Structural and Functional Analysis of SHP Promoter and Its Transcriptional Response to FXR in Zn-Induced Changes to Lipid Metabolism
Abstract
:1. Introduction
2. Results
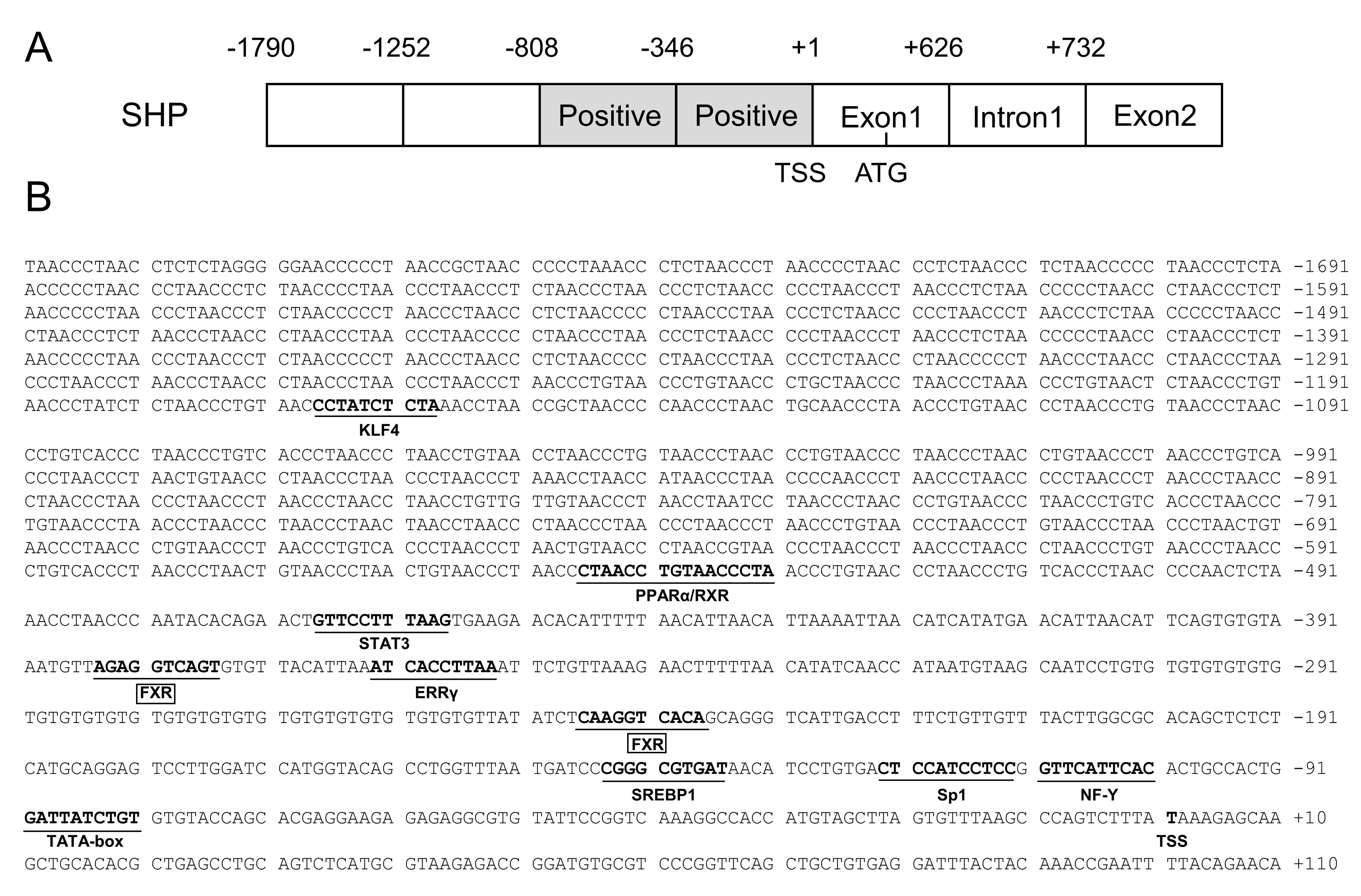
2.1. Cloning and Sequence Analysis of the SHP Promoter
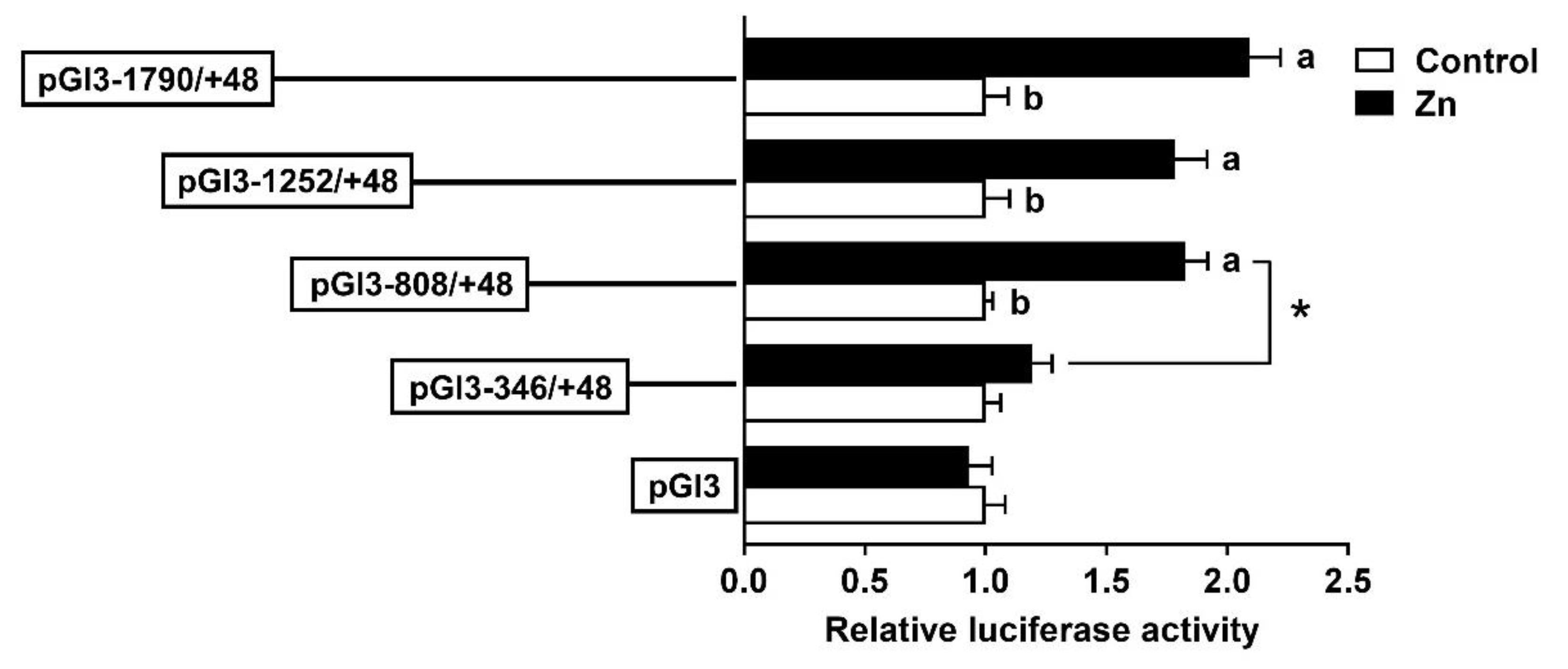
2.2. The 5′-Deletion Assay of the SHP Promoter
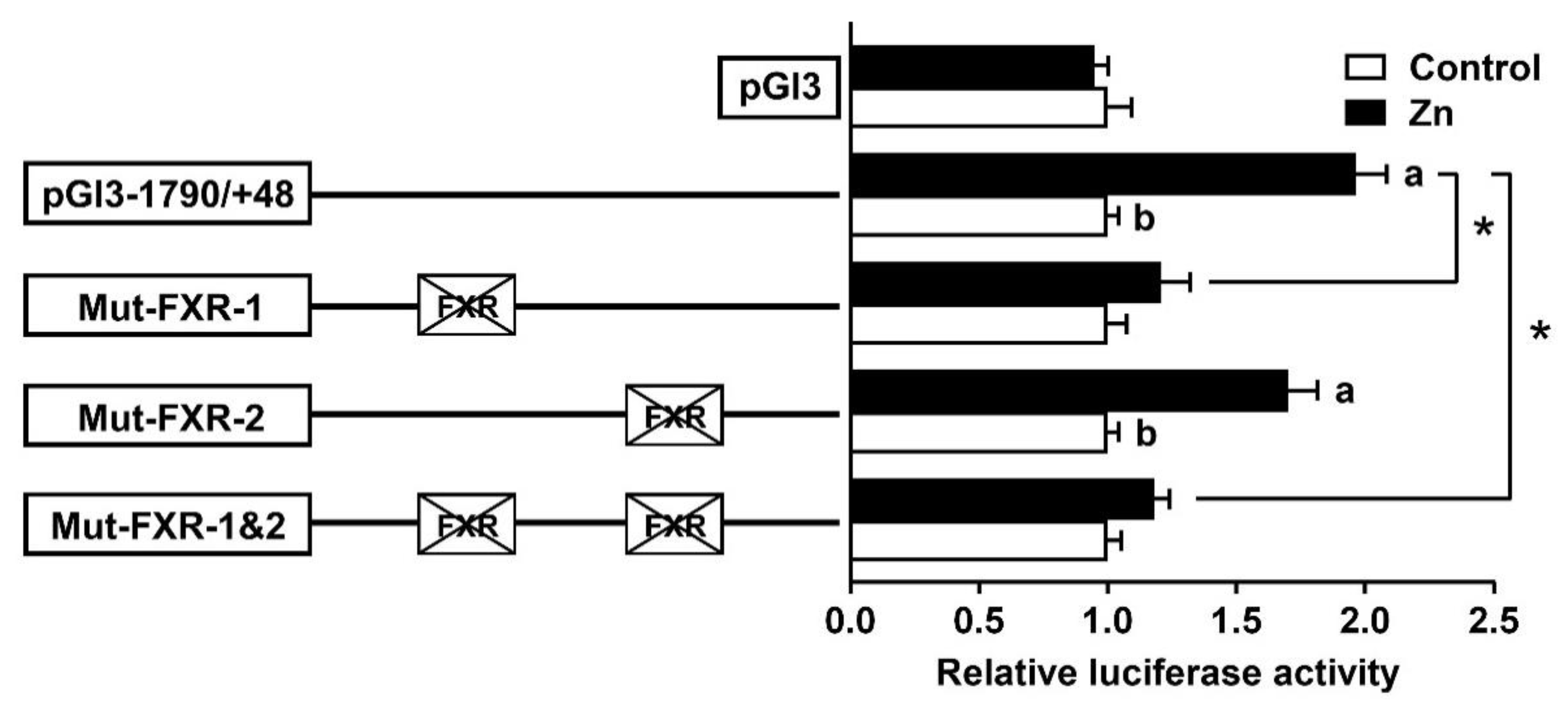
2.3. Site Mutation Analysis of FXR Binding Sites on the SHP Promoter
2.4. FXR Bind with the SHP Promoter
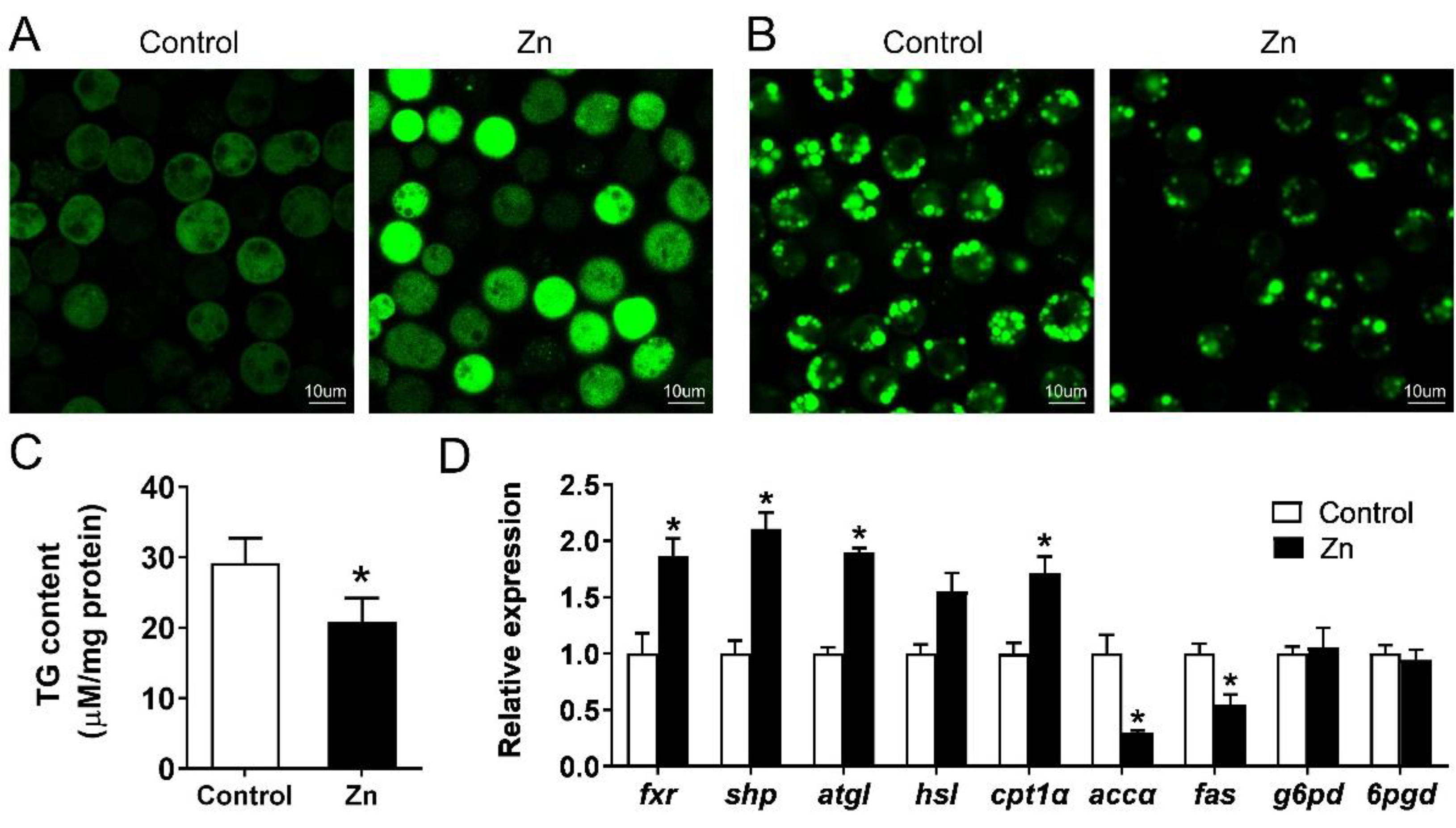
2.5. Effect of Zn on Lipid Metabolism of Yellow Catfish Hepatocytes
3. Discussion
4. Materials and Methods
4.1. Experimental Animals and Reagents
4.2. Promoter Cloning and Plasmid Construction
4.3. Sequence Analysis and Activities Assays of Luciferase
4.4. Site Mutation Assays of FXR Binding Sites on the SHP Promoter
4.5. Analysis of Functional Binding Sites of FXR on the SHP Promoter
4.6. Zn, LD and TG Contents in Hepatocytes and Gene Expression
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bury, N.R.; Walker, P.A.; Glover, C.N. Nutritive metal uptake in teleost fish. J. Exp. Biol. 2003, 206, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Hambidge, K.M.; Krebs, N.F. Zinc deficiency: A special challenge. J. Nutr. 2007, 137, 1101–1105. [Google Scholar] [CrossRef] [PubMed]
- Qu, R.; Feng, M.; Wang, X.; Qin, L.; Wang, C.; Wang, Z.; Wang, L. Metal accumulation and oxidative stress biomarkers in liver of freshwater fish Carassius auratus following in vivo exposure to waterborne zinc under different pH values. Aquat. Toxicol. 2014, 150, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, P.; Wathurapatha, W.S.; Ishara, M.H.; Jayawardana, R.; Galappatthy, P.; Katulanda, P.; Constantine, G.R. Ef-fects of zinc supplementation on serum lipids: A systematic review and meta-analysis. Nutr. Metab. 2015, 12, 26. [Google Scholar] [CrossRef]
- Kang, X.; Zhong, W.; Liu, J.; Song, Z.; McClain, C.J.; Kang, Y.J.; Zhou, Z. Zinc supplementation reverses alcohol-induced steatosis in mice through reactivating hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha. Hepatology 2009, 50, 1241–1250. [Google Scholar] [CrossRef]
- Zhang, J.J.; Hao, J.J.; Zhang, Y.R.; Wang, Y.L.; Li, M.Y.; Miao, H.L.; Zou, X.J.; Liang, B. Zinc mediates the SREBP-SCD axis to regulate lipid metabolism in Caenorhabditis elegans. J. Lipid Res. 2017, 58, 1845–1854. [Google Scholar] [CrossRef]
- Wu, K.; Huang, C.; Shi, X.; Chen, F.; Xu, Y.H.; Pan, Y.X.; Luo, Z.; Liu, X. Role and mechanism of the AMPK pathway in waterborne Zn exposure influencing the hepatic energy metabolism of Synechogobius hasta. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Wei, C.C.; Luo, Z.; Hogstrand, C.; Xu, Y.H.; Wu, L.X.; Chen, G.H.; Pan, Y.X.; Song, Y.F. Zinc reduces hepatic lipid deposition and activates lipophagy via Zn2+/MTF-1/PPARα and Ca2+/CaMKKβ/AMPK pathways. FASEB J. 2018, 32, 6666–6680. [Google Scholar] [CrossRef]
- Wu, K.; Luo, Z.; Hogstrand, C.; Chen, G.H.; Wei, C.C.; Li, D.D. Zn stimulates the phospholipids biosynthesis via the pathways of oxidative and endoplasmic reticulum stress in the intestine of freshwater teleost yellow catfish. Environ. Sci. Technol. 2018, 52, 9206–9214. [Google Scholar] [CrossRef]
- Han, S.L.; Qian, Y.C.; Limbu, S.M.; Wang, J.; Chen, L.Q.; Zhang, M.L.; Du, Z.Y. Lipolysis and lipophagy play individual and interactive roles in regulating triacylglycerol and cholesterol homeostasis and mitochondrial form in zebrafish. BBA-Mol. Cell Biol. L. 2021, 1866, 158988. [Google Scholar] [CrossRef]
- Zou, A.; Magee, N.; Deng, F.; Lehn, S.; Zhong, C.; Zhang, Y. Hepatocyte nuclear receptor SHP suppresses inflammation and fibrosis in a mouse model of nonalcoholic steatohepatitis. J. Biol. Chem. 2018, 293, 8656–8671. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhao, T.; Hogstrand, C.; Xu, Y.C.; Ling, S.C.; Chen, G.H.; Luo, Z. FXR-mediated inhibition of autophagy contributes to FA-induced TG accumulation and accordingly reduces FA-induced lipotoxicity. Cell Commun. Signal. 2020, 18, 1–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Hagedorn, C.H.; Wang, L. Role of nuclear receptor SHP in metabolism and cancer. BBA-Mol. Basis Dis. 2011, 1812, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.W.; Lv, W.H.; Wu, K.; Chen, G.H.; Chen, F.; Song, C.C.; Luo, Z. Dietary nano-ZnO is absorbed via endocytosis and ZIP pathways, upregulates lipogenesis, and induces lipotoxicity in the intestine of yellow catfish. Int. J. Mol. Sci. 2021, 22, 12047. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.L.; Singer, D.S. Core promoters in transcription: Old problem, new insights. Trends Biochem. Sci. 2015, 40, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Basehoar, A.D.; Zanton, S.J.; Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116, 699–709. [Google Scholar] [CrossRef]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Wu, C.; Sun, M.; Liu, L.; Zhou, G.W. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene 2003, 306, 1–12. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.Y.; Kim, J.Y.; Park, S.K.; Seo, J.H.; Kim, J.B.; Choi, H.S. Differential regulation of human and mouse orphan nuclear receptor small heterodimer partner promoter by sterol regulatory element binding protein-1. J. Biol. Chem. 2004, 279, 28122–28131. [Google Scholar] [CrossRef]
- Kassam, A.; Capone, J.P.; Rachubinski, R.A. The short heterodimer partner receptor differentially modulates peroxisome proliferator-activated receptor alphamediated transcription from the peroxisome proliferator-response elements of the genes encoding the peroxisomal beta-oxidation enzymes acyl-CoA oxidase and hydratase-dehydrogenase. Mol. Cell Endocrinol. 2001, 176, 49–56. [Google Scholar]
- Frank, C.; Makkonen, H.; Dunlop, T.W.; Matilainen, M.; Vaisanen, S.; Carlberg, C. Identification of pregnane X receptor binding sites in the regulatory regions of genes involved in bile acid homeostasis. J. Mol. Biol. 2005, 346, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Karagianni, P.; Talianidis, I. Transcription factor networks regulating hepatic fatty acid metabolism. BBA-Mol. Cell Biol. L. 2015, 1851, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Norouzi, S.; Adulcikas, J.; Sohal, S.S.; Myers, S. Zinc stimulates glucose oxidation and glycemic control by modulating the insulin signaling pathway in human and mouse skeletal muscle cell lines. PLoS ONE 2018, 13, e0191727. [Google Scholar] [CrossRef] [PubMed]
- Hoeke, M.O.; Heegsma, J.; Hoekstra, M.; Moshage, H.; Faber, K.N. Human FXR regulates SHP expression through direct binding to an LRH-1 binding site, independent of an IR-1 and LRH-1. PLoS ONE 2014, 9, e88011. [Google Scholar] [CrossRef]
- Huang, X.F.; Zhao, W.Y.; Huang, W.D. FXR and liver carcinogenesis. Acta Pharmacol. Sin. 2015, 36, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Chanda, D.; Sim, J.; Park, Y.Y.; Choi, H.S. Structure and function of the atypical orphan nuclear receptor small heterodimer partner. Int. Rev. Cytol. 2007, 261, 117–158. [Google Scholar] [PubMed]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Mangelsdorf, D.J.; Heyman, R.A.; Auwerx, J. Bile acids lower tri-glyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef]
- Primers, P. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2007, 43, 649–656. [Google Scholar]
- Wu, K.; Tan, X.Y.; Xu, Y.H.; Chen, G.H.; Zhuo, M.Q. Functional analysis of promoters of genes in lipid metabolism and their transcriptional response to STAT3 under leptin signals. Genes 2018, 9, 334. [Google Scholar] [CrossRef]
- Chen, S.W.; Wu, K.; Lv, W.H.; Chen, F.; Song, C.C.; Luo, Z. Functional analysis of two zinc (Zn) transporters (ZIP3 and ZIP8) promoters and their distinct response to MTF1 and RREB1 in the regulation of Zn metabolism. Int. J. Mol. Sci. 2020, 21, 6135. [Google Scholar] [CrossRef]
- Wu, K.; Chen, G.H.; Hogstrand, C.; Ling, S.C.; Wu, L.X.; Luo, Z. Methionine-chelated Zn promotes anabolism by integrating mTOR signal and autophagy pathway in juvenile yellow catfish. J. Trace Elem. Med. Bio. 2021, 65, 126732. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Fan, X.; Wu, Q.-C.; Chen, C.; Xiao, F.; Wu, K. Structural and Functional Analysis of SHP Promoter and Its Transcriptional Response to FXR in Zn-Induced Changes to Lipid Metabolism. Int. J. Mol. Sci. 2022, 23, 6523. https://doi.org/10.3390/ijms23126523
Gao H, Fan X, Wu Q-C, Chen C, Xiao F, Wu K. Structural and Functional Analysis of SHP Promoter and Its Transcriptional Response to FXR in Zn-Induced Changes to Lipid Metabolism. International Journal of Molecular Sciences. 2022; 23(12):6523. https://doi.org/10.3390/ijms23126523
Chicago/Turabian StyleGao, Han, Xing Fan, Qi-Chun Wu, Chuan Chen, Fei Xiao, and Kun Wu. 2022. "Structural and Functional Analysis of SHP Promoter and Its Transcriptional Response to FXR in Zn-Induced Changes to Lipid Metabolism" International Journal of Molecular Sciences 23, no. 12: 6523. https://doi.org/10.3390/ijms23126523





