Selective Elimination of Senescent Fibroblasts by Targeting the Cell Surface Protein ACKR3
Abstract
:1. Introduction
2. Results
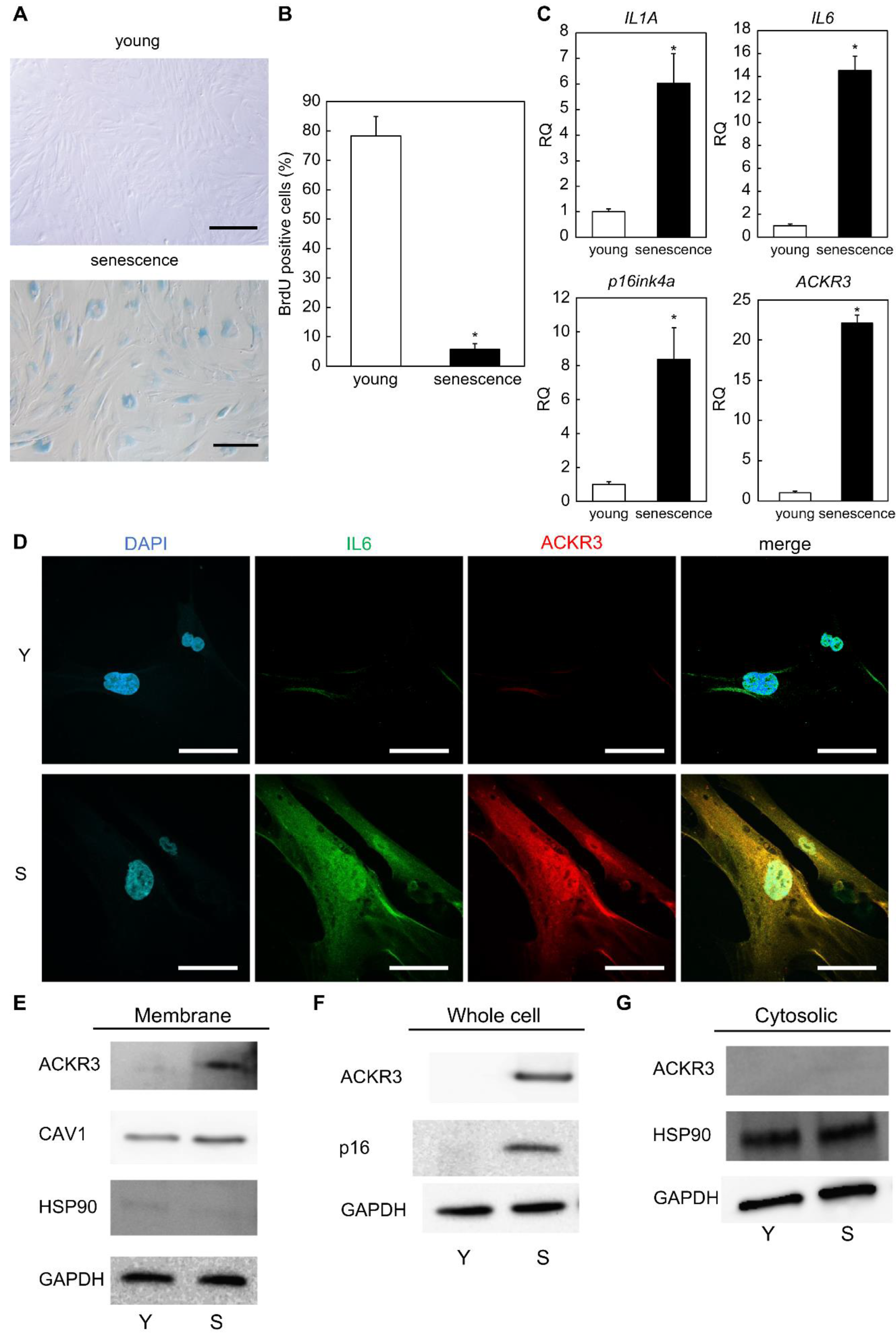
2.1. ACKR3 Is Specifically Expressed in Senescent Human Dermal Fibroblasts
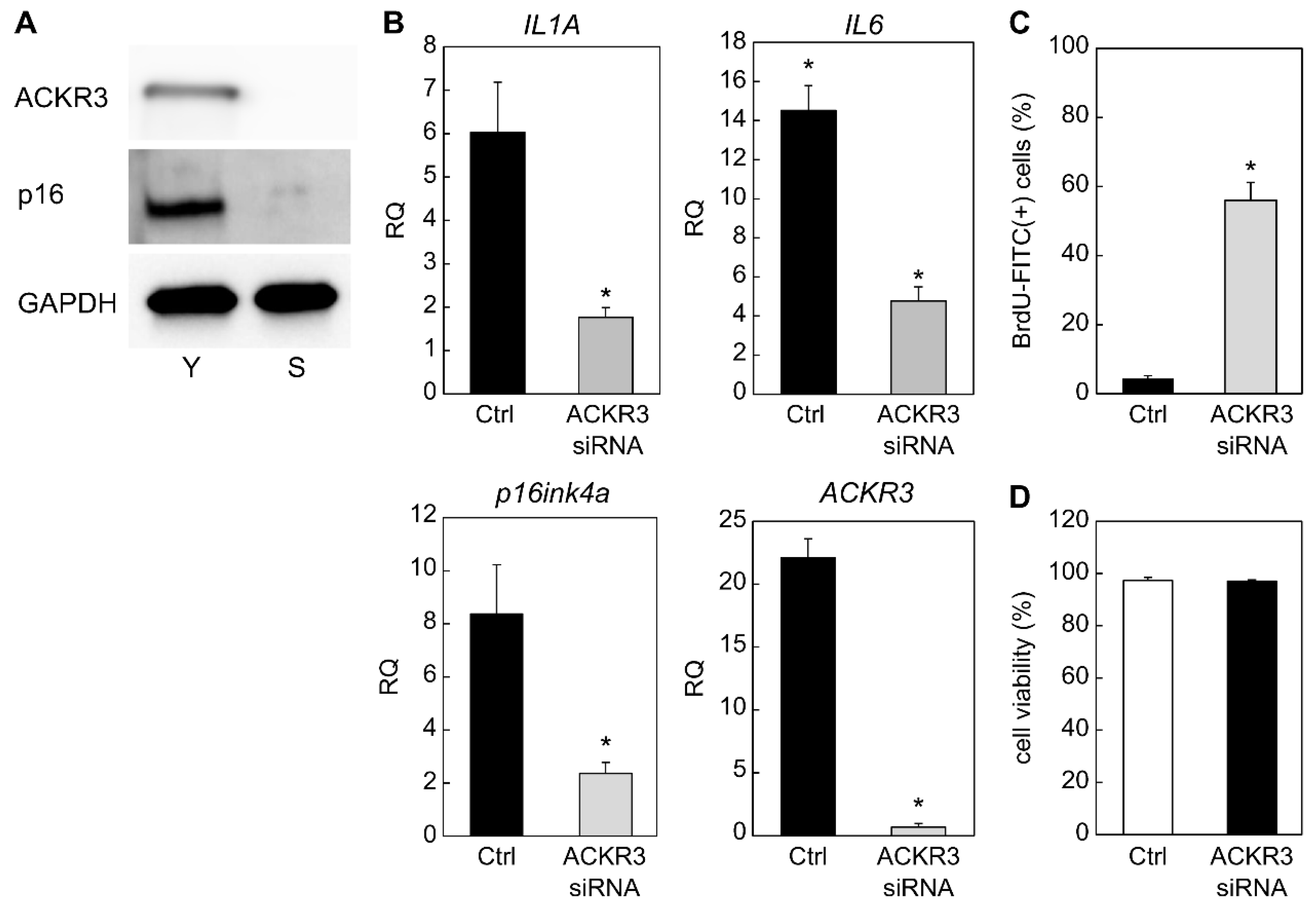
2.2. Contribution of ACKR3 to Senescence
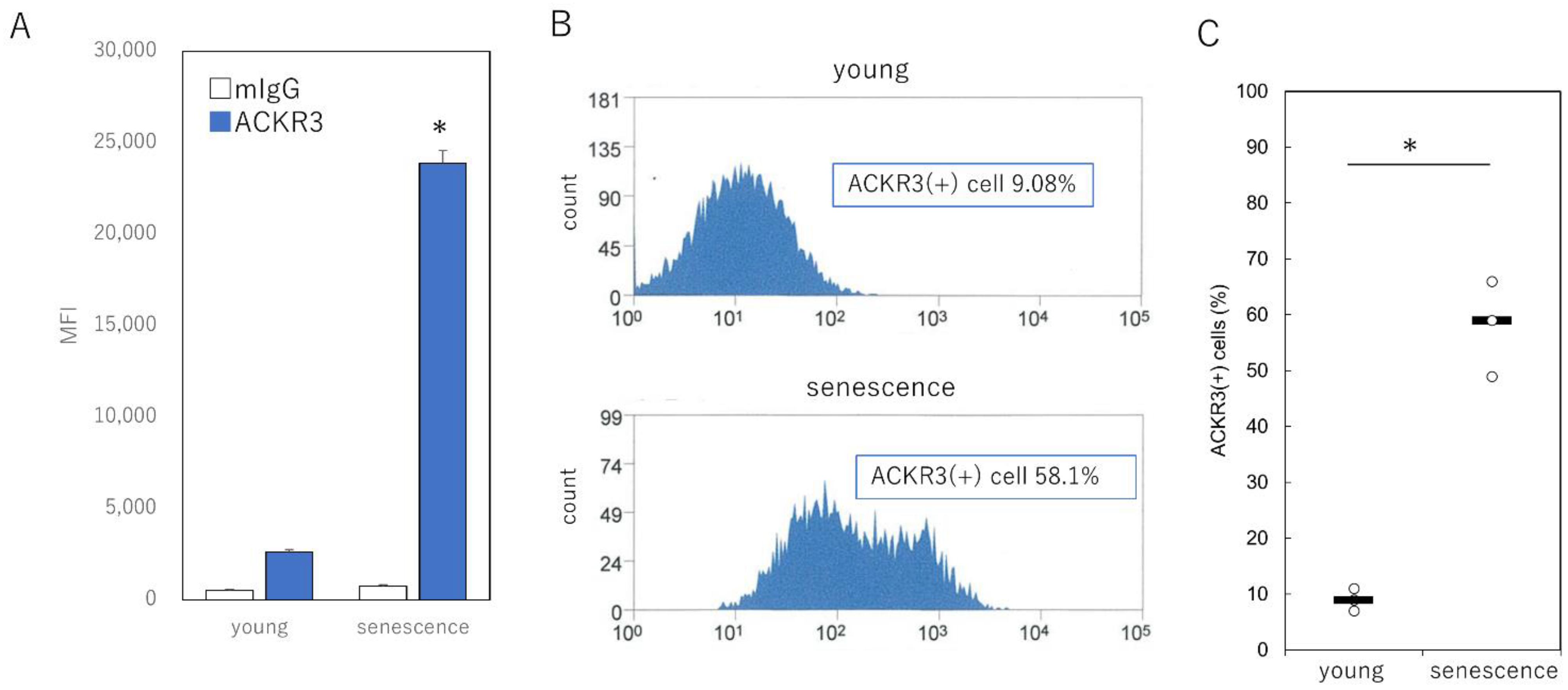
2.3. Specific Selection of Senescent Cells Using Cell Surface ACKR3
2.4. Selective Elimination of ACKR3-Positive Senescent Cells Using the Antibody-Dependent Cellular Cytotoxicity Assay
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Assessment of BrdU Incorporation in Fibroblasts by Flow Cytometry
4.3. Immunocytochemistry
4.4. RNA Interference and Transfection Method
4.5. RNA Extraction and Reverse Transcription
4.6. Real-Time Quantitative Polymerase Chain Reaction
4.7. Western Blotting
4.8. Fluorescence-Activated Cell Sorting (FACS)
4.9. Antibody-Dependent Cellular Cytotoxicity Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, S.; Sharpless, N.E. Senescence in health and disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Sharpless, N.E.; Sherr, C.J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 2015, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, A.; Sufian, M.A.; Uddin, M.S.; Sumsuzzman, D.M.; Jeandet, P.; Islam, M.S.; Zhang, H.J.; Kong, A.N.; Sarwar, M.S. Exploring the role of senescence inducers and senotherapeutics as targets for anticancer natural products. Eur. J. Pharmacol. 2022, 45, 174991. [Google Scholar] [CrossRef]
- Collado, M.; Blasco, M.A.; Serrano, M. Cellular senescence in cancer and aging. Cell 2007, 130, 223–233. [Google Scholar] [CrossRef]
- Stoczynska-Fidelus, E.; Węgierska, M.; Kierasińska, A.; Ciunowicz, D.; Rieske, P. Role of senescence in tumorigenesis and anticancer therapy. J. Oncol. 2022, 2022, 5969536. [Google Scholar] [CrossRef]
- Han, X.; Lei, Q.; Xie, J.; Liu, H.; Sun, H.; Jing, L.; Zhang, X.; Zhang, T.; Gou, X. Potential regulators of the senescence-associated secretory phenotype during senescence and ageing. J. Gerontol. A Biol. Sci. Med. Sci. 2022, 11, glac097. [Google Scholar] [CrossRef]
- Baker, D.J.; Childs, B.G.; Durik, M.; Wijers, M.E.; Sieben, C.J.; Zhong, J.; Saltness, R.A.; Jeganathan, K.B.; Verzosa, G.C.; Pezeshki, A.; et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 2016, 530, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Johmura, Y.; Yamanaka, T.; Omori, S.; Wang, T.W.; Sugiura, Y.; Matsumoto, M.; Suzuki, N.; Kumamoto, S.; Yamaguchi, K.; Hatakeyama, S.; et al. Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science 2021, 371, 265–270. [Google Scholar] [CrossRef]
- Xu, M.; Pirtskhalava, T.; Farr, J.N.; Weigand, B.M.; Palmer, A.K.; Weivoda, M.M.; Inman, C.L.; Ogrodnik, M.B.; Hachfeld, C.M.; Fraser, D.G.; et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 2018, 24, 1246–1256. [Google Scholar] [CrossRef]
- Koch, C.; Engele, J. Functions of the CXCL12 receptor ACKR3/CXCR7-What has been perceived and what has been overlooked. Mol. Pharmacol. 2020, 98, 577–585. [Google Scholar] [CrossRef]
- Ware, M.B.; El-Rayes, B.F.; Lesinski, G.B. Mirage or long-awaited oasis: Reinvigorating T-cell responses in pancreatic cancer. J. Immunother. Cancer 2020, 8, e001100. [Google Scholar] [CrossRef] [PubMed]
- Melk, A.; Schmidt, B.M.; Takeuchi, O.; Sawitzki, B.; Rayner, D.C.; Halloran, P.F. Expression of p16INK4a and other cell cycle regulator and senescence associated genes in aging human kidney. Kidney Int. 2004, 65, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Rodier, F.; Muñoz, D.P.; Teachenor, R.; Chu, V.; Le, O.; Bhaumik, D.; Coppé, J.P.; Campeau, E.; Beauséjour, C.M.; Kim, S.H.; et al. DNA-SCARS: Distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011, 124, 68–81. [Google Scholar] [CrossRef]
- Cahu, J.; Bustany, S.; Sola, B. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 2012, 3, e446. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, J.; Torrice, C.; Ramsey, M.R.; Kovalev, G.I.; Al-Regaiey, K.; Su, L.; Sharpless, N.E. Ink4a/Arf expression is a biomarker of aging. J. Clin. Investig. 2004, 114, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Malumbres, M.; Barbacid, M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim. Biophys. Acta 2002, 1602, 73–87. [Google Scholar] [CrossRef]
- Waldera Lupa, D.M.; Kalfalah, F.; Safferling, K.; Boukamp, P.; Poschmann, G.; Volpi, E.; Götz-Rösch, C.; Bernerd, F.; Haag, L.; Huebenthal, U.; et al. Characterization of skin aging-associated secreted proteins (SAASP) produced by dermal fibroblasts isolated from intrinsically aged human skin. J. Investig. Dermatol. 2015, 135, 1954–1968. [Google Scholar] [CrossRef]
- Yosef, R.; Pilpel, N.; Tokarsky-Amiel, R.; Biran, A.; Ovadya, Y.; Cohen, S.; Vadai, E.; Dassa, L.; Shahar, E.; Condiotti, R.; et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 2016, 7, 11190. [Google Scholar] [CrossRef]
- Zhu, Y.; Tchkonia, T.; Fuhrmann-Stroissnigg, H.; Dai, H.M.; Ling, Y.Y.; Stout, M.B.; Pirtskhalava, T.; Giorgadze, N.; Johnson, K.O.; Giles, C.B.; et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 2016, 15, 428–435. [Google Scholar] [CrossRef]
- Sharma, A.K.; Roberts, R.L.; Benson, R.D.; Pierce, J.L.; Yu, K.; Hamrick, M.W.; McGee-Lawrence, M.E. The Senolytic drug navitoclax (ABT-263) causes trabecular bone loss and impaired osteoprogenitor function in aged mice. Front. Cell Dev. Biol. 2020, 8, 354. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Anti-aging: Senolytics or gerostatics (unconventional view). Oncotarget 2021, 12, 1821–1835. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, N.; Ditzel, H.J. Identification of the specificity of isolated phage display single-chain antibodies using yeast two-hybrid screens. Methods Mol. Biol. 2009, 562, 165–176. [Google Scholar] [PubMed]
- Kim, K.M.; Noh, J.H.; Bodogai, M.; Martindale, J.L.; Yang, X.; Indig, F.E.; Basu, S.K.; Ohnuma, K.; Morimoto, C.; Johnson, P.F.; et al. Identification of senescent cell surface targetable protein DPP4. Genes Dev. 2017, 31, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Amor, C.; Feucht, J.; Leibold, J.; Ho, Y.J.; Zhu, C.; Alonso-Curbelo, D.; Mansilla-Soto, J.; Boyer, J.A.; Li, X.; Giavridis, T.; et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 2020, 583, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Althubiti, M.; Lezina, L.; Carrera, S.; Jukes-Jones, R.; Giblett, S.M.; Antonov, A.; Barlev, N.; Saldanha, G.S.; Pritchard, C.A.; Cain, K.; et al. Characterization of novel markers of senescence and their prognostic potential in cancer. Cell Death Dis. 2014, 5, e1528. [Google Scholar] [CrossRef]
- Rossi, M.; Abdelmohsen, K. The Emergence of Senescent Surface Biomarkers as Senotherapeutic Targets. Cells. 2021, 10, 1740. [Google Scholar] [CrossRef]
- Worthen, C.A.; Cui, Y.; Orringer, J.S.; Johnson, T.M.; Voorhees, J.J.; Fisher, G.J. CD26 Identifies a Subpopulation of Fibroblasts that Produce the Majority of Collagen during Wound Healing in Human Skin. J. Investig. Dermatol. 2020, 140, 2515–2524. [Google Scholar] [CrossRef]
- Burns, J.M.; Summers, B.C.; Wang, Y.; Melikian, A.; Berahovich, R.; Miao, Z.; Penfold, M.E.; Sunshine, M.J.; Littman, D.R.; Kuo, C.J.; et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 2006, 203, 2201–2213. [Google Scholar] [CrossRef]
- Balabanian, K.; Lagane, B.; Infantino, S.; Chow, K.Y.; Harriague, J.; Moepps, B.; Arenzana-Seisdedos, F.; Thelen, M.; Bachelerie, F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 2005, 280, 35760–35766. [Google Scholar] [CrossRef]
- Luker, K.E.; Lewin, S.A.; Mihalko, L.A.; Schmidt, B.T.; Winkler, J.S.; Coggins, N.L.; Thomas, D.G.; Luker, G.D. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene 2012, 31, 4750–4758. [Google Scholar] [CrossRef]
- Rajagopal, S.; Kim, J.; Ahn, S.; Craig, S.; Lam, C.M.; Gerard, N.P.; Gerard, C.; Lefkowitz, R.J. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc. Natl Acad. Sci. USA 2010, 107, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Levoye, A.; Balabanian, K.; Baleux, F.; Bachelerie, F.; Lagane, B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood 2009, 113, 6085–6093. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Mei, X.; Wang, J.; Zhang, X.; Yuan, Y. ShRNA-mediated knock-down of CXCR7 increases TRAIL-sensitivity in MCF-7 breast cancer cells. Tumour Biol. 2015, 36, 7243–7250. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Jia, L.T.; Liu, N.N.; Zhu, X.S.; Liu, Q.Q.; Wang, X.L.; Yu, F.; Liu, Y.L.; Yang, A.G.; Gao, C.F. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget 2015, 6, 30818–30830. [Google Scholar] [CrossRef] [PubMed]
- Begley, L.; Monteleon, C.; Shah, R.B.; Macdonald, J.W.; Macoska, J.A. CXCL12 overexpression and secretion by aging fibroblasts enhance human prostate epithelial proliferation in vitro. Aging Cell 2005, 4, 291–298. [Google Scholar] [CrossRef]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef]
- Gargett, T.; Brown, M.P. The inducible caspase-9 suicide gene system as a “safety switch” to limit on-target, off-tumor toxicities of chimeric antigen receptor T cells. Front. Pharmacol. 2014, 5, 235. [Google Scholar] [CrossRef]
- Sierro, F.; Biben, C.; Martínez-Muñoz, L.; Mellado, M.; Ransohoff, R.M.; Li, M.; Woehl, B.; Leung, H.; Groom, J.; Batten, M.; et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc. Natl Acad. Sci. USA 2007, 104, 14759–14764. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takaya, K.; Asou, T.; Kishi, K. Selective Elimination of Senescent Fibroblasts by Targeting the Cell Surface Protein ACKR3. Int. J. Mol. Sci. 2022, 23, 6531. https://doi.org/10.3390/ijms23126531
Takaya K, Asou T, Kishi K. Selective Elimination of Senescent Fibroblasts by Targeting the Cell Surface Protein ACKR3. International Journal of Molecular Sciences. 2022; 23(12):6531. https://doi.org/10.3390/ijms23126531
Chicago/Turabian StyleTakaya, Kento, Toru Asou, and Kazuo Kishi. 2022. "Selective Elimination of Senescent Fibroblasts by Targeting the Cell Surface Protein ACKR3" International Journal of Molecular Sciences 23, no. 12: 6531. https://doi.org/10.3390/ijms23126531





