007, 366, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Tsai, M.D. Nucleoside monophosphate kinases: Structure, mechanism, and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol. 1999, 73, 103–134. [Google Scholar] [PubMed]
- Dahnke, T.; Tsai, M.D. Mechanism of adenylate kinase. The conserved aspartates 140 and 141 are important for transition state stabilization instead of substrate-induced conformational changes. J. Biol. Chem. 1994, 269, 8075–8081. [Google Scholar] [CrossRef]
- Brokaw, J.B.; Chu, J.W. On the roles of substrate binding and hinge unfolding in conformational changes of adenylate kinase. Biophys. J. 2010, 99, 3420–3429. [Google Scholar] [CrossRef]
- Klibanov, A.M. Improving enzymes by using them in organic solvents. Nature 2001, 409, 241–246. [Google Scholar] [CrossRef]
- Behrends, H.W.; Folkers, G.; Beck-Sickinger, A.G. A new approach to secondary structure evaluation: Secondary structure prediction of porcine adenylate kinase and yeast guanylate kinase by CD spectroscopy of overlapping synthetic peptide segments. Biopolymers 1997, 41, 213–231. [Google Scholar] [CrossRef]
- Beck-Sickinger, A.G.; Kiess, M.; Kern, P.; Folkers, G. Secondary structure prediction of adenylate kinase by circular dichroism spectroscopy of synthetic peptides. Pharm. Acta Helv. 1995, 70, 33–41. [Google Scholar] [CrossRef]
- Chen, L.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef]
- Garcia-Cruz, A.; Ahmad, O.S.; Alanazi, K.; Piletska, E.; Piletsky, S.A. Generic sensor platform based on electro-responsive molecularly imprinted polymer nanoparticles (e-NanoMIPs). Microsyst. Nanoeng. 2020, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Clegg, J.R.; Peppas, N.A. Molecular recognition with soft biomaterials. Soft Matter 2020, 16, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.F.; Ho, Y.F.; Wu, C.H.; Lin, T.C.; Lu, K.H.; Lin, K.S. Artificial-epitope mapping for CK-MB assay. Analyst 2011, 136, 2230–2233. [Google Scholar] [CrossRef] [PubMed]
- Kanubaddi, K.R.; Huang, P.Y.; Chang, Y.L.; Wu, C.H.; Li, W.; Kankala, R.K.; Tai, D.F.; Lee, C.H. Deviation of Trypsin Activity Using Peptide Conformational Imprints. Nanomaterials 2021, 11, 334. [Google Scholar] [CrossRef] [PubMed]
- Maragakis, P.; Karplus, M. Large amplitude conformational change in proteins explored with a plastic network model: Adenylate kinase. J. Mol. Biol. 2005, 352, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.F.; Jhang, M.H.; Chen, G.Y.; Wang, S.C.; Lu, K.H.; Lee, Y.D.; Liu, H.T. Epitope-cavities generated by molecularly imprinted films measure the coincident response to anthrax protective antigen and its segments. Anal. Chem. 2010, 82, 2290–2293. [Google Scholar] [CrossRef]
- Emir Diltemiz, S.; Keçili, R.; Ersöz, A.; Say, R. Molecular Imprinting Technology in Quartz Crystal Microbalance (QCM) Sensors. Sensors 2017, 17, 454. [Google Scholar] [CrossRef]
- Bartold, K.; Pietrzyk-Le, A.; Lisowski, W.; Golebiewska, K.; Siklitskaya, A.; Borowicz, P.; Shao, S.; D’Souza, F.; Kutner, W. Promoting bioanalytical concepts in genetics: A TATA box molecularly imprinted polymer as a small isolated fragment of the DNA damage repairing system. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 100, 1–10. [Google Scholar] [CrossRef]
- Thompson, A.A.; Zou, A.; Yan, J.; Duggal, R.; Hao, W.; Molina, D.; Cronin, C.N.; Wells, P.A. Biochemical characterization of recombinant hepatitis C virus nonstructural protein 4B: Evidence for ATP/GTP hydrolysis and adenylate kinase activity. Biochemistry 2009, 48, 906–916. [Google Scholar] [CrossRef]
- Jambovane, S.; Duin, E.C.; Kim, S.K.; Hong, J.W. Determination of kinetic parameters, Km and kcat, with a single experiment on a chip. Anal. Chem. 2009, 81, 3239–3245. [Google Scholar] [CrossRef]
- Xu, J.; Miao, H.; Zou, L.; Tse Sum Bui, B.; Haupt, K.; Pan, G. Evolution of Molecularly Imprinted Enzyme Inhibitors: From Simple Activity Inhibition to Pathological Cell Regulation. Angew. Chem. Int. Ed. Engl. 2021, 60, 24526–24533. [Google Scholar] [CrossRef]
- Orädd, F.; Ravishankar, H.; Goodman, J.; Rogne, P.; Backman, L.; Duelli, A.; Nors Pedersen, M.; Levantino, M.; Wulff, M.; Wolf-Watz, M.; et al. Tracking the ATP-binding response in adenylate kinase in real time. Sci. Adv. 2021, 7, eabi5514. [Google Scholar] [CrossRef] [PubMed]
- Hetmann, A.; Wujak, M.; Bolibok, P.; Zięba, W.; Wiśniewski, M.; Roszek, K. Novel biocatalytic systems for maintaining the nucleotide balance based on adenylate kinase immobilized on carbon nanostructures. Mater. Sci. Eng. C Mater. Biol. Appl. 2018, 88, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.Y.; Lin, C.Y.; Wu, C.H.; Tai, D.F. Sensing HIV Protease and Its Inhibitor Using “Helical Epitope”-Imprinted Polymers. Sensors 2020, 20, 3592. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Tsai, S.H.; Tai, D.F. Detection of oxytocin, atrial natriuretic peptide, and brain natriuretic peptide using novel imprinted polymers produced with amphiphilic monomers. J. Pept. Sci. 2019, 25, e3150. [Google Scholar] [CrossRef] [PubMed]
| QCM Cp a | Template | CD Conformation/ MIP Condition b | Kd (nM) | Flow System b |
|---|---|---|---|---|
| AK19-23 CpAB | KIIFVVGGPGSGKGT MW 1417 pI 9.2 | RC/AB | 5 | AB |
| AK19-23 CpMB | Sheet/MB | 7 | MB | |
| AK19-23 Cp3T7B | Sheet/3T7B | 10 | MB | |
| AK137-51 CpAB | LSTGDLLRSEVSSGS MW 1506 pI 4.2 | RC/AB | 4 | AB |
| AK137-51 CpMB | Helix/MB | 3 | MB | |
| AK137-51 Cp7T3B | Helix/7T3B | 5 | MB | |
| AK169-83 CpAB | LSTGDLLRSEVSSGS MW 1506 pI 4.2 | RC/AB | 3 | AB |
| AK169-83 CpMB | Helix/MB | 4 | MB | |
| AK169-83 Cp7T3B | Helix/7T3B | 3 | MB | |
| AK1107-121 Cp2A8B | RRIGQPTLLLYVDAG MW 1672 pI 8.2 | RC/2A8B | 3 | AB |
| AK1107-121 Cp7M3B | Sheet/7M3B | 6 | MB | |
| AK1107-121 Cp3T7B | Sheet/3T7B | 2 | MB | |
| AK1107-121 Cp7T3B | Helix/7T3B | 3 | MB | |
| AK1133-144 CpAB | GETSGRVDNEE | RC/AB | <5 | AB |
| AK1141-155 Cp7M3B | DNEETIKKRUETYYK | Helix/7M3B | <5 | MB |
| AK1157-171 Cp3T7B | TEPVIAFYEKRGIVR | Sheet/3T7B | <5 | MB |
| NIP | - | - | - | PBS |
| QCM Cp | CD Structure | Kd (pM) | Conversion a (%) | Activity b (nM⋅min−1⋅ng−1) |
|---|---|---|---|---|
| AK19-23 CpAB | RC | - | - | - |
| AK19-23 CpMB | Sheet | 9000 | 0.2 | 0.051 |
| AK19-23 Cp3T7B | Sheet | 50 | 0.39 | 0.099 |
| AK137-51 CpAB | RC | 3000 | 41.42 | 10.5 |
| AK137-51 CpMB | Helix | 1000 | 41.58 | 10.5 |
| AK137-51 Cp7T3B | Helix | 70 | 42.36 | 10.7 |
| AK169-83 CpAB | RC | 2000 | 41.82 | 10.6 |
| AK169-83 CpMB | Helix | 400 | 42.49 | 10.7 |
| AK169-83 Cp7T3B | Helix | 40 | 43.35 | 10.9 |
| AK1107-121 Cp2A8B | RC | 200 | 42.21 | 10.7 |
| AK1107-121 Cp7M3B | Sheet | 50 | 42.61 | 10.8 |
| AK1107-121 Cp3T7B | Sheet | 60 | 42.55 | 10.7 |
| AK1107-121 Cp7T3B | Helix | 200 | 42.20 | 10.7 |
| AK1133-144 CpAB | RC | 70 | 0.13 | 0.033 |
| AK1141-155 Cp7M3B | Helix | 50 | 42.85 | 10.8 |
| AK1157-171 Cp3T7B | Sheet | 60 | 42.42 | 10.7 |
| NIP c | - | - | 0.32 | 0.081 |
| Free AK1 d | - | - | 12.01 | 3.03 |
| QCM Cp | Va M⋅S−1 | Vmax M⋅S−1 | Km M | Kcat S−1 | Kcat/Km M−1⋅S−1 |
|---|---|---|---|---|---|
| AK19-23 CpAB | - | - | - | - | - |
| AK19-23 CpMB | 6.55 × 10−11 | 3.36 × 10−11 | 2.57 × 10−5 | 0.007 | 283.55 |
| AK19-23 Cp3T7B | 1.01 × 10−10 | 9.33 × 10−9 | 4.62 × 10−3 | 2.02 | 437.23 |
| AK137-51 CpAB | 1.58 × 10−10 | 2.43 × 10−10 | 7.70 × 10−5 | 0.053 | 683.98 |
| AK137-51 CpMB | 1.60 × 10−10 | 7.39 × 10−10 | 2.31 × 10−4 | 0.16 | 692.64 |
| AK137-51 Cp7T3B | 4.83 × 10−10 | 3.19 × 10−8 | 3.30 × 10−3 | 6.90 | 2090.91 |
| AK169-83 CpAB | 1.08 × 10−10 | 2.49 × 10−10 | 1.16 × 10−4 | 0.054 | 467.53 |
| AK169-83 CpMB | 8.21 × 10−10 | 9.48 × 10−9 | 5.78 × 10−4 | 2.053 | 3554.11 |
| AK169-83 Cp7T3B | 1.22 × 10−9 | 1.41 × 10−7 | 5.77 × 10−3 | 30.50 | 5281.38 |
| AK1107-121 Cp2A8B | 1.20 × 10−10 | 2.77 × 10−9 | 1.12 × 10−3 | 0.60 | 519.48 |
| AK1107-121 Cp7M3B | 5.42 × 10−10 | 5.01 × 10−8 | 4.62 × 10−3 | 10.84 | 2346.32 |
| AK1107-121 Cp3T7B | 3.12 × 10−10 | 2.40 × 10−8 | 3.85 × 10−3 | 5.20 | 1350.65 |
| AK1107-121 Cp7T3B | 1.42 × 10−10 | 3.28 × 10−9 | 1.12 × 10−3 | 0.71 | 614.72 |
| NIP b | 6.52 × 10−11 | - | - | - | - |
| Free AK1 c | 1.29 × 10−11 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-H.; Lin, C.-Y.; Lin, T.-C.; Tai, D.-F. Integration of Adenylate Kinase 1 with Its Peptide Conformational Imprint. Int. J. Mol. Sci. 2022, 23, 6521. https://doi.org/10.3390/ijms23126521
Wu C-H, Lin C-Y, Lin T-C, Tai D-F. Integration of Adenylate Kinase 1 with Its Peptide Conformational Imprint. International Journal of Molecular Sciences. 2022; 23(12):6521. https://doi.org/10.3390/ijms23126521
Chicago/Turabian StyleWu, Cheng-Hsin, Chung-Yin Lin, Tzu-Chieh Lin, and Dar-Fu Tai. 2022. "Integration of Adenylate Kinase 1 with Its Peptide Conformational Imprint" International Journal of Molecular Sciences 23, no. 12: 6521. https://doi.org/10.3390/ijms23126521
APA StyleWu, C.-H., Lin, C.-Y., Lin, T.-C., & Tai, D.-F. (2022). Integration of Adenylate Kinase 1 with Its Peptide Conformational Imprint. International Journal of Molecular Sciences, 23(12), 6521. https://doi.org/10.3390/ijms23126521






