Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa
Abstract
1. Introduction
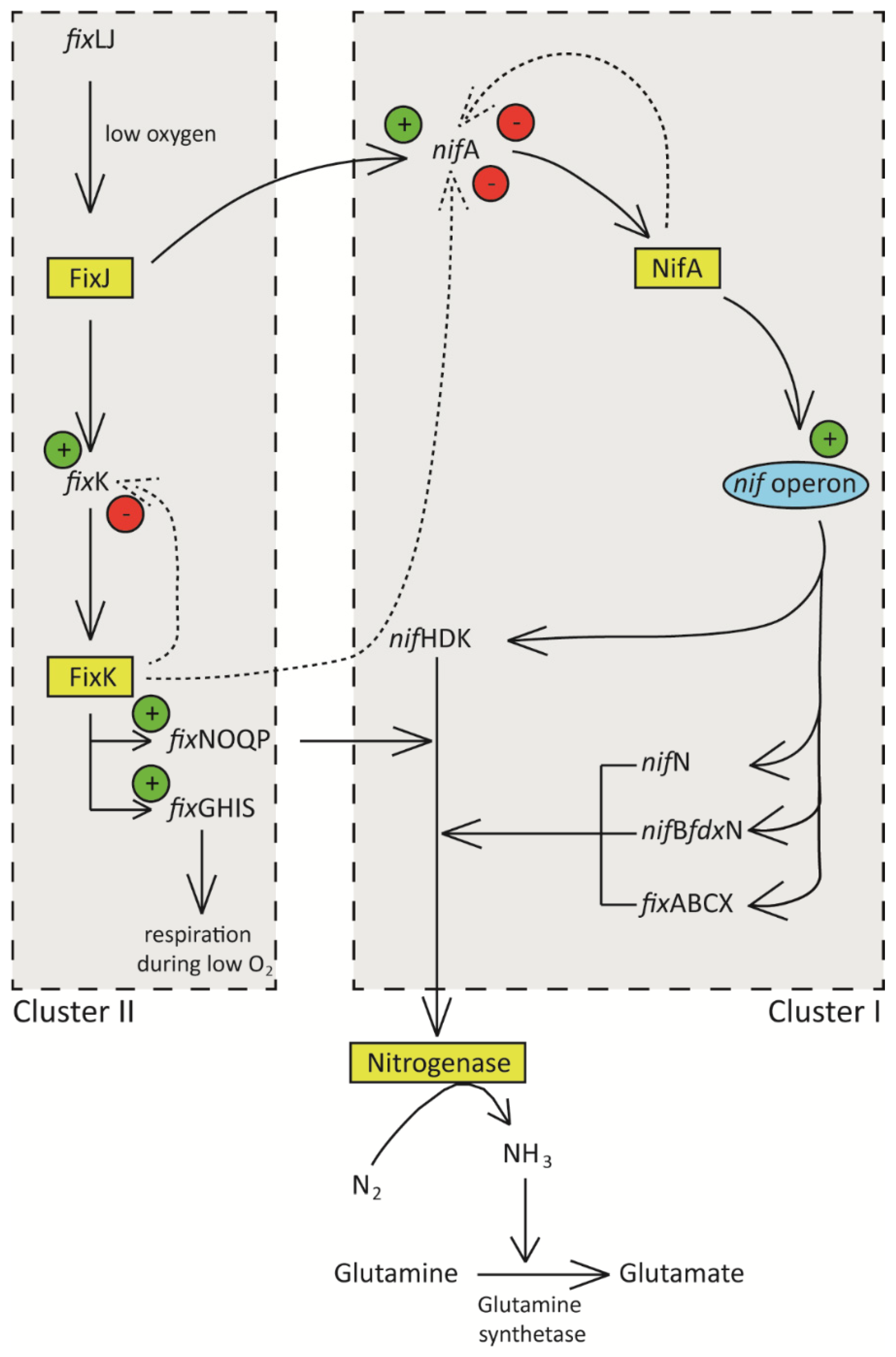
2. Nodulation and Nitrogen Fixation Processes
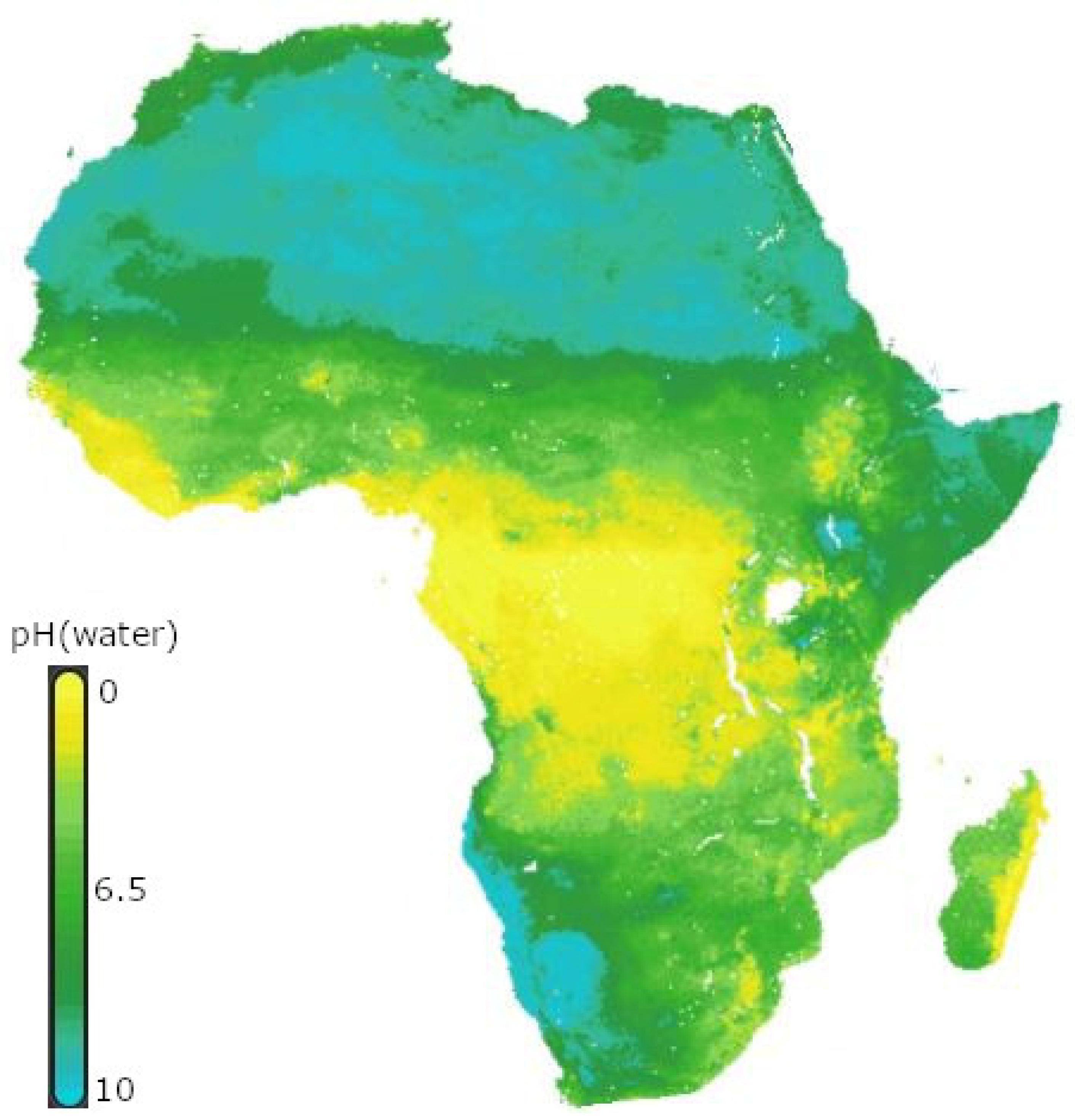
3. Distribution of Rhizobia in African Soil
| Strain | Legume | Reference |
|---|---|---|
| R. phaseoli | P. vulagaris | [17,66] |
| R. paranaense, | P. vulagaris | [66] |
| R. sophoriradicis | P. vulagaris | [66] |
| R. leucaenae | P. vulagaris | [66] |
| R. aegyptiacum | P. vulagaris | [66] |
| R. tropicii | P. vulagaris, Sesbania sesban | [67,68,69] |
| R. etli | P. vulagaris | [67,68] |
| R. leguminosarum | Sesbania sesban, V. faba | [69,70] |
| M. amorphae | S. sesban, C. arietinum | [69,71] |
| M. plurifarium | S. sesban | [69] |
| R. huautlense | S. sesban, C. arietinum | [69,71] |
| M. plurifarium | S. sesban, C. arietinum | [69,71] |
| B. elkanii | Vigna subterranea, Glycine max, A. hypogaea | [72,73,74] |
| B. japonicum | V. subterranea, Glycine max, A. hypogaea | [72,73] |
| M. ciceri | Cicer arietinum | [71] |
| M. mediterraneum | C. arietinum | [71,75] |
| M. loti | C. arietinum | [71] |
| M. opportunistum | C. arietinum | [71] |
| M. haukuii | C. arietinum | [71] |
| M. tianshanense | C. arietinum | [71] |
| M. cicero | C. arietinum | [75] |
| B. yuanmingense | A. hypogaea | [73] |
| B. canariense | A. hypogaea | [73] |
| B. liaoningense | A. hypogaea | [73] |
| R. pisi | V. faba | [70] |
| R. anhuiense | V. faba | [70] |
| R. laguerreae | V. faba | [70] |
| R. binae | V. faba | [70] |
| R. bangladeshense | V. faba | [70] |
| R. lentis | V. faba | [70] |
| R. aethiopicum | V. faba | [70] |
| R. aegypticum | V. faba | [70] |
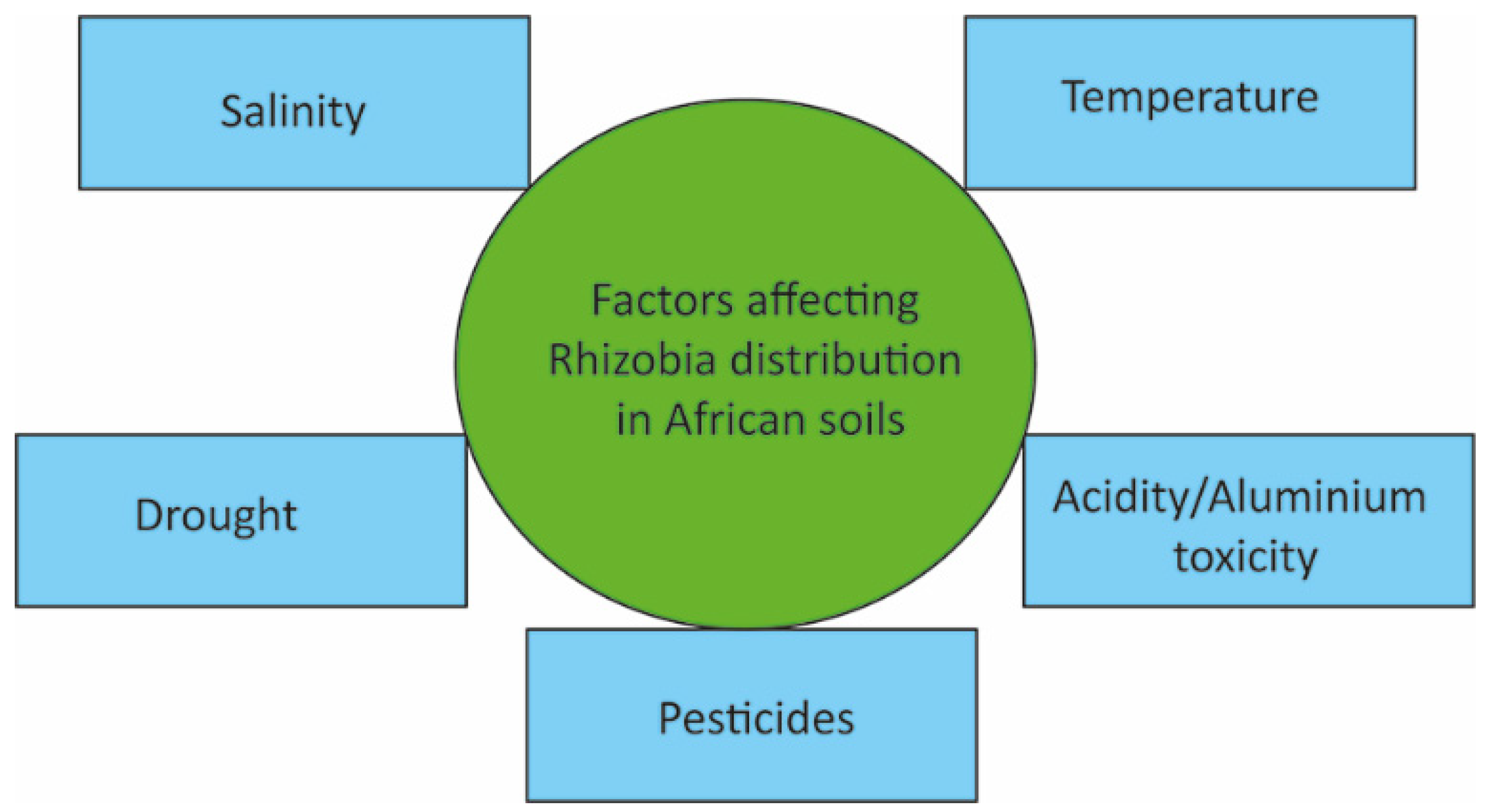
4. Factors Affecting the Distribution of Rhizobia in Africa
5. Methods Used in Rhizobia Characterization in Africa
5.1. Phenotypic and Biochemical Characterization
5.2. Molecular Characterization
6. Potential and Challenges to Commercialization of Rhizobia-Based Inoculants
| No. | Name of Inoculant | Rhizobia Strain | Manufacturer, Country | Target Crops | Reference(s) |
|---|---|---|---|---|---|
| 1 | Biofix | Various strains | MEA Ltd., Kenya | Common bean, soybean, lucerne, peas, cowpea, groundnuts | [185,194] |
| 2 | Nodumax | Bradyrhizobia | IITA, Nigeria | Soybean | [195] |
| 3 | Sojapak | B. japonicum | Soygro, South Africa | Soybean | [13] |
| 4 | Soyflo | B. japonicum | Soygro, South Africa | Soybean | [196] |
| 5 | Peanutflo | Bradyrhizobium sp. | Soygro, South Africa | Peanut and Bambara nuts | [196] |
| 6 | Rhizoflo | Bradyrhizobium japonicum | BASF South Africa (Pty) Ltd. | Soybean | [186] |
| 7 | SeedQuestR | Rhizobium | Soygro, South Africa | Soybean | [197] |
| 8 | Organo | Rhizobium | Microbial solution Ltd., South Africa | Soybean | [197] |
| 9 | Likuiq Semia | Bradyrhizobium elkanii | Microbial Solution Ltd., South Africa | Soybean | [198] |
| 10 | Histick | B. japonicum | BASF South Africa Ltd., South Africa | Soybean | [197] |
| 11 | Organico | Rhizobium | Amka Products | Soybean | [197] |
| 12 | Nitrasec Alfalfa (Lucerne) | Sinorhizobium meliloti | Microbial solution (Pty) Ltd., South Africa | Lucerne | [199] |
| 13 | RAB inoculant | Rhizobium tropici | Rwanda Agricultural Board, Rwanda | Common bean | [187] |
| 14 | N-Soy | B. japonicum | BioControl Products SA (Pty) Ltd., South Africa | Soybean | [197] |
| 15 | N-Bean | Rhizobium phaseolus | BioControl Products SA (Pty) Ltd., South Africa | Common bean | [197] |
| 16 | Nitrosua | B. japonicum | Sokoine University of Agriculture, Tanzania | Soybean | [187,189] |
| 17 | Nitrozam | Various rhizobia strains | Mt. Makulu Research Station, Zambia | soybean, lucerne and common bean | [189] |
| 18 | Bio-N fix | B. japonicum and Rhizobium tropici | Madhavani Ltd. and Makerere University, Uganda | Soybean and common bean | [187] |
| 19 | Kefrifix | Rhizobium | Kenya Forestry Research Institute (KEFRI), Kenya | Leguminous trees, common bean | [186] |
7. Future Prospects
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef] [PubMed]
- Simbine, M.G.; Mohammed, M.; Jaiswal, S.K.; Dakora, F.D. Functional and genetic diversity of native rhizobial isolates nodulating cowpea (Vigna unguiculata L. Walp.) in Mozambican soils. Sci. Rep. 2021, 11, 12747. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Taxonomic Outline of the Prokaryotes. Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2004. [Google Scholar] [CrossRef]
- Namkeleja, Y.; Mtei, K.; Ndakidemi, P.A. Isolation and molecular characterization of elite indigenous rhizobia nodulating Phaseolus bean (P. vulagaris L.). Am. J. Plant Sci. 2016, 7, 1905. [Google Scholar] [CrossRef]
- Kawaka, F.; Makonde, H.; Dida, M.; Opala, P.; Ombori, O.; Maingi, J.; Muoma, J. Genetic diversity of symbiotic bacteria nodulating common bean (P. vulagaris) in western Kenya. PLoS ONE 2018, 13, e0207403. [Google Scholar] [CrossRef]
- Jalloh, A.A. Potential of native rhizobia isolates to improve production of legume crops in small holder farms. Biosci. Res. 2020, 17, 1498–1510. [Google Scholar]
- Vanlauwe, B.; Hungria, M.; Kanampiu, F.; Giller, K.E. The role of legumes in the sustainable intensification of African smallholder agriculture: Lessons learnt and challenges for the future. Agric. Ecosyst. Environ. 2019, 284, 106583. [Google Scholar] [CrossRef]
- Kawaka, F.; Dida, M.M.; Opala, P.A.; Ombori, O.; Maingi, J.; Osoro, N.; Muthini, M.; Amoding, A.; Mukaminega, D.; Muoma, J. Symbiotic efficiency of native rhizobia nodulating common bean (P. vulagaris L.) in soils of Western Kenya. Int. Sch. Res. Notices 2014, 2014, 258497. [Google Scholar]
- Mutuma, S.; Okello, J.; Karanja, N.; Woomer, P. Smallholder farmers’ use and profitability of legume inoculants in western Kenya. Afr. Crop Sci. J. 2014, 22, 205–214. [Google Scholar]
- Mungai, N.; Karubiu, N. Effectiveness of rhizobia isolates from Njoro soils (Kenya) and commercial inoculants in nodulation of common beans (P. vulagaris). J. Agric. Sci. 2011, 12, 47–59. [Google Scholar]
- Abubakar, F.J.; Yusuf, A.A. Relative efficiency and response of promiscuous soybean to rhizobia inoculant in Savanna region of Nigeria. Afr. J. Microbiol. Res. 2016, 10, 1187–1193. [Google Scholar]
- Musiyiwa, K.; Mpepereki, S.; Giller, K. Symbiotic effectiveness and host ranges of indigenous rhizobia nodulating promiscuous soyabean varieties in Zimbabwean soils. Soil Biol. Biochem. 2005, 37, 1169–1176. [Google Scholar] [CrossRef]
- Thuita, M.; Pypers, P.; Herrmann, L.; Okalebo, R.J.; Othieno, C.; Muema, E.; Lesueur, D. Commercial rhizobial inoculants significantly enhance growth and nitrogen fixation of a promiscuous soybean variety in Kenyan soils. Biol. Fertil. Soils 2012, 48, 87–96. [Google Scholar] [CrossRef]
- Sivparsad, B.J.; Chiuraise, N.; Laing, M.D. Comparative evaluation of commercial rhizobial inoculants of soybean. S. Afr. J. Plant Soil 2016, 33, 157–160. [Google Scholar] [CrossRef]
- Waswa, M.N. Identifying Elite Rhizobia for Commercial Soybean (Glycine Max) Inoculants; University of Nairobi: Nairobi, Kenya, 2013. [Google Scholar]
- Tena, W.; Wolde-Meskel, E.; Walley, F. Symbiotic efficiency of native and exotic Rhizobium strains nodulating lentil (Lens culinaris Medik.) in soils of Southern Ethiopia. Agronomy 2016, 6, 11. [Google Scholar] [CrossRef]
- Wekesa, C.S.; Furch, A.C.; Oelmüller, R. Isolation and Characterization of High-Efficiency Rhizobia From Western Kenya Nodulating With Common Bean. Front. Microbiol. 2021, 12, 697567. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Njeru, E.M.; Kimiti, J.M.; Ombori, O.; Maingi, J.M. Potential of native rhizobia in enhancing nitrogen fixation and yields of climbing beans (P. vulagaris L.) in contrasting environments of Eastern Kenya. Front. Plant Sci. 2017, 443. [Google Scholar] [CrossRef]
- Stacey, G. The Rhizobium-legume nitrogen-fixing symbiosis. In Biology of the Nitrogen Cycle; Elsevier: Amsterdam, The Netherlands, 2007; pp. 147–163. [Google Scholar]
- Wulandari, D.; Songwattana, P.; Gressent, F.; Piromyou, P.; Teamtisong, K.; Boonkerd, N.; Giraud, E.; Tittabutr, P.; Teaumroong, N. Nod-Factor structure and functional redundancy of nod genes contribute the broad host range Bradyrhizobium sp. DOA9. Rhizosphere 2022, 22, 100503. [Google Scholar] [CrossRef]
- Goss, M.J.; Carvalho, M.; Kadir, S.; Brito, I.; Alho, L. Impacts on Host Plants of Interactions between AMF and Other Soil Organisms in the Rhizosphere. In Functional Diversity of Mycorrhiza and Sustainable Agriculture; Academic Press: Cambridge, MA, USA, 2017; pp. 81–109. [Google Scholar]
- Nap, J.-P.; Bisseling, T. Developmental biology of a plant-prokaryote symbiosis: The legume root nodule. Science 1990, 250, 948–954. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zhu, H. Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef]
- Appleby, C.A. Leghemoglobin and Rhizobium respiration. Annu. Rev. Plant Physiol. 1984, 35, 443–478. [Google Scholar] [CrossRef]
- Brear, E.M.; Day, D.A.; Smith, P.M.C. Iron: An essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci. 2013, 4, 359. [Google Scholar] [CrossRef] [PubMed]
- Ott, T.; van Dongen, J.T.; Gu, C.; Krusell, L.; Desbrosses, G.; Vigeolas, H.; Bock, V.; Czechowski, T.; Geigenberger, P.; Udvardi, M.K. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 2005, 15, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Strodtman, K.N.; Emerich, D.W. Nodule metabolism. Nitrogen Fixation in Crop Production; Crop Science Society of America: Madison, WI, USA, 2009; pp. 95–124. [Google Scholar]
- Udvardi, M.K.; Day, D.A. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Biol. 1997, 48, 493–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Contador, C.A.; Fan, K.; Lam, H.-M. Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front. Plant Sci. 2018, 1860. [Google Scholar] [CrossRef] [PubMed]
- Tajima, S.; Nomura, M.; Kouchi, H. Ureide biosynthesis in legume nodules. Front. Biosci. 2004, 9, 1374–1381. [Google Scholar] [CrossRef]
- Chandler, M.R.; Date, R.; Roughley, R. Infection and root-nodule development in Stylosanthes species by Rhizobium. J. Exp. Bot. 1982, 33, 47–57. [Google Scholar] [CrossRef]
- Alazard, D.; Duhoux, E. Development of stem nodules in a tropical forage legume, Aeschynomene afraspera. J. Exp. Bot. 1990, 41, 1199–1206. [Google Scholar] [CrossRef]
- Chandler, M.R. Some observations on infection of A. hypogaea L. by Rhizobium. J. Exp. Bot. 1978, 29, 749–755. [Google Scholar] [CrossRef]
- Boogerd, F.C.; van Rossum, D. Nodulation of groundnut by Bradyrhizobium: A simple infection process by crack entry. FEMS Microbiol. Rev. 1997, 21, 5–27. [Google Scholar] [CrossRef]
- Fischinger, S.A.; Drevon, J.-J.; Claassen, N.; Schulze, J. Nitrogen from senescing lower leaves of common bean is re-translocated to nodules and might be involved in a N-feedback regulation of nitrogen fixation. J. Plant Physiol. 2006, 163, 987–995. [Google Scholar] [CrossRef]
- Nishida, H.; Ito, M.; Miura, K.; Kawaguchi, M.; Suzaki, T. Autoregulation of nodulation pathway is dispensable for nitrate-induced control of rhizobial infection. Plant Signal. Behav. 2020, 15, 1733814. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.H.; Lin, Y.H.; Reid, D.E.; Gresshoff, P.M. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.; Lin, M.-H.; Gresshoff, P.M. Regulation of legume nodulation by acidic growth conditions. Plant Signal. Behav. 2013, 8, e23426. [Google Scholar] [CrossRef]
- Schnabel, E.; Journet, E.-P.; de Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 1994, 58, 352–386. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.M.; Ludden, P.W. Maturation of nitrogenase: A biochemical puzzle. J. Bacteriol. 2005, 187, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Poudel, S.; Colman, D.R.; Fixen, K.R.; Ledbetter, R.N.; Zheng, Y.; Pence, N.; Seefeldt, L.C.; Peters, J.W.; Harwood, C.S.; Boyd, E.S. Electron transfer to nitrogenase in different genomic and metabolic backgrounds. J. Bacteriol. Res. 2018, 200, e00757-17. [Google Scholar] [CrossRef]
- McGlynn, S.E.; Boyd, E.S.; Peters, J.W.; Orphan, V.J. Classifying the metal dependence of uncharacterized nitrogenases. Front. Microbiol. 2013, 3, 419. [Google Scholar] [CrossRef]
- Shah, V.K.; Allen, J.R.; Spangler, N.J.; Ludden, P.W. In vitro synthesis of the iron-molybdenum cofactor of nitrogenase. Purification and characterization of NifB cofactor, the product of NIFB protein. J. Biol. Chem. 1994, 269, 1154–1158. [Google Scholar] [CrossRef]
- Curatti, L.; Ludden, P.W.; Rubio, L.M. NifB-dependent in vitro synthesis of the iron–molybdenum cofactor of nitrogenase. Proc. Natl. Acad. Sci. USA 2006, 103, 5297–5301. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.; Maillet, F.; Plazanet, C.; Debellé, F.; Ferro, M.; Truchet, G.; Promé, J.-C.; Dénarié, J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc. Natl. Acad. Sci. USA 1996, 93, 15305–15310. [Google Scholar] [CrossRef] [PubMed]
- Masepohl, B.; Kutsche, M.; Riedel, K.-U.; Schmehl, M.; Klipp, W.; Pühler, A. Functional analysis of the cysteine motifs in the ferredoxin-like protein FdxN of Rhizobium meliloti involved in symbiotic nitrogen fixation. Mol. Gen. Genet. 1992, 233, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Edgren, T.; Nordlund, S. The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J. Bacteriol. Res. 2004, 186, 2052–2060. [Google Scholar] [CrossRef]
- Webb, I.U.; Xu, J.; Sánchez-Cañizares, C.; Karunakaran, R.; Ramachandran, V.K.; Rutten, P.J.; East, A.K.; Huang, W.E.; Watmough, N.J.; Poole, P.S. Regulation and characterization of mutants of fixABCX in Rhizobium leguminosarum. Mol. Plant Microbe Interact. 2021, 34, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, A.; Patschkowski, T.; Quandt, J.; Selinger, L.B.; Weidner, S.; Krämer, M.; Zhou, L.; Hynes, M.F.; Priefer, U.B. Functional and regulatory analysis of the two copies of the fixNOQP operon of Rhizobium leguminosarum strain VF39. Mol. Plant Microbe Interact. 1997, 10, 605–616. [Google Scholar] [CrossRef]
- Rutten, P.J.; Poole, P.S. Oxygen regulatory mechanisms of nitrogen fixation in rhizobia. Adv. Microb. Physiol. 2019, 75, 325–389. [Google Scholar]
- Cherkasov, N.; Ibhadon, A.; Fitzpatrick, P. A review of the existing and alternative methods for greener nitrogen fixation. Chem. Eng. Process. Process Intensif. 2015, 90, 24–33. [Google Scholar] [CrossRef]
- Burris, R.H.; Roberts, G.P. Biological nitrogen fixation. Annu. Rev. Nutr. 1993, 13, 317–335. [Google Scholar] [CrossRef]
- Lopez, O.; Morera, C.; Miranda-Rios, J.; Girard, L.; Romero, D.; Soberόn, M. Regulation of gene expression in response to oxygen in Rhizobium etli: Role of FnrN in fixNOQP expression and in symbiotic nitrogen fixation. J. Bacteriol. Res. 2001, 183, 6999–7006. [Google Scholar] [CrossRef]
- Halbleib, C.M.; Ludden, P.W. Regulation of biological nitrogen fixation. J. Nutr. 2000, 130, 1081–1084. [Google Scholar] [CrossRef]
- Allen, O.N.; Allen, E.K. The Leguminosae, a Source Book of Characteristics, Uses, and Nodulation; University of Wisconsin Press: Madison, WI, USA, 1981. [Google Scholar]
- Gepts, P. Biochemical evidence bearing on the domestication of Phaseolus (Fabaceae) beans. Econ. Bot. 1990, 44, 28–38. [Google Scholar] [CrossRef]
- Logozzo, G.; Donnoli, R.; Macaluso, L.; Papa, R.; Knüpffer, H.; Zeuli, P.S. Analysis of the contribution of Mesoamerican and Andean gene pools to European common bean (P. vulagaris L.) germplasm and strategies to establish a core collection. Genet. Resour. Crop Evol. 2007, 54, 1763–1779. [Google Scholar] [CrossRef]
- Lie, T. Symbiotic specialisation in pea plants: The requirement of specific Rhizobium strains for peas from Afghanistan. Ann. Appl. Biol. 1978, 88, 462–465. [Google Scholar] [CrossRef]
- Keyser, H.H.; Bohlool, B.B.; Hu, T.; Weber, D.F. Fast-growing rhizobia isolated from root nodules of soybean. Science 1982, 215, 1631–1632. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, K.; Paulin, L.; Roos, C.; Suominen, L. Nodulation genes of Rhizobium galegae. In Nitrogen Fixation: Fundamentals and Applications; Springer: Berlin/Heidelberg, Germany, 1995; pp. 365–370. [Google Scholar]
- D’Andrea, A.C.; Kahlheber, S.; Logan, A.L.; Watson, D.J. Early domesticated cowpea (Vigna unguiculata) from Central Ghana. Antiquity 2007, 81, 686–698. [Google Scholar] [CrossRef]
- Rachie, K.O. Tropical legumes: Resources for the future. In Tropical Legumes: Resources for the Future; National Academy of Sciences: Washington, DC, USA, 1979. [Google Scholar]
- Martínez-Romero, E.; Caballero-Mellado, J. Rhizobium phylogenies and bacterial genetic diversity. CRC Crit Rev. Plant Sci. 1996, 15, 113–140. [Google Scholar] [CrossRef]
- Mwenda, G.M.; O’Hara, G.W.; De Meyer, S.E.; Howieson, J.G.; Terpolilli, J.J. Genetic diversity and symbiotic effectiveness of Phaseolus vulgaris-nodulating rhizobia in Kenya. Syst. Appl. Microbiol. 2018, 41, 291–299. [Google Scholar] [CrossRef]
- Anyango, B.; Wilson, K.J.; Beynon, J.L.; Giller, K.E. Diversity of rhizobia nodulating P. vulagaris L. in two Kenyan soils with contrasting pHs. Appl. Environ. Microbiol. 1995, 61, 4016–4021. [Google Scholar] [CrossRef]
- Diouf, A.; de Lajudie, P.; Neyra, M.; Kersters, K.; Gillis, M.; Martinez-Romero, E.; Gueye, M. Polyphasic characterization of rhizobia that nodulate Phaseolus vulgaris in West Africa (Senegal and Gambia). Int. J. Syst. Evol. Microbiol. 2000, 50, 159–170. [Google Scholar] [CrossRef][Green Version]
- Bala, A.; Murphy, P.; Giller, K.E. Occurrence and genetic diversity of rhizobia nodulating S. sesban in African soils. Soil Biol. Biochem. 2002, 34, 1759–1768. [Google Scholar] [CrossRef]
- Asfaw, B.; Aserse, A.A.; Asefa, F.; Yli-Halla, M.; Lindström, K. Genetically diverse lentil-and faba bean-nodulating rhizobia are present in soils across Central and Southern Ethiopia. FEMS Microbiol. Ecol. 2020, 96, fiaa015. [Google Scholar] [CrossRef] [PubMed]
- Gunnabo, A.; van Heerwaarden, J.; Geurts, R.; Wolde-Meskel, E.; Degefu, T.; Giller, K. Symbiotic interactions between chickpea (Cicer arietinum L.) genotypes and Mesorhizobium strains. Symbiosis 2020, 82, 235–248. [Google Scholar] [CrossRef]
- Adjei, J.A.; Aserse, A.A.; Yli-Halla, M.; Ahiabor, B.D.; Abaidoo, R.C.; Lindstrom, K. Phylogenetically diverse Bradyrhizobium genospecies nodulate Bambara groundnut (Vigna. subterranea L. Verdc) and soybean (Glycine max L. Merril) in the northern savanna zones of Ghana. FEMS Microbiol. Ecol. 2022, 98, fiac043. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, E.T.; Stępkowski, T.; Przymusiak, A.; Botha, W.J.; Law, I.J. Cowpea and peanut in southern Africa are nodulated by diverse Bradyrhizobium strains harboring nodulation genes that belong to the large pantropical clade common in Africa. Mol. Phylogen. Evol. 2008, 48, 1131–1144. [Google Scholar] [CrossRef] [PubMed]
- Gyogluu, C.; Jaiswal, S.K.; Kyei-Boahen, S.; Dakora, F.D. Identification and distribution of microsymbionts associated with soybean nodulation in Mozambican soils. Syst. Appl. Microbiol. 2018, 41, 506–515. [Google Scholar] [CrossRef]
- Ben Romdhane, S.; Aouani, M.; Trabelsi, M.; De Lajudie, P.; Mhamdi, R. Selection of high nitrogen-fixing rhizobia nodulating chickpea (C. arietinum) for semi-arid Tunisia. J. Agron. Crop Sci. 2008, 194, 413–420. [Google Scholar] [CrossRef]
- Pérez-Ramírez, N.O.; Rogel, M.A.; Wang, E.; Castellanos, J.Z.; Martínez-Romero, E. Seeds of Phaseolus vulgaris bean carry Rhizobium etli. FEMS Microbiol. Ecol. 1998, 26, 289–296. [Google Scholar] [CrossRef]
- Beukes, C.W.; Boshoff, F.S.; Phalane, F.L.; Hassen, A.I.; Le Roux, M.M.; Stȩpkowski, T.; Venter, S.N.; Steenkamp, E.T. Both alpha-and beta-rhizobia occupy the root nodules of Vachellia karroo in South Africa. Front. Microbiol. 2019, 10, 1195. [Google Scholar] [CrossRef]
- Zahran, H.H. Rhizobia from wild legumes: Diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J. Biotechnol. 2001, 91, 143–153. [Google Scholar] [CrossRef]
- Pule-Meulenberg, F.; Belane, A.K.; Krasova-Wade, T.; Dakora, F.D. Symbiotic functioning and bradyrhizobial biodiversity of cowpea (Vigna unguiculata L. Walp.) in Africa. BMC Microbiol. 2010, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Muindi, M.M.; Muthini, M.; Njeru, E.M.; Maingi, J. Symbiotic efficiency and genetic characterization of rhizobia and non rhizobial endophytes associated with cowpea grown in semi-arid tropics of Kenya. Heliyon 2021, 7, e06867. [Google Scholar] [CrossRef] [PubMed]
- Nyaga, J.W.; Njeru, E.M. Potential of native rhizobia to improve cowpea growth and production in semiarid regions of Kenya. Front. Agron. 2020, 28, 1–13. [Google Scholar] [CrossRef]
- Kebede, E.; Amsalu, B.; Argaw, A.; Tamiru, S. Symbiotic effectiveness of cowpea (Vigna unguiculata (L.) Walp.) nodulating rhizobia isolated from soils of major cowpea producing areas in Ethiopia. Cogent Food Agric. 2020, 6, 1763648. [Google Scholar] [CrossRef]
- Argaw, A. Development of environmental friendly bioinoculate for peanut (Arachis hypogea L.) production in Eastern Ethiopia. Environ. Syst. Res. 2017, 6, 23. [Google Scholar] [CrossRef]
- Hassen, A.I.; Bopape, F.L.; Trytsman, M. Nodulation study and characterization of rhizobial microsymbionts of forage and pasture legumes in South Africa. World J. Agric. Res. 2014, 2, 93–100. [Google Scholar] [CrossRef][Green Version]
- Jaiswal, S.K.; Dakora, F.D. Widespread distribution of highly adapted Bradyrhizobium species nodulating diverse legumes in Africa. Front. Microbiol. 2019, 310, 1–16. [Google Scholar] [CrossRef]
- Puozaa, D.K.; Jaiswal, S.K.; Dakora, F.D. Phylogeny and distribution of Bradyrhizobium symbionts nodulating cowpea (Vigna unguiculata L. Walp) and their association with the physicochemical properties of acidic African soils. Syst. Appl. Microbiol. 2019, 42, 403–414. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Beyan, S.M.; Dakora, F.D. Distribution, diversity and population composition of soybean-nodulating bradyrhizobia from different agro-climatic regions in Ethiopia. Biol. Fertil. Soils 2016, 52, 725–738. [Google Scholar] [CrossRef]
- Goyal, R.K.; Mattoo, A.K.; Schmidt, M.A. Rhizobial–host interactions and symbiotic nitrogen fixation in legume crops toward agriculture sustainability. Front. Microbiol. 2021, 12, 1290. [Google Scholar] [CrossRef]
- Ormeño-Orrillo, E.; Gomes, D.F.; Del Cerro, P.; Vasconcelos, A.T.R.; Canchaya, C.; Almeida, L.G.P.; Mercante, F.M.; Ollero, F.J.; Megías, M.; Hungria, M. Genome of Rhizobium leucaenae strains CFN 299 T and CPAO 29.8: Searching for genes related to a successful symbiotic performance under stressful conditions. BMC Genom. 2016, 17, 534. [Google Scholar] [CrossRef] [PubMed]
- Hungria, M.; Franco, A.A.; Sprent, J.I. New sources of high-temperature tolerant rhizobia for P. vulagaris L. Plant Soil. 1993, 149, 103–109. [Google Scholar] [CrossRef]
- Liu, S.; Liu, W.; Shi, X.; Li, S.; Hu, T.; Song, L.; Wu, C. Dry-hot stress significantly reduced the nitrogenase activity of epiphytic cyanolichen. Sci. Total Environ. 2018, 619, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Hungria, M.; Boddey, L.; Santos, M.; Vargas, M. Nitrogen fixation capacity and nodule occupancy by Bradyrhizobium japonicum and B. elkanii strains. Biol. Fertil. 1998, 27, 393–399. [Google Scholar] [CrossRef]
- Trevors, J. Plasmid curing in bacteria. FEMS Microbiol. Rev. 1986, 1, 149–157. [Google Scholar] [CrossRef]
- Soberón-Chávez, G.; Nájera, R.; Olivera, H.; Segovia, L. Genetic rearrangements of a Rhizobium phaseoli symbiotic plasmid. J. Bacteriol. Res. 1986, 167, 487–491. [Google Scholar] [CrossRef]
- Hungria, M.; Stacey, G. Molecular signals exchanged between host plants and rhizobia: Basic aspects and potential application in agriculture. Soil Biol. Biochem. 1997, 29, 819–830. [Google Scholar] [CrossRef]
- Hungria, M.; Franco, A.A. Effects of high temperature on nodulation and nitrogen fixation by P. vulagaris L. Plant Soil 1993, 149, 95–102. [Google Scholar] [CrossRef]
- Yuan, K.; Reckling, M.; Ramirez, M.D.A.; Djedidi, S.; Fukuhara, I.; Ohyama, T.; Yokoyama, T.; Bellingrath-Kimura, S.D.; Halwani, M.; Egamberdieva, D. Characterization of rhizobia for the improvement of soybean cultivation at cold conditions in central Europe. Microbes Environ. 2020, 35, ME19124. [Google Scholar] [CrossRef]
- Simon, Z.; Mtei, K.; Gessesse, A.; Ndakidemi, P.A. Isolation and characterization of nitrogen fixing rhizobia from cultivated and uncultivated soils of northern Tanzania. Am. J. Plant Sci. 2014, 5, 4050. [Google Scholar] [CrossRef]
- Swelim, D.; Ali, M.; El-Khatib, E. Some tree-legume-rhizobia are meagerly arising in Egyptian soil. Aust. J. Basic Appl. Sci. 2010, 4, 1297–1304. [Google Scholar]
- Mulama, S.; Onamu, R.; Odongo, F.; Muoma, J. Abundance and Symbiotic Rhizobia Colonizing Soybean (Glycine max) in Soils of Kakamega County, Western Kenya. Int. J. Agron. 2021, 2021, 6627541. [Google Scholar] [CrossRef]
- Matsumoto, S.; Shimada, H.; Sasaoka, T.; Miyajima, I.; Kusuma, G.J.; Gautama, R.S. Effects of acid soils on plant growth and successful revegetation in the case of mine site. In Soil pH for Nutrient Availability and Crop Performance; IntechOpen: London, UK, 2017; pp. 9–27. [Google Scholar]
- Gentili, R.; Ambrosini, R.; Montagnani, C.; Caronni, S.; Citterio, S. Effect of soil pH on the growth, reproductive investment and pollen allergenicity of Ambrosia artemisiifolia L. Front. Plant Sci. 2018, 1335, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.; Cooper, J.; Bjourson, A. Response of Lotus rhizobia to acidity and aluminium in liquid culture and in soil. Plant Soil 1988, 107, 227–231. [Google Scholar] [CrossRef]
- Keyser, H.; Munns, D. Tolerance of rhizobia to acidity, aluminum, and phosphate. Soil Sci. Soc. Am. J. 1979, 43, 519–523. [Google Scholar] [CrossRef]
- Wekesa, C.; Muoma, J.O.; Reichelt, M.; Asudi, G.O.; Furch, A.C.; Oelmüller, R. The Cell Membrane of a Novel Rhizobium phaseoli Strain Is the Crucial Target for Aluminium Toxicity and Tolerance. Cells 2022, 11, 873. [Google Scholar] [CrossRef]
- Wood, M.; Cooper, J. Aluminium toxicity and multiplication of Rhizobium trifolii in a defined growth medium. Soil Biol. Biochem. 1984, 16, 571–576. [Google Scholar] [CrossRef]
- Johnson, A.; Wood, M. DNA, a possible site of action of aluminum in Rhizobium spp. Appl. Environ. Microbiol. 1990, 56, 3629–3633. [Google Scholar] [CrossRef]
- Flis, S.; Glenn, A.; Dilworth, M. The interaction between aluminium and root nodule bacteria. Soil Biol. Biochem. 1993, 25, 403–417. [Google Scholar] [CrossRef]
- Cunningham, S.D.; Munns, D.N. Effects of rhizobial extracellular polysaccharide on pH and aluminum activity. Soil Sci. Soc. Am. J. 1984, 48, 1276–1280. [Google Scholar] [CrossRef]
- Shahid, S.A.; Zaman, M.; Heng, L. Soil salinity: Historical perspectives and a world overview of the problem. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer: Berlin/Heidelberg, Germany, 2018; pp. 43–53. [Google Scholar]
- De Villiers, M.; Nell, J.; Barnard, R.; Henning, A. Salt-affected soils: South Africa. In Food Agricultural Organization Contract; No. GW/B/2004/01; ISCW: Pretoria, South Africa, 2003; Volume 26897. [Google Scholar]
- Ballhorn, D.J.; Wolfe, E.R.; Tyler, J.; Ronan, W.; Sands-Gouner, S.; Shaw, C.; Balkan, M.A.; Kautz, S. Quantitative effects of soil salinity on the symbiosis of wild lima bean (Phaseolus lunatus L.) and Bradyrhizobium in Costa Rica. J. Appl. Bot. Food Qual. 2018, 91, 304–309. [Google Scholar] [CrossRef]
- Tu, J. Effect of salinity on Rhizobium-root-hair interaction, nodulation and growth of soybean. Can. J. Plant Sci. 1981, 61, 231–239. [Google Scholar] [CrossRef]
- Burghardt, L.T. Evolving together, evolving apart: Measuring the fitness of rhizobial bacteria in and out of symbiosis with leguminous plants. New Phytol. 2020, 228, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Zahran, H.; Abdel-Fattah, M.; Yasser, M.; Mahmoud, A.; Bedmar, E. Diversity and environmental stress responses of rhizobial bacteria from Egyptian grain legumes. Aust. J. Basic Appl. Sci. 2012, 6, 571–583. [Google Scholar]
- Botha, W.J.; Jaftha, J.B.; Bloem, J.F.; Habig, J.H.; Law, I.J. Effect of soil bradyrhizobia on the success of soybean inoculant strain CB 1809. Microbiol. Res. 2004, 159, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Muleta, D.; Ryder, M.H.; Denton, M.D. The potential for rhizobial inoculation to increase soybean grain yields on acid soils in Ethiopia. Soil Sci. Plant Nutr. 2017, 63, 441–451. [Google Scholar] [CrossRef]
- Marquez-Garcia, B.; Shaw, D.; Cooper, J.W.; Karpinska, B.; Quain, M.D.; Makgopa, E.M.; Kunert, K.; Foyer, C.H. Redox markers for drought-induced nodule senescence, a process occurring after drought-induced senescence of the lowest leaves in soybean (Glycine max). Ann. Bot. 2015, 116, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Hungria, M.; Vargas, M.A. Environmental factors affecting N2 fixation in grain legumes in the tropics, with an emphasis on Brazil. Field Crops Res. 2000, 65, 151–164. [Google Scholar] [CrossRef]
- Abdel-Wahab, A.; Shabeb, M.; Younis, M. Studies on the effect of salinity, drought stress and soil type on nodule activities of Lablab purpureus (L.) sweet (Kashrangeeg). J. Arid Environ. 2002, 51, 587–602. [Google Scholar] [CrossRef]
- Salzenstein, L. East Africa Deploys Huge Volumes of ‘Highly Hazardous’ Pesticides against Locust Plague. Available online: https://news.mongabay.com/2021/04/east-africa-deploys-huge-volumes-of-highly-hazardous-pesticides-against-locust-plague/#:~:text=Language-,East%20Africa%20deploys%20huge%20volumes%20of,hazardous’%20pesticides%20against%20locust%20plague&text=More%20than%2095%25%20of%20pesticides,such%20as%20birds%20and%20fish (accessed on 12 April 2022).
- Moorman, T. A review of pesticide effects on microorganisms and microbial processes related to soil fertility. J. Prod. Agric. 1989, 2, 14–23. [Google Scholar] [CrossRef]
- Fox, J.E.; Gulledge, J.; Engelhaupt, E.; Burow, M.E.; McLachlan, J.A. Pesticides reduce symbiotic efficiency of nitrogen-fixing rhizobia and host plants. Proc. Natl. Acad. Sci. USA 2007, 104, 10282–10287. [Google Scholar] [CrossRef] [PubMed]
- Poggio, L.; De Sousa, L.M.; Batjes, N.H.; Heuvelink, G.; Kempen, B.; Ribeiro, E.; Rossiter, D. SoilGrids 2.0: Producing soil information for the globe with quantified spatial uncertainty. Soil 2021, 7, 217–240. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Kimiti, J.M.; Ombori, O.; Maingi, J.M.; Njeru, E.M. Genetic characterization and diversity of Rhizobium isolated from root nodules of mid-altitude climbing bean (P. vulagaris L.) varieties. Front. Microbiol. 2018, 9, 968. [Google Scholar] [CrossRef] [PubMed]
- Naik, V.; Rahi, P. Methods for Isolation and Identification of Rhizobia. In Practical Handbook on Agricultural Microbiology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 3–14. [Google Scholar]
- Johnson, G. High throughput DNA extraction of legume root nodules for rhizobial metagenomics. AMB Express 2019, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Iyer, B.; Rajkumar, S. A Metagenomic Approach to Identify Distinct Rhizospheric and Endophytic Bacterial Communities from Roots and Root Nodules of Vigna radiata. In Understanding Host-Microbiome Interactions—An Omics Approach; Springer: Berlin/Heidelberg, Germany, 2017; pp. 173–191. [Google Scholar]
- Jia, R.Z.; Zhang, R.J.; Wei, Q.; Chen, W.F.; Cho, I.K.; Chen, W.X.; Li, Q.X. Identification and classification of rhizobia by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Proteom. Bioinform. 2015, 8, 98. [Google Scholar]
- Somasegaran, P.; Hoben, H.J. Handbook for Rhizobia: Methods in Legume-Rhizobium Technology; Springer Science & Business Media: New York, NY, USA, 2012; pp. 1–443. [Google Scholar]
- Geetha, H. Rhizobium: Isolation and Authentication|Microbial Biotechnology. Available online: https://www.biotechnologynotes.com/microbial-biotechnology/rhizobium-isolation-and-authentication-microbial-biotechnology (accessed on 12 April 2022).
- Hamza, T.A.; Alebejo, A.L. Isolation and characterization of rhizobia from rhizospher and root nodule of cowpea, elephant and lab lab plants. Int. J. Nov. Res. Interdiscip. Stud. 2017, 4, 1–7. [Google Scholar]
- Park, J.-M.; Yang, C.-Y.; Park, H.; Kim, J.-M. Development of a Genus-specific PCR combined with ARDRA for the identification of Leuconostoc species in kimchi. Food Sci. Biotechnol. 2014, 23, 511–516. [Google Scholar] [CrossRef]
- Onyango, B.; Anyango, B.; Nyunja, R.; Koech, P.K.; Skilton, R.A.; Stomeo, F. Morphological, Genetic and Symbiotic Characterization of Root Nodule Bacteria Isolated from Bambara Groundnuts (Vigna subterranea L. Verdc) from Soils of Lake Victoria Basin, Western Kenya. J. Appl. Biol. 2015, 3, 1–10. [Google Scholar]
- Boakye, E.Y.; Lawson, I.Y.D.; Danso, S.K.A. Characterization and diversity of rhizobia nodulating selected tree legumes in Ghana. Symbiosis 2016, 69, 89–99. [Google Scholar] [CrossRef]
- Brusetti, L.; Malkhazova, I.; Gtari, M.; Tamagnini, I.; Borin, S.; Merabishvili, M.; Chanishvili, N.; Mora, D.; Cappitelli, F.; Daffonchio, D. Fluorescent-BOX-PCR for resolving bacterial genetic diversity, endemism and biogeography. BMC Microbiol. 2008, 8, 220. [Google Scholar] [CrossRef]
- Mohammed, M.; Jaiswal, S.K.; Dakora, F.D. Insights into the phylogeny, nodule function, and biogeographic distribution of microsymbionts nodulating the orphan Kersting’s groundnut [Macrotyloma geocarpum (Harms) Marechal & Baudet] in African soils. Appl. Environ. Microbiol. 2019, 85, e00342-19. [Google Scholar] [PubMed]
- Karthik, C.; Oves, M.; Sathya, K.; Sri Ramkumar, V.; Arulselvi, P.I. Isolation and characterization of multi-potential Rhizobium strain ND2 and its plant growth-promoting activities under Cr (VI) stress. Arch. Acker Pflanzenbau Bodenkd. 2017, 63, 1058–1069. [Google Scholar]
- Grönemeyer, J.L.; Kulkarni, A.; Berkelmann, D.; Hurek, T.; Reinhold-Hurek, B. Rhizobia indigenous to the Okavango region in Sub-Saharan Africa: Diversity, adaptations, and host specificity. Appl. Environ. Microbiol. 2014, 80, 7244–7257. [Google Scholar] [CrossRef]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P., Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Evol. Microbiol. 1992, 42, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Pei, A.Y.; Oberdorf, W.E.; Nossa, C.W.; Agarwal, A.; Chokshi, P.; Gerz, E.A.; Jin, Z.; Lee, P.; Yang, L.; Poles, M. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 2010, 76, 3886–3897. [Google Scholar] [CrossRef] [PubMed]
- Maroniche, G.A.; García, J.E.; Salcedo, F.; Creus, C.M. Molecular identification of Azospirillum spp.: Limitations of 16S rRNA and qualities of rpoD as genetic markers. Microbiol. Res. 2017, 195, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Maiden, M.C. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006, 60, 561–588. [Google Scholar] [CrossRef]
- Tounsi-Hammami, S.; Le Roux, C.; Dhane-Fitouri, S.; De Lajudie, P.; Duponnois, R.; Jeddi, F.B. Genetic diversity of rhizobia associated with root nodules of white lupin (Lupinus albus L.) in Tunisian calcareous soils. Syst. Appl. Microbiol. 2019, 42, 448–456. [Google Scholar] [CrossRef]
- Aserse, A.A.; Räsänen, L.A.; Assefa, F.; Hailemariam, A.; Lindström, K. Phylogeny and genetic diversity of native rhizobia nodulating common bean (P. vulagaris L.) in Ethiopia. Syst. Appl. Microbiol. 2012, 35, 120–131. [Google Scholar] [CrossRef]
- Belhadi, D.; De Lajudie, P.; Ramdani, N.; Le Roux, C.; Boulila, F.; Tisseyre, P.; Boulila, A.; Benguedouar, A.; Kaci, Y.; Laguerre, G.V. Vicia faba L. in the Bejaia region of Algeria is nodulated by Rhizobium leguminosarum sv. viciae, Rhizobium laguerreae and two new genospecies. Syst. Appl. Microbiol. 2018, 41, 122–130. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Fraser, C.; Alm, E.J.; Polz, M.F.; Spratt, B.G.; Hanage, W.P. The bacterial species challenge: Making sense of genetic and ecological diversity. Science 2009, 323, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Beukes, C.W.; Steenkamp, E.T.; Van Zyl, E.; Avontuur, J.; Chan, W.Y.; Hassen, A.I.; Palmer, M.; Mthombeni, L.S.; Phalane, F.L.; Sereme, T.K. Paraburkholderia strydomiana sp. nov. and Paraburkholderia steynii sp. nov.: Rhizobial symbionts of the fynbos legume Hypocalyptus sophoroides. Antonie Leeuwenhoek. 2019, 112, 1369–1385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Tian, C.F.; Sui, X.H.; Chen, W.F.; Chen, W.X. Robust markers reflecting phylogeny and taxonomy of rhizobia. PLoS ONE 2012, e44936. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; de Peer, Y.V.; Vandamme, P.; Thompson, F.L. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.-J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Klenk, H.-P.; Göker, M. Taxonomic use of DNA G+ C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef]
- Henz, S.R.; Huson, D.H.; Auch, A.F.; Nieselt-Struwe, K.; Schuster, S.C. Whole-genome prokaryotic phylogeny. Bioinformatics 2005, 21, 2329–2335. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, E.T.; van Zyl, E.; Beukes, C.W.; Avontuur, J.R.; Chan, W.Y.; Palmer, M.; Mthombeni, L.S.; Phalane, F.L.; Sereme, T.K.; Venter, S.N. Burkholderia kirstenboschensis sp. nov. nodulates papilionoid legumes indigenous to South Africa. Syst. Appl. Microbiol. 2015, 38, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Diouf, F.; Diouf, D.; Klonowska, A.; Le Quere, A.; Bakhoum, N.; Fall, D.; Neyra, M.; Parrinello, H.; Diouf, M.; Ndoye, I. Genetic and genomic diversity studies of Acacia symbionts in Senegal reveal new species of Mesorhizobium with a putative geographical pattern. PLoS ONE 2015, 10, e0117667. [Google Scholar] [CrossRef] [PubMed]
- Fossou, R.K.; Pothier, J.F.; Zézé, A.; Perret, X. Bradyrhizobium ivorense sp. nov. as a potential local bioinoculant for Cajanus cajan cultures in Côte d’Ivoire. Int. J. Syst. Evol. Microbiol. 2020, 70, 1421. [Google Scholar] [CrossRef] [PubMed]
- Mavima, L.; Beukes, C.W.; Palmer, M.; De Meyer, S.E.; James, E.K.; Maluk, M.; Muasya, M.A.; Avontuur, J.R.; Chan, W.Y.; Venter, S.N. Delineation of Paraburkholderia tuberum sensu stricto and description of Paraburkholderia podalyriae sp. nov. nodulating the South African legume Podalyria calyptrata. Syst. Appl. Microbiol. 2022, 45, 126316. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, N.S. Metagenome analysis and interpretation. In Data Processing Handbook for Complex Biological Data Sources; Elsevier: Berlin/Heidelberg, Germany, 2019; pp. 139–160. [Google Scholar]
- WHO. The State of Food Security and Nutrition in the World 2020: Transforming Food Systems for Affordable Healthy Diets; Food & Agriculture Organization: Rome, Italy, 2020; Volume 2020. [Google Scholar]
- Koskey, G.; Mburu, S.W.; Awino, R.; Njeru, E.M.; Maingi, J.M. Potential Use of Beneficial Microorganisms for Soil Amelioration, Phytopathogen Biocontrol, and Sustainable Crop Production in Smallholder Agroecosystems. Front. Sustain. Food Syst. 2021, 130, 1–20. [Google Scholar] [CrossRef]
- Gunnabo, A.; Geurts, R.; Wolde-meskel, E.; Degefu, T.; Giller, K.; Van Heerwaarden, J. Genetic interaction studies reveal superior performance of Rhizobium tropici CIAT899 on a range of diverse East African common bean (P. vulagaris L.) genotypes. Appl. Environ. Microbiol. 2019, 85, e01763-19. [Google Scholar] [CrossRef]
- Tairo, E.V.; Ndakidemi, P.A. Possible benefits of rhizobial inoculation and phosphorus supplementation on nutrition, growth and economic sustainability in grain legumes. Am. J. Res. Commun. 2013, 1, 532–556. [Google Scholar]
- Nyoki, D.; Ndakidemi, P.A. Yield response of intercropped soybean and maize under rhizobia (Bradyrhizobium japonicum) inoculation and P and K fertilization. Commun. Soil Sci. Plant Anal. 2018, 49, 1168–1185. [Google Scholar] [CrossRef]
- Del Barrio-Duque, A.; Ley, J.; Samad, A.; Antonielli, L.; Sessitsch, A.; Compant, S. Beneficial endophytic bacteria-Serendipita indica interaction for crop enhancement and resistance to phytopathogens. Front. Microbiol. 2019, 10, 2888. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Lindström, K.; Murwira, M.; Willems, A.; Altier, N. The biodiversity of beneficial microbe-host mutualism: The case of rhizobia. Res. Microbiol. 2010, 161, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Dey, R.; Pal, K.; Bhatt, D.; Chauhan, S. Growth promotion and yield enhancement of peanut (A. hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 2004, 159, 371–394. [Google Scholar] [CrossRef]
- Mathu, S.; Herrmann, L.; Pypers, P.; Matiru, V.; Mwirichia, R.; Lesueur, D. Potential of indigenous bradyrhizobia versus commercial inoculants to improve cowpea (Vigna unguiculata L. walp.) and green gram (Vigna radiata L. wilczek.) yields in Kenya. Soil Sci. Plant Nutr. 2012, 58, 750–763. [Google Scholar] [CrossRef]
- Ouma, E.W.; Asango, A.M.; Maingi, J.; Njeru, E.M. Elucidating the potential of native rhizobial isolates to improve biological nitrogen fixation and growth of common bean and soybean in smallholder farming systems of Kenya. Int. J. Agron. 2016, 2016, 4569241. [Google Scholar] [CrossRef]
- Korir, H.; Mungai, N.W.; Thuita, M.; Hamba, Y.; Masso, C. Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front. Plant Sci. 2017, 8, 141. [Google Scholar] [CrossRef]
- Checcucci, A.; DiCenzo, G.C.; Bazzicalupo, M.; Mengoni, A. Trade, diplomacy, and warfare: The quest for elite rhizobia inoculant strains. Front. Microbiol. 2017, 8, 2207. [Google Scholar] [CrossRef]
- Maingi, J.M.; Shisanya, C.A.; Gitonga, N.M.; Hornetz, B. Nitrogen fixation by common bean (P. vulagaris L.) in pure and mixed stands in semi-arid south-east Kenya. Eur. J. Agron. 2001, 14, 1–12. [Google Scholar] [CrossRef]
- Soumare, A.; Diedhiou, A.G.; Thuita, M.; Hafidi, M.; Ouhdouch, Y.; Gopalakrishnan, S.; Kouisni, L. Exploiting biological nitrogen fixation: A route towards a sustainable agriculture. Plants 2020, 9, 1011. [Google Scholar] [CrossRef]
- Babalola, O.O.; Glick, B.R. Indigenous African agriculture and plant associated microbes: Current practice and future transgenic prospects. J. Sci. Res. Essay. 2012, 7, 2431–2439. [Google Scholar]
- Matiru, V.N.; Dakora, F.D. Potential use of rhizobial bacteria as promoters of plant growth for increased yield in landraces of African cereal crops. Afr. J. Biotechnol. 2004, 3, 1–7. [Google Scholar]
- Jaiswal, S.K.; Mohammed, M.; Ibny, F.Y.; Dakora, F.D. Rhizobia as a source of plant growth-promoting molecules: Potential applications and possible operational mechanisms. Front. Sustain. Food Syst. 2021, 4, 619676. [Google Scholar] [CrossRef]
- Raimi, A.; Adeleke, R.; Roopnarain, A. Soil fertility challenges and Biofertiliser as a viable alternative for increasing smallholder farmer crop productivity in sub-Saharan Africa. Cogent Food Agric. 2017, 3, 1400933. [Google Scholar] [CrossRef]
- Lesueur, D.; Herrmann, L.; Thuita, M.; Atieno, M.; Mutegi, E.; Ndung’u, K.; Faye, A.; Kamaa, M.M.; Pypers, P.; Okalebo, J.R. How commercial rhizobial inoculants can contribute to improved livelihoods of resource poor African farmers. In Proceedings of the International Conference on Nitrogen Fixation, Perth, Australia, 27 November–1 December 2011. [Google Scholar]
- Keya, S.; Imbamba, S. The East AfricanRhizobium MIRCEN. A framework for promoting regionally co-ordinated biological nitrogen fixation. MIRCEN J. Appl. Microbiol. 1986, 2, 237–251. [Google Scholar] [CrossRef]
- Odame, H. Biofertilizer in Kenya: Research, production and extension dilemmas. Biotechnol. Dev. Monit. 1997, 30, 20–23. [Google Scholar]
- Raimi, A.; Roopnarain, A.; Adeleke, R. Biofertilizer production in Africa: Current status, factors impeding adoption and strategies for success. Sci. Afr. 2021, 11, e00694. [Google Scholar] [CrossRef]
- Chianu, J.; Nkonya, E.M.; Mairura, F.; Chianu, J.; Akinnifesi, F. Biological nitrogen fixation and socioeconomic factors for legume production in sub-Saharan Africa: A review. Agron. Sustain. Dev. 2011, 31, 139–154. [Google Scholar] [CrossRef]
- Raimi, A.R.; Ezeokoli, O.T.; Adeleke, R.A. High-throughput sequence analysis of bacterial communities in commercial biofertiliser products marketed in South Africa: An independent snapshot quality assessment. 3 Biotech 2019, 9, 108. [Google Scholar] [CrossRef]
- Bala, A.; Karanja, N.; Murwira, M.; Lwimbi, L.; Abaidoo, R.; Giller, K. Production and Use of Rhizobial Inoculants in Africa; Milestone Report 3.4.1; N2Africa Report: Nairobi, Kenya, 2011; pp. 1–21. [Google Scholar]
- Musyoka, D.M.; Njeru, E.M.; Nyamwange, M.M.e.; Maingi, J.M. Arbuscular mycorrhizal fungi and Bradyrhizobium co-inoculation enhances nitrogen fixation and growth of green grams (Vigna radiata L.) under water stress. J. Plant Nutr. 2020, 43, 1036–1047. [Google Scholar] [CrossRef]
- Gabasawa, A. Prospects for developing effective and competitive native strains of rhizobium inoculants in Nigeria. In Current Microbiological Research in Africa; Springer: Berlin/Heidelberg, Germany, 2020; pp. 223–256. [Google Scholar]
- Ogbuehi, H.C. Effect of nodumax inoculant on morpho-physiological parameters, nutrient content and yield of soybean (Glycine max. L). Agric. Food Sci. 2020, 18, 54–72. [Google Scholar] [CrossRef]
- Abdullahi, A.A.; Howieson, J.; O’Hara, G.; Terpolilli, J.; Tiwari, R.; Yusuf, A.A. History of Rhizobia inoculants use for improving performance of grain legumes based on experience from Nigeria. In Just Enough Nitrogen; Springer: Berlin/Heidelberg, Germany, 2020; pp. 101–113. [Google Scholar]
- Balume, I. Assessment of Quality Control of Inoculants Used on Bean and Soybean in Eastern and Central Africa; University of Nairobi: Nairobi, Kenya, 2013. [Google Scholar]
- Woomer, P.; Huising, J.; Giller, K.E.; Baijukya, F.P.; Kantengwa, S.; Vanlauwe, B.; Boahen, S.; Wolf, J.d.; Franke, L.; Abaidoo, R.C. N2Africa Final Report of the First Phase: 2009–2013; 2014. [Google Scholar]
- Atieno, M.; Herrmann, L.; Okalebo, R.; Lesueur, D. Efficiency of different formulations of Bradyrhizobium japonicum and effect of co-inoculation of Bacillus subtilis with two different strains of Bradyrhizobium japonicum. World J. Microbiol. Biotechnol. 2012, 28, 2541–2550. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, R.A.; Raimi, A.R.; Roopnarain, A.; Mokubedi, S.M. Status and prospects of bacterial inoculants for sustainable management of agroecosystems. In Biofertilizers for Sustainable Agriculture and Environment; Springer: Berlin/Heidelberg, Germany, 2019; pp. 137–172. [Google Scholar]
- Raimi, A. Quality Assessment of Commercial Biofertilisers and the Awareness of Smallholder Farmers in Gauteng Province, South Africa. Master’s Thesis, University of South Africa, Gauteng, South Africa, 2018. [Google Scholar]
- Villegas, M.D.C.; Rome, S.; Mauré, L.; Domergue, O.; Gardan, L.; Bailly, X.; Cleyet-Marel, J.-C.; Brunel, B. Nitrogen-fixing sinorhizobia with Medicago laciniata constitute a novel biovar (bv. medicaginis) of S. meliloti. Syst. Appl. Microbiol. 2006, 29, 526–538. [Google Scholar] [CrossRef]
- Muthini, M.; Awino, R.; Kirui, K.C.; Koech, K.; Jalloh, A.A.; Njeru, E.M. Optimizing Rhizobium-Legume Symbiosis in Smallholder Agroecosystems. In Sustainable Agriculture Reviews 45; Springer: Berlin/Heidelberg, Germany, 2020; pp. 159–177. [Google Scholar]
- Jefwa, J.M.; Pypers, P.; Jemo, M.; Thuita, M.; Mutegi, E.; Laditi, M.; Faye, A.; Kavoo, A.; Munyahali, W.; Herrmann, L. Do commercial biological and chemical products increase crop yields and economic returns under smallholder farmer conditions? In Challenges and Opportunities for Agricultural Intensification of the Humid Highland Systems of Sub-Saharan Africa; Springer: Berlin/Heidelberg, Germany, 2014; pp. 81–96. [Google Scholar]
- Masso, C.; Jefwa, J.; Jemo, M.; Thuita, M.; Tarus, D.; Vanlauwe, B. Impact of inadequate regulatory frameworks on the adoption of bio-fertilizer (eg PGPR) technologies: A case study of sub-Saharan Africa. In Proceedings of the Recent Advances in Biofertilizers and Biofungicides (PGPR) for Sustainable Agriculture. Proceedings of 3rd Asian Conference on Plant Growth-Promoting Rhizobacteria (PGPR) and other Microbials, Manila, Philippines, 21–24 April 2013; pp. 276–286. [Google Scholar]
- Njira, K.O.W. Microbial contributions in alleviating decline in soil fertility. Br. Microbiol. Res. J. 2013, 3, 724. [Google Scholar] [CrossRef]
- Ofori, P. Yield Response of Soybean and Cowpea to Rock Phosphate Fertilizer Blend and Rhizobial Inoculation on Two Benchmark Soils of Northern Ghana; 2016. [Google Scholar]
- Tarus, D.; Watiti, J.; Nang’ayo, F. Effective Regulation of bio-fertilizers and bio-pesticides: A potential avenue to increase agricultural productivity. In Compro II Project Policy Series; 2013. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wekesa, C.; Jalloh, A.A.; Muoma, J.O.; Korir, H.; Omenge, K.M.; Maingi, J.M.; Furch, A.C.U.; Oelmüller, R. Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa. Int. J. Mol. Sci. 2022, 23, 6599. https://doi.org/10.3390/ijms23126599
Wekesa C, Jalloh AA, Muoma JO, Korir H, Omenge KM, Maingi JM, Furch ACU, Oelmüller R. Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa. International Journal of Molecular Sciences. 2022; 23(12):6599. https://doi.org/10.3390/ijms23126599
Chicago/Turabian StyleWekesa, Clabe, Abdul A. Jalloh, John O. Muoma, Hezekiah Korir, Keziah M. Omenge, John M. Maingi, Alexandra C. U. Furch, and Ralf Oelmüller. 2022. "Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa" International Journal of Molecular Sciences 23, no. 12: 6599. https://doi.org/10.3390/ijms23126599
APA StyleWekesa, C., Jalloh, A. A., Muoma, J. O., Korir, H., Omenge, K. M., Maingi, J. M., Furch, A. C. U., & Oelmüller, R. (2022). Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa. International Journal of Molecular Sciences, 23(12), 6599. https://doi.org/10.3390/ijms23126599







