Importance of Tyrosine Phosphorylation in Hormone-Regulated Plant Growth and Development
Abstract
:1. Introduction
2. PTKs and Dual-Specificity PTKs
3. PTP and Dual-Specificity Protein Phosphatases
4. The Role of Tyr Phosphorylation in Plant Growth Development
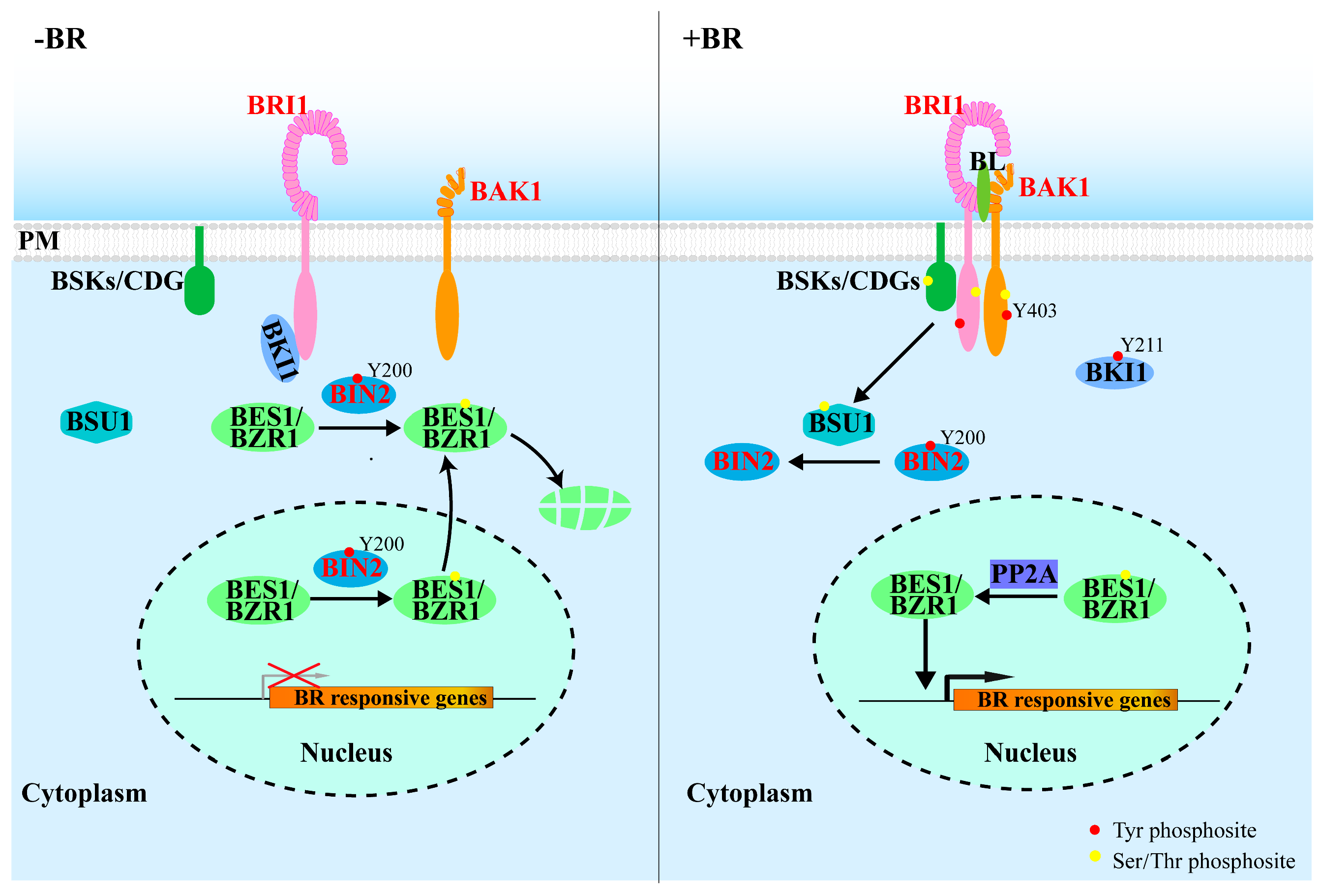
4.1. BR
4.2. GA
4.3. Auxin
4.4. Cytokinin
4.5. Ethylene
4.6. ABA
4.7. Other Plant Growth Processes
5. Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krebs, E.G.; Kent, A.B.; Fischer, E.H. The Muscle Phosphorylase b Kinase reaction. J. Biol. Chem. 1958, 231, 73–83. [Google Scholar] [CrossRef]
- Gnad, F.; Forner, F.; Zielinska, D.F.; Birney, E.; Gunawardena, J.; Mann, M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol. Cell Proteom. 2010, 9, 2642–2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, T. Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Tonks, N.K. The coordinated action of protein tyrosine phosphatases and kinases in cell signaling. Trends Biochem. Sci. 1994, 19, 480–485. [Google Scholar] [CrossRef]
- Nishi, H.; Shaytan, A.; Panchenko, A.R. Physicochemical mechanisms of protein regulation by phosphorylation. Front. Genet. 2014, 5, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offringa, R.; Huang, F. Phosphorylation-dependent Trafficking of Plasma Membrane Proteins in Animal and Plant Cells. J. Integr. Plant Biol. 2013, 55, 789–808. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Rudrabhatla, P.; Reddy, M.M.; Rajasekharan, R. Genome-wide analysis and experimentation of plant serine/ threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 2006, 60, 293–319. [Google Scholar] [CrossRef]
- Shankar, A.; Agrawal, N.; Sharma, M.; Pandey, A.; Girdhar, K.P.M. Role of Protein Tyrosine Phosphatases in Plants. Curr. Genom. 2015, 16, 224–236. [Google Scholar] [CrossRef]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef] [Green Version]
- Ahsan, N.; Wilson, R.S.; Rao, R.S.P.; Salvato, F.; Sabila, M.; Ullah, H.; Miernyk, J.A. Mass Spectrometry-Based Identification of Phospho-Tyr in Plant Proteomics. J. Proteome. Res. 2020, 19, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Wang, X.; Kota, U.; Goshe, M.B.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 658–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perraki, A.; DeFalco, T.A.; Derbyshire, P.; Avila, J.; Sere, D.; Sklenar, J.; Qi, X.; Stransfeld, L.; Schwessinger, B.; Kadota, Y.; et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 2018, 561, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Hothorn, M.; Belkhadir, Y.; Dabi, T.; Nimchuk, Z.L.; Meyerowitz, E.M.; Chory, J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011, 25, 232–237. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef]
- Nemoto, K.; Ramadan, A.; Arimura, G.I.; Imai, K.; Tomii, K.; Shinozaki, K.; Sawasaki, T. Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation. Nat. Commun. 2017, 8, 1004. [Google Scholar] [CrossRef]
- Strader, L.C.; Monroe-Augustus, M.; Bartel, B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant. Biol. 2008, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Monroe-Augustus, M.; Zolman, B.K.; Bartel, B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 2003, 15, 2979–2991. [Google Scholar] [CrossRef] [Green Version]
- Muhlenbeck, H.; Bender, K.W.; Zipfel, C. Importance of tyrosine phosphorylation for transmembrane signaling in plants. Biochem. J. 2021, 478, 2759–2774. [Google Scholar] [CrossRef]
- Ghelis, T. Signal processing by protein tyrosine phosphorylation in plants. Plant Signal. Behav. 2011, 6, 942–951. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Q.L.; Bi, L.; Ren, Y.D.; Song, S.L.; Wang, Q.; Wang, Y.S. Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol. Cancer 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [Green Version]
- Hubbard, S.R.; Till, J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000, 69, 373–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drake, J.M.; Lee, J.K.; Witte, O.N. Clinical Targeting of Mutated and Wild-Type Protein Tyrosine Kinases in Cancer. Mol. Cell. Biol. 2014, 34, 1722–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knosel, T.; Kampmann, E.; Kirchner, T.; Altendorf-Hofmann, A. Tyrosine kinases in soft tissue tumors. Pathologe 2014, 35, 198–201. [Google Scholar]
- Macho, A.P.; Lozano-Duran, R.; Zipfel, C. Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci. 2015, 20, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Elangovan, A.; Dalal, M.; Krishna, G.K.; Devika, S.; Kumar, R.R.; Sathee, L.; Chinnusamy, V. Characterization of Atypical Protein Tyrosine Kinase (PTK) Genes and Their Role in Abiotic Stress Response in Rice. Plants 2020, 9, 664. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, K.; Takemori, N.; Seki, M.; Shinozaki, K.; Sawasaki, T. Members of the Plant CRK Superfamily Are Capable of Trans- and Autophosphorylation of Tyrosine Residues. J. Biol. Chem. 2015, 290, 16665–16677. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.L.; Wang, L.J.; Shigley, C.; Yang, W.T. Protein tyrosine phosphatases in skeletal development and diseases. Bone Res. 2022, 10, 10. [Google Scholar] [CrossRef]
- Hendriks, W.J.A.J.; Elson, A.; Harroch, S.; Stoker, A.W. Protein tyrosine phosphatases: Functional inferences from mouse models and human diseases. Febs J. 2008, 275, 816–830. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.T. Protein phosphatase 1--targeted in many directions. J. Cell Sci. 2002, 115, 241–256. [Google Scholar] [CrossRef]
- Alonso, A.; Pulido, R. The extended human PTPome: A growing tyrosine phosphatase family. Febs J. 2016, 283, 2197–2201. [Google Scholar] [CrossRef]
- Conrad, C.A.; Steck, P.A. Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 1995, 9, 193–204. [Google Scholar]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein Tyrosine Phosphatases in the Human Genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Fu, H.H.; Gupta, R.; Luan, S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 1998, 10, 849–857. [Google Scholar] [CrossRef] [Green Version]
- Kerk, D.; Templeton, G.; Moorhead, G.B. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008, 146, 351–367. [Google Scholar] [CrossRef] [Green Version]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Barsam, B.; Veretnik, S.; Gribskov, M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002, 129, 908–925. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 435. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Ozawa, A.; Nemoto, K.; Nozawa, A.; Seki, M.; Shinozaki, K.; Takeda, H.; Endo, Y.; Sawasaki, T. Genome-wide biochemical analysis of Arabidopsis protein phosphatase using a wheat cell-free system. Febs Lett. 2012, 586, 3134–3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhrig, R.G.; Labandera, A.M.; Muhammad, J.; Samuel, M.; Moorhead, G.B. Rhizobiales-like Phosphatase 2 from Arabidopsis thaliana Is a Novel Phospho-tyrosine-specific Phospho-protein Phosphatase (PPP) Family Protein Phosphatase. J. Biol. Chem. 2016, 291, 5926–5934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, K.; Pan, S. Maize protein phosphatase gene family: Identification and molecular characterization. BMC Genom. 2014, 15, 773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Pandey, G.K. Protein phosphatases: A genomic outlook to understand their function in plants. J. Plant Biochem Biot 2012, 21, S100–S107. [Google Scholar] [CrossRef]
- Mao, J.; Li, J. Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 4340. [Google Scholar] [CrossRef] [PubMed]
- Li, J. Brassinosteroid signaling: From receptor kinases to transcription factors. Curr. Opin. Plant Biol. 2005, 8, 526–531. [Google Scholar] [CrossRef]
- Guo, H.; Li, L.; Aluru, M.; Aluru, S.; Yin, Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 2013, 16, 545–553. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Planas-Riverola, A.; Gupta, A.; Betegon-Putze, I.; Bosch, N.; Ibanes, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.W.; Guan, S.; Burlingame, A.L.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Bojar, D.; Martinez, J.; Santiago, J.; Rybin, V.; Bayliss, R.; Hothorn, M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014, 78, 31–43. [Google Scholar] [CrossRef]
- Wang, Y.L.; Li, Z.C.; Liu, D.; Xu, J.H.; Wei, X.C.; Yan, L.M.; Yang, C.; Lou, Z.Y.; Shui, W.Q. Assessment of BAK1 activity in different plant receptor-like kinase complexes by quantitative profiling of phosphorylation patterns. J. Proteom. 2014, 108, 484–493. [Google Scholar] [CrossRef]
- Macho, A.P.; Schwessinger, B.; Ntoukakis, V.; Brutus, A.; Segonzac, C.; Roy, S.; Kadota, Y.; Oh, M.H.; Sklenar, J.; Derbyshire, P.; et al. A Bacterial Tyrosine Phosphatase Inhibits Plant Pattern Recognition Receptor Activation. Science 2014, 343, 1509–1512. [Google Scholar] [CrossRef]
- Mitra, S.K.; Chen, R.Q.; Dhandaydham, M.; Wang, X.F.; Blackburn, R.K.; Kota, U.; Goshe, M.B.; Schwartz, D.; Huber, S.C.; Clouse, S.D. An autophosphorylation site database for leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. Plant J. 2015, 82, 1042–1060. [Google Scholar] [CrossRef]
- Oh, M.H.; Wang, X.; Xia, W.; Zhao, Y.; Clouse, S.D.; Huber, S.C. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 17827–17832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.; Perraki, A.; Kim, S.Y.; Shrivastava, S.; Lee, J.H.; Zhao, Y.F.; Schwessinger, B.; Oh, M.H.; Marshall-Colon, A.; Zipfel, C.; et al. Tyrosine-610 in the Receptor Kinase BAK1 Does Not Play a Major Role in Brassinosteroid Signaling or Innate Immunity. Front. Plant Sci. 2017, 8, 1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuda, S.; Okada, K.; Saijo, Y. A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr. Opin. Plant Biol. 2017, 38, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Shan, L.; He, P.; de Vries, S.; Kemmerling, B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009, 14, 535–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshammari, S.O.; Dakshanamurthy, S.; Ullah, H. Small compounds targeting tyrosine phosphorylation of Scaffold Protein Receptor for Activated C Kinase1A (RACK1A) regulate auxin mediated lateral root development in Arabidopsis. Plant Signal. Behav. 2021, 16, 1899488. [Google Scholar] [CrossRef]
- Dautel, R.; Wu, X.N.; Heunemann, M.; Schulze, W.X.; Harter, K. The Sensor Histidine Kinases AHK2 and AHK3 Proceed into Multiple Serine/Threonine/Tyrosine Phosphorylation Pathways in Arabidopsis thaliana. Mol. Plant 2016, 9, 182–186. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Yang, D.; Ha, Y.; Lee, J.Y.; Kim, J.Y.; Oh, M.H.; Nam, K.H. Open stomata 1 exhibits dual serine/threonine and tyrosine kinase activity in regulating abscisic acid signaling. J. Exp. Bot. 2021, 72, 5494–5507. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids Regulate Dissociation of BKI1, a Negative Regulator of BRI1 Signaling, from the Plasma Membrane. Sci.ence 2006, 313, 1118–1122. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Hoffmeister, L.; Diekmann, M.; Brand, K.; Huber, R. GSK3: A Kinase Balancing Promotion and Resolution of Inflammation. Cells 2020, 9, 820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, K.; Nikolakaki, E.; Plyte, S.E.; Totty, N.F.; Woodgett, J.R. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 1993, 12, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Dajani, R.; Fraser, E.; Roe, S.M.; Young, N.; Good, V.; Dale, T.C.; Pearl, L.H. Crystal structure of glycogen synthase kinase 3 beta: Structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 2001, 105, 721–732. [Google Scholar] [CrossRef]
- Buescher, J.L.; Phiel, C.J. A Noncatalytic Domain of Glycogen Synthase Kinase-3 (GSK-3) Is Essential for Activity. J. Biol. Chem. 2010, 285, 7957–7963. [Google Scholar] [CrossRef] [Green Version]
- Itoh, K.; Tang, T.L.; Neel, B.G.; Sokol, S.Y. Specific modulation of ectodermal cell fates in Xenopus embryos by glycogen synthase kinase. Development 1995, 121, 3979–3988. [Google Scholar] [CrossRef]
- Maselli, G.A.; Slamovits, C.H.; Bianchi, J.I.; Vilarrasa-Blasi, J.; Cano-Delgado, A.I.; Mora-Garcia, S. Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 2014, 164, 1527–1541. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Genschik, P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 2009, 60, 1085–1092. [Google Scholar] [CrossRef] [Green Version]
- Daviere, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [Green Version]
- Ueguchi-Tanaka, M.; Nakajima, M.; Motoyuki, A.; Matsuoka, M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 2007, 58, 183–198. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Itoh, H.; Gomi, K.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Jeong, D.H.; An, G.; Kitano, H.; Ashikari, M.; et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 2003, 299, 1896–1898. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Cao, D.; Cheng, H.; Wen, Z.; Peng, J. Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 2005, 44, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Gomi, K.; Sasaki, A.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Kitano, H.; Matsuoka, M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004, 37, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Cao, D.; Peng, J. Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein. Planta 2007, 226, 475–483. [Google Scholar] [CrossRef]
- Fu, X.; Richards, D.E.; Ait-Ali, T.; Hynes, L.W.; Ougham, H.; Harberd, P. Gibberellin-Mediated Proteasome-Dependent Degradation of the Barley DELLA Protein SLN1 Repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamins, R.; Scheres, B. Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef]
- Quint, M.; Gray, W.M. Auxin signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Wang, S.; Sritubtim, S.; Chen, J.G.; Ellis, B.E. Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 2009, 57, 975–985. [Google Scholar] [CrossRef]
- Pearson, G.; Robinson, F.; Gibson, T.B.; Xu, B.E.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153. [Google Scholar]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef]
- Bartels, S.; Anderson, J.C.; Gonzalez Besteiro, M.A.; Carreri, A.; Hirt, H.; Buchala, A.; Metraux, J.P.; Peck, S.C.; Ulm, R. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 2009, 21, 2884–2897. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Bucio, J.S.; Raya-Gonzalez, J.; Ravelo-Ortega, G.; Ruiz-Herrera, L.F.; Ramos-Vega, M.; Leon, P.; Lopez-Bucio, J.; Guevara-Garcia, A.A. Mitogen activated protein kinase 6 and MAP kinase phosphatase 1 are involved in the response of Arabidopsis roots to L-glutamate. Plant Mol. Biol. 2018, 96, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Meng, T.; Li, P.; Yu, Y.; Cui, Y.; Wang, Y.; Gong, Q.; Wang, N.N. A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 2011, 157, 2131–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Hwang, I. Cytokinin: Perception, signal transduction, and role in plant growth and development. J. Plant Biol. 2007, 50, 98–108. [Google Scholar] [CrossRef]
- Kroll, C.K.; Brenner, W.G. Cytokinin Signaling Downstream of the His-Asp Phosphorelay Network: Cytokinin-Regulated Genes and Their Functions. Front. Plant Sci. 2020, 11, 604489. [Google Scholar] [CrossRef]
- Li, S.Y.; Zhao, B.R.; Yuan, D.Y.; Duan, M.J.; Qian, Q.; Tang, L.; Wang, B.; Liu, X.Q.; Zhang, J.; Wang, J.; et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 2013, 110, 3167–3172. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Lu, Z.Q.; Shan, J.X.; Ye, W.W.; Lin, H.X. ERECTA1 Acts Upstream of the OsMKKK10-OsMKK4-OsMPK6 Cascade to Control Spikelet Number by Regulating Cytokinin Metabolism in Rice. Plant Cell 2020, 32, tpc.00351.02020. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.Q.; Shi, C.L.; Ye, W.W.; Gao, J.P.; Shan, J.X.; Lin, H.X. GRAIN SIZE AND NUMBER1 Negatively Regulates the OsMKKK10-OsMKK4-OsMPK6 Cascade to Coordinate the Trade-off between Grain Number per Panicle and Grain Size in Rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Mohanta, T.; K.Sinha, A.K. Unraveling the Intricate Nexus of Molecular Mechanisms Governing Rice Root Development: OsMPK3/6 and Auxin-Cytokinin Interplay. PLoS ONE 2015, 10, e0123620. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.R.; Diederich, L.; John, P.C.L. The cytokinin requirement for cell division in cultured Nicotiana plumbaginifolia cells can be satisfied by yeast cdc25 protein tyrosine phosphatase. Implications for mechanisms of cytokinin response and plant development. Plant Physiol. 2005, 137, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, jbc.REV120.010854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, C.L.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.H.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L.; et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Yin, C.C.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant Biol. 2021, 63, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Li, H.; Hutchison, C.E.; Laskey, J.; Kieber, J.J. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2003, 33, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Ouaked, F.; Rozhon, W.; Lecourieux, D.; Hirt, H. A MAPK pathway mediates ethylene signaling in plants. Embo J. 2003, 22, 1282–1288. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Tena, G.; Xiong, Y.; Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 2008, 451, 789–795. [Google Scholar] [CrossRef]
- Zhao, Q.; Guo, H.W. Paradigms and Paradox in the Ethylene Signaling Pathway and Interaction Network. Mol. Plant 2011, 4, 626–634. [Google Scholar] [CrossRef] [Green Version]
- Hahn, A.; Harter, K. Mitogen-Activated Protein Kinase Cascades and Ethylene: Signaling, Biosynthesis, or Both? Plant Physiol. 2009, 149, 1207–1210. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.X.; Lei, L.; Yang, H.L.; Liu, G.Q.; Ren, D.T. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [Green Version]
- An, F.Y.; Zhao, Q.O.; Ji, Y.S.; Li, W.Y.; Jiang, Z.Q.; Yu, X.C.; Zhang, C.; Han, Y.; He, W.R.; Liu, Y.D.; et al. Ethylene-Induced Stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 Is Mediated by Proteasomal Degradation of EIN3 Binding F-Box 1 and 2 That Requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Li, G.J.; Yang, K.Y.; Mao, G.; Wang, R.; Liu, Y.; Zhang, S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.D.; Zhang, S.Q. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, S.W.; Sang, L.N.; Zhao, Z.X.; Deng, Y.; Liu, H.T.; Yu, Y.X.; Liu, J.X. Phosphoproteome analysis reveals the involvement of protein dephosphorylation in ethylene-induced corolla senescence in petunia. Bmc Plant Biol. 2021, 21, 512. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Guo, G.Y.; Zhang, M.Y.; Liu, C.Y.; Hu, Q.; Lam, H.; Cheng, H.; Xue, Y.; Li, J.Y.; Li, N. Stable Isotope Metabolic Labeling-based Quantitative Phosphoproteomic Analysis of Arabidopsis Mutants Reveals Ethylene-regulated Time-dependent Phosphoproteins and Putative Substrates of Constitutive Triple Response 1 Kinase. Mol. Cell. Proteom. 2013, 12, 3559–3582. [Google Scholar] [CrossRef] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef]
- Ma, Y. Regulators of PP2C phosphatase activity function as abscisic acid sensors (vol 324, pg 1064, 2009). Science 2009, 324, 1266. [Google Scholar]
- Rubio, S.; Rodrigues, A.; Saez, A.; Dizon, M.B.; Galle, A.; Kim, T.H.; Santiago, J.; Flexas, J.; Schroeder, J.I.; Rodriguez, P.L. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009, 150, 1345–1355. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.C.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; Van Oosten, M.J.; Zhang, H.M.; Tao, W.A.; Zhu, J.K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Knetsch, M.; Wang, M.; Snaar-Jagalska, B.E.; Heimovaara-Dijkstra, S. Abscisic Acid Induces Mitogen-Activated Protein Kinase Activation in Barley Aleurone Protoplasts. Plant Cell 1996, 8, 1061–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacRobbie, E.A. Evidence for a role for protein tyrosine phosphatase in the control of ion release from the guard cell vacuole in stomatal closure. Proc. Natl. Acad. Sci. USA 2002, 99, 11963–11968. [Google Scholar] [CrossRef] [Green Version]
- Reyes, D.; Rodriguez, D.; Nicolas, G.; Nicolas, C. Evidence of a role for tyrosine dephosphorylation in the control of postgermination arrest of development by abscisic acid in Arabidopsis thaliana L. Planta 2006, 223, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Ghelis, T.; Bolbach, G.; Clodic, G.; Habricot, Y.; Miginiac, E.; Sotta, B.; Jeannette, E. Protein Tyrosine Kinases and Protein Tyrosine Phosphatases Are Involved in Abscisic Acid-Dependent Processes in Arabidopsis Seeds and Suspension Cells. Plant Physiol. 2008, 148, 1668–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quettier, A.L.; Bertrand, C.; Habricot, Y.; Miginiac, E.; Agnes, C.; Jeannette, E.; Maldiney, R. The phs1-3 mutation in a putative dual-specificity protein tyrosine phosphatase gene provokes hypersensitive responses to abscisic acid in Arabidopsis thaliana. Plant J. 2006, 47, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xiang, Y.; He, N.; Liu, X.; Liu, H.; Fang, L.; Zhang, F.; Sun, X.; Zhang, D.; Li, X.; et al. Enhanced Vitamin C Production Mediated by an ABA-Induced PTP-like Nucleotidase Improves Plant Drought Tolerance in Arabidopsis and Maize. Mol. Plant 2020, 13, 760–776. [Google Scholar] [CrossRef]
- Liu, R.; Liu, Y.G.; Ye, N.H.; Zhu, G.H.; Chen, M.X.; Jia, L.G.; Xia, Y.J.; Shi, L.; Jia, W.S.; Zhang, J.H. AtDsPTP1 acts as a negative regulator in osmotic stress signalling during Arabidopsis seed germination and seedling establishment. J. Exp. Bot. 2015, 66, 1339–1353. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Fan, J.Q.; Zhang, Y.; Mu, P.Q.; Wang, P.; Su, J.B.; Lai, H.H.; Li, S.W.; Feng, D.R.; Wang, J.F.; et al. OsPFA-DSP1, a rice protein tyrosine phosphatase, negatively regulates drought stress responses in transgenic tobacco and rice plants. Plant Cell Rep. 2012, 31, 1021–1032. [Google Scholar] [CrossRef]
- Barizza, E.; Lo Schiavo, F.; Terzi, M.; Filippini, F. Evidence suggesting protein tyrosine phosphorylation in plants depends on the developmental conditions. Febs Lett. 1999, 447, 191–194. [Google Scholar] [CrossRef]
- Corellou, F.; Potin, P.; Brownlee, C.; Kloareg, B.; Bouget, F.Y. Inhibition of the establishment of zygotic polarity by protein tyrosine kinase inhibitors leads to an alteration of embryo pattern in Fucus. Dev. Biol. 2000, 219, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Maehama, T.; Dixon, J.E. PTEN: A tumor suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999, 9, 125–128. [Google Scholar] [CrossRef]
- Gupta, R.; Ting, J.T.L.; Sokolov, L.N.; Johnson, S.A.; Luan, S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 2002, 14, 2495–2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, S.; Zhou, L.Z.; Fox, E.; Pao, J.; Sun, W.; Zhou, C.; McCormick, S. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 2011, 68, 1081–1092. [Google Scholar] [CrossRef]
- Kameyama, K.; Kishi, Y.; Yoshimura, M.; Kanzawa, N.; Sameshima, M.; Tsuchiya, T. Tyrosine phosphorylation in plant bending. Nature 2000, 407, 37. [Google Scholar] [CrossRef]
- Aparicio-Fabre, R.; Guillen, G.; Estrada, G.; Olivares-Grajales, J.; Gurrola, G.; Sanchez, F. Profilin tyrosine phosphorylation in poly-L-proline-binding regions inhibits binding to phosphoinositide 3-kinase in Phaseolus vulgaris. Plant J. 2006, 47, 491–500. [Google Scholar] [CrossRef]
- Guillén, G.; Valdés-López, V.; Noguez, R.; Olivares, J.; Rodríguez-Zapata, L.C.; Pérez, H.; Vidali, L.; Villanueva, M.A.; Sánchez, F. Profilin in Phaseolus vulgaris is encoded by two genes (only one expressed in root nodules) but multiple isoforms are generated in vivo by phosphorylation on tyrosine residues. Plant J. 1999, 19, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Yemets, A.; Sheremet, Y.; Vissenberg, K.; Van Orden, J.; Verbelen, J.P.; Blume, Y.B. Effects of tyrosine kinase and phosphatase inhibitors on microtubules in Arabidopsis root cells. Cell Biol. Int. 2008, 32, 630–637. [Google Scholar] [CrossRef]
- Nuhse, T.S.; Bottrill, A.R.; Jones, A.M.; Peck, S.C. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007, 51, 931–940. [Google Scholar] [CrossRef] [Green Version]
- Blume, Y.; Yemets, A.; Sulimenko, V.; Sulimenko, T.; Chan, J.; Lloyd, C.; Draber, P. Tyrosine phosphorylation of plant tubulin. Planta 2008, 229, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Karpov, P.; Raevsky, A.; Korablyov, M.; Blume, Y. Identification of Plant Homologues of Dual Specificity Yak1-Related Kinases. Comput. Biol. J. 2014, 2014, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Furuya, T.; Shinkawa, H.; Kajikawa, M.; Nishihama, R.; Kohchi, T.; Fukuzawa, H.; Tsukaya, H. A plant-specific DYRK kinase DYRKP coordinates cell morphology in Marchantia polymorpha. J. Plant Res. 2021, 134, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-Regulated Plant Growth and Development. Curr. Top. Dev. Biol. 2010, 91, 29–66. [Google Scholar]
- Nito, K.; Wong, C.C.; Yates, J.R., 3rd; Chory, J. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep. 2013, 3, 1970–1979. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.J.; Ntui, V.O.; Zhang, N.S.; Xiong, L.M. Arabidopsis Yak1 protein (AtYak1) is a dual specificity protein kinase. Febs Lett. 2015, 589, 3321–3327. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.Y.; Wu, Y.C.; Pu, H.Y.; Wang, Y.; Jang, G.J.; Wu, S.H. Plant dual-specificity tyrosine phosphorylation-regulated kinase optimizes light-regulated growth and development in Arabidopsis. Plant Cell Environ. 2017, 40, 1735–1747. [Google Scholar] [CrossRef]
- Barrada, A.; Djendli, M.; Desnos, T.; Mercier, R.; Robaglia, C.; Montane, M.H.; Menand, B. A TOR-YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis. Development 2019, 146, dev171298. [Google Scholar] [CrossRef] [Green Version]
- Forzani, C.; Duarte, G.T.; Van Leene, J.; Clement, G.; Huguet, S.; Paysant-Le-Roux, C.; Mercier, R.; De Jaeger, G.; Leprince, A.S.; Meyer, C. Mutations of the AtYAK1 Kinase Suppress TOR Deficiency in Arabidopsis. Cell Rep. 2019, 27, 3696. [Google Scholar] [CrossRef] [Green Version]
- Ryabova, L.A.; Robaglia, C.; Meyer, C. Target of Rapamycin kinase: Central regulatory hub for plant growth and metabolism. J. Exp. Bot. 2019, 70, 2211–2216. [Google Scholar] [CrossRef]
- Frohner, I.E.; Mudrak, I.; Schüchner, S.; Anrather, D.; Hartl, M.; Sontag, J.M.; Sontag, E.; Wadzinski, B.E.; Preglej, T.; Ellmeier, W. PP2AC Phospho-Tyr307 Antibodies Are Not Specific for this Modification but Are Sensitive to Other PP2AC Modifications Including Leu309 Methylation—ScienceDirect. Cell Rep. 2020, 30, 3171–3182.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnabel, J.; Hombach, P.; Waksman, T.; Giuriani, G.; Petersen, J.; Christie, J.M. A chemical genetic approach to engineer phototropin kinases for substrate labeling. J. Biol. Chem. 2018, 293, 5613–5623. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Hormones | Protein | Residue | Auto/Trans- Phosphorylation | Kinase | MS * | Site and p Tyr -Specific Antibody | Site-Directed Mutagenesis | Effects | Reference |
|---|---|---|---|---|---|---|---|---|---|
| BR | BRI1 | Y831 | Auto | BRI1 | No | Yes | Yes | Reduced kinase activity | [12] |
| Y956 | Auto | BRI1 | No | Yes | Yes | Abolished kinase activity | [12] | ||
| Y1072 | Auto | BRI1 | No | Yes | Yes | Abolished kinase activity | [12] | ||
| BKI1 | Y211 | Trans | BRI1 | No | No | Yes | Interaction with BRI1 | [14] | |
| BAK1 | Y403 | Auto | BAK1 | No | Yes | Yes | Immunity response | [13] | |
| Y610 | Auto | BAK1 | No | Yes | Yes | Unclear | [61] | ||
| BIN2 | Y200 | Auto | BIN2 | Yes | Yes | Yes | Reduced kinase activity | [15] | |
| GA | GARU | Y321 | Trans | TAGK2 | No | No | Yes | Interaction with GID1 | [16] |
| Auxin | RACK1A | Y248 | Trans | Unknown | No | No | Yes | Homo- dimerization | [65] |
| Cytokinin | CKX2 | Y213 | Trans | Unknown | Yes | No | No | Unclear | [66] |
| Ethylene | ERF13 | Y16/207 | Trans | CRK3 | No | No | Yes | Unclear | [29] |
| ABA | OST1 | Y163 | Auto and Trans | OST1 and BAK1 | Yes | No | Yes | Reduced kinase activity | [67] |
| Y182 | Auto | OST1 | Yes | Yes | Yes | Reduced kinase activity | [67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, W.; Hu, L.; Ma, Z.; Yang, L.; Li, J. Importance of Tyrosine Phosphorylation in Hormone-Regulated Plant Growth and Development. Int. J. Mol. Sci. 2022, 23, 6603. https://doi.org/10.3390/ijms23126603
Song W, Hu L, Ma Z, Yang L, Li J. Importance of Tyrosine Phosphorylation in Hormone-Regulated Plant Growth and Development. International Journal of Molecular Sciences. 2022; 23(12):6603. https://doi.org/10.3390/ijms23126603
Chicago/Turabian StyleSong, Weimeng, Li Hu, Zhihui Ma, Lei Yang, and Jianming Li. 2022. "Importance of Tyrosine Phosphorylation in Hormone-Regulated Plant Growth and Development" International Journal of Molecular Sciences 23, no. 12: 6603. https://doi.org/10.3390/ijms23126603
APA StyleSong, W., Hu, L., Ma, Z., Yang, L., & Li, J. (2022). Importance of Tyrosine Phosphorylation in Hormone-Regulated Plant Growth and Development. International Journal of Molecular Sciences, 23(12), 6603. https://doi.org/10.3390/ijms23126603






