Coronin1C Is a GDP-Specific Rab44 Effector That Controls Osteoclast Formation by Regulating Cell Motility in Macrophages
Abstract
:1. Introduction
2. Results
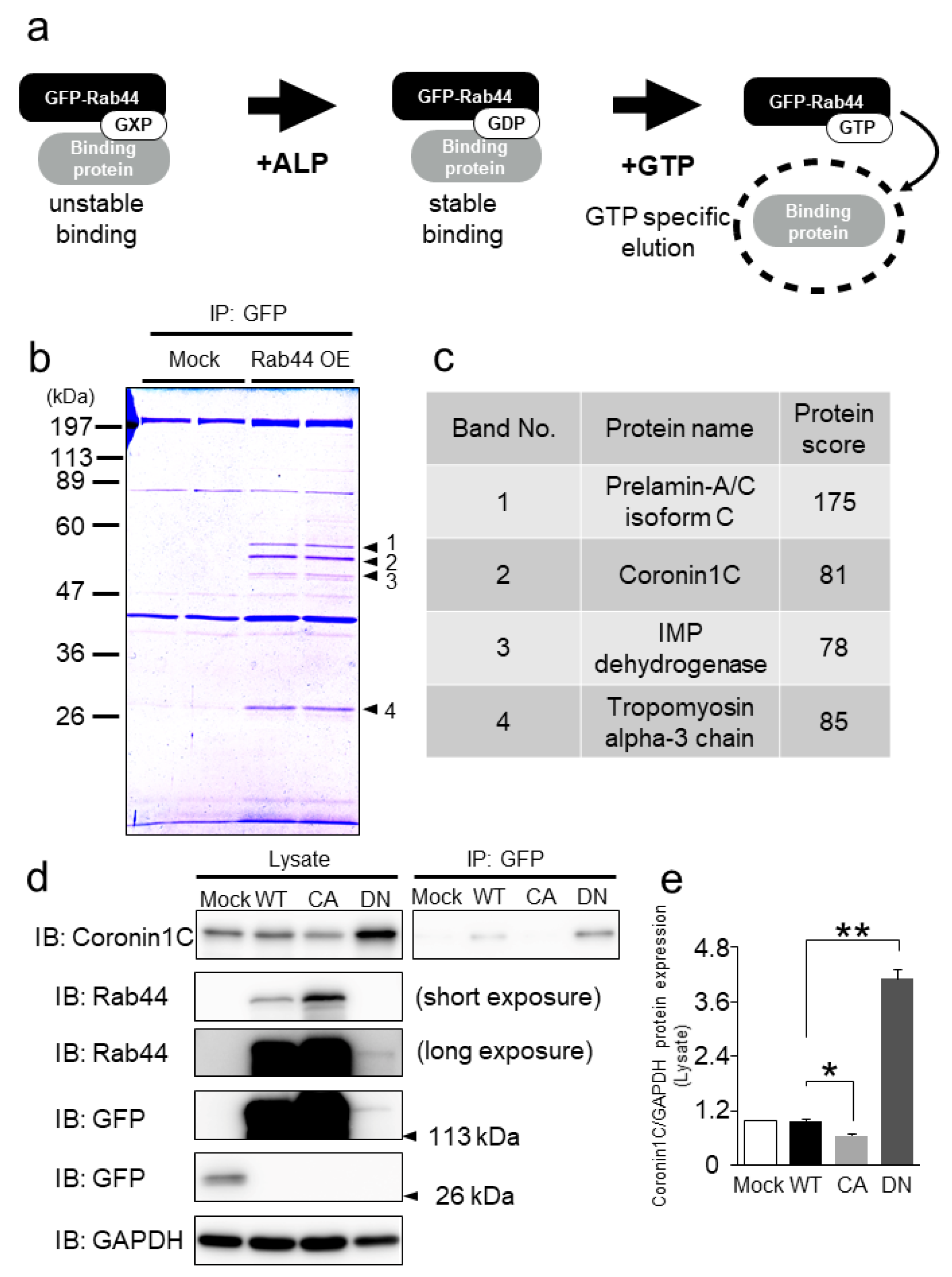
2.1. Rab44 Interacts with the Actin-Binding Protein, Coronin1C, in RAW-D Macrophages
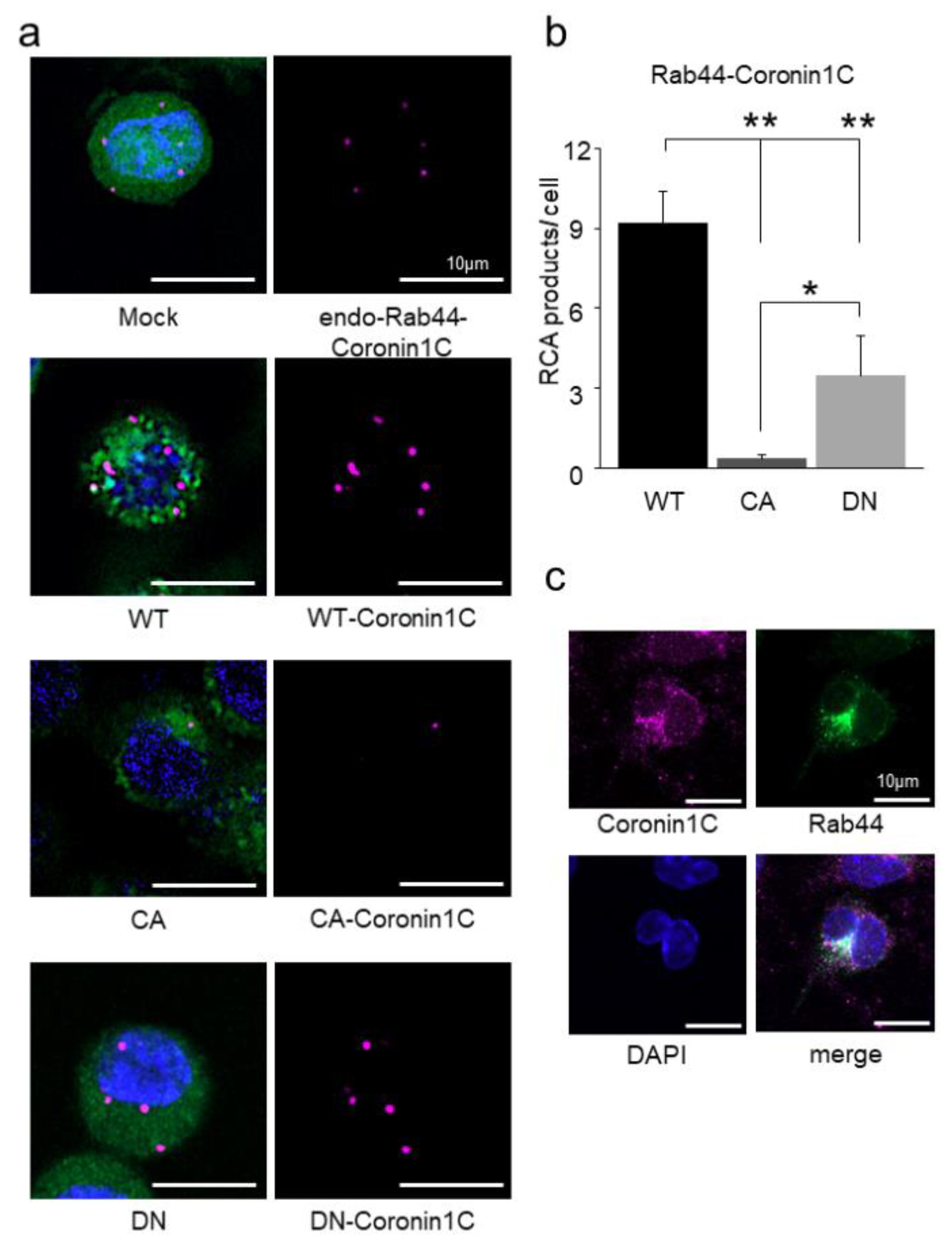
2.2. Coronin1C Exclusively Binds to the GDP form of Rab44
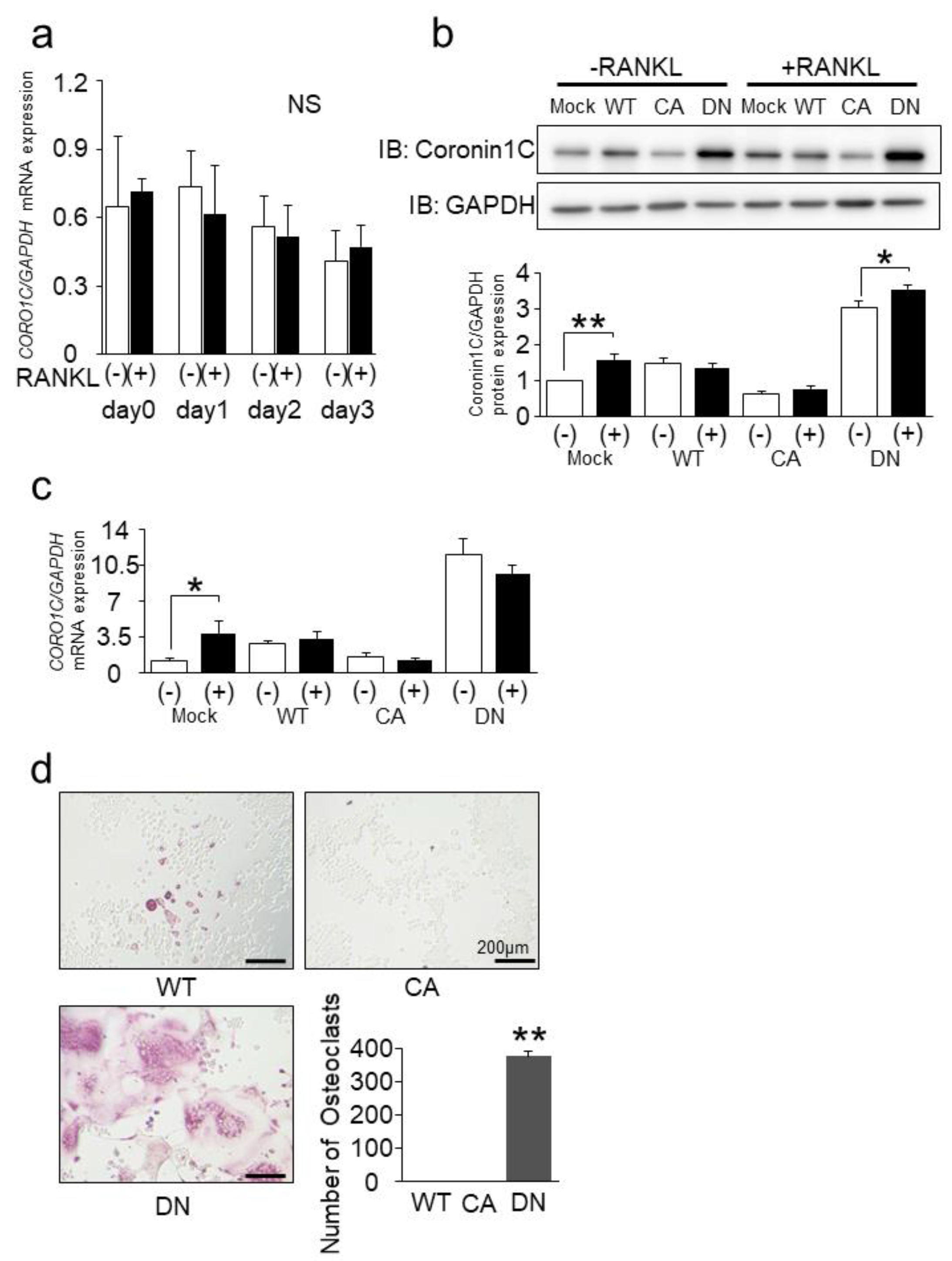
2.3. Rab44 Constitutively Active (CA) Mutant Inhibits Osteoclast Differentiation, Whereas Dominant-Negative (DN) Mutant Enhances This Differentiation
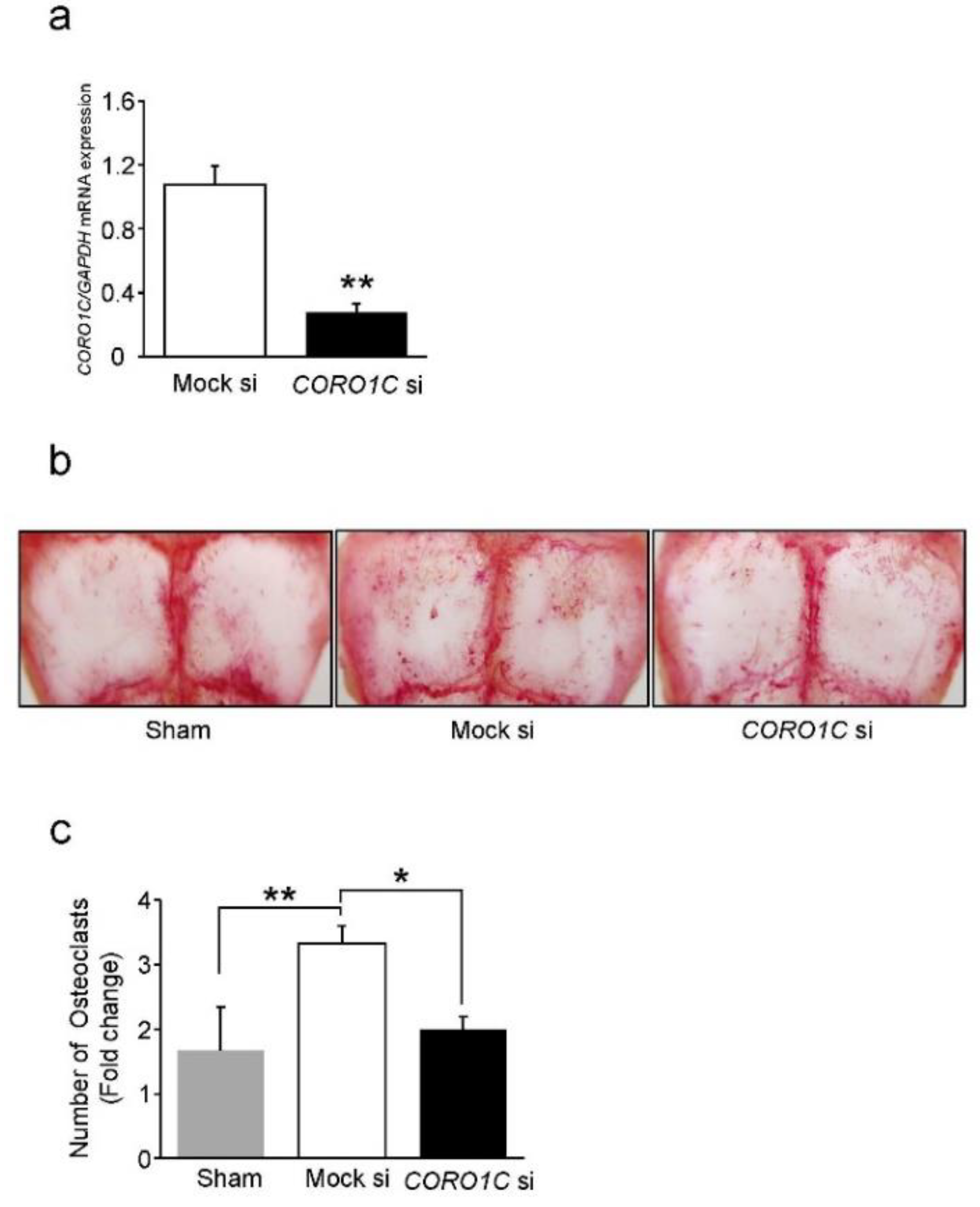
2.4. Knockdown of Coronin1C Inhibits Osteoclast Differentiation through Osteoclast Formation and Fusion
2.5. Coronin1C Knockdown Impairs the Migration and Chemotaxis of RAW-D Macrophages
2.6. Coronin1C Knockdown Suppresses Osteoclastogenesis In Vivo
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Culture
4.3. Retrovirus Construction and Expression of a Constitutively Active (CA) and a Dominant-Negative (DN) Mutants of Mouse Rab44 in RAW-D Cells
4.4. GTP-Specific Pull-Down Assay
4.5. Western Blotting Analysis
4.6. Matrix Associated Laser Desorption/Ionization (MALDI)-Time of Flight Mass Spectrometry (TOF-MS)
4.7. Proximity Ligation Assay (PLA)
4.8. Quantitative Real-Time Polymerase Chain Reaction (QPCR) Analysis
4.9. Immunocytochemistry
4.10. Small Interfering RNA (siRNA)
4.11. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
4.12. Chemotaxis Assay
4.13. Live Cell Imaging
4.14. In Vivo Experiments with Mouse Calvaria
4.15. Animal Experiments and Care
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.; et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Lacombe, J.; Karsenty, G.; Ferron, M. Regulation of lysosome biogenesis and functions in osteoclasts. Cell Cycle 2013, 12, 2744–2752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oikawa, T.; Kuroda, Y.; Matsuo, K. Regulation of osteoclasts by membrane-derived lipid mediators. Cell. Mol. Life Sci. 2013, 70, 3341–3353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okusha, Y.; Tran, M.T.; Itagaki, M.; Sogawa, C.; Eguchi, T.; Okui, T.; Kadowaki, T.; Sakai, E.; Tsukuba, T.; Okamoto, K. Rab11A Functions as a Negative Regulator of Osteoclastogenesis through Dictating Lysosome-Induced Proteolysis of c-fms and RANK Surface Receptors. Cells 2020, 9, 2384. [Google Scholar] [CrossRef]
- Tran, M.T.; Okusha, Y.; Feng, Y.; Morimatsu, M.; Wei, P.; Sogawa, C.; Eguchi, T.; Kadowaki, T.; Sakai, E.; Okamura, H.; et al. The Inhibitory Role of Rab11b in Osteoclastogenesis through Triggering Lysosome-Induced Degradation of c-Fms and RANK Surface Receptors. Int. J. Mol. Sci. 2020, 21, 9352. [Google Scholar] [CrossRef]
- Tran, M.T.; Okusha, Y.; Feng, Y.; Sogawa, C.; Eguchi, T.; Kadowaki, T.; Sakai, E.; Tsukuba, T.; Okamoto, K. A novel role of HSP90 in regulating osteoclastogenesis by abrogating Rab11b-driven transport. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119096. [Google Scholar] [CrossRef]
- Shimada-Sugawara, M.; Sakai, E.; Okamoto, K.; Fukuda, M.; Izumi, T.; Yoshida, N.; Tsukuba, T. Rab27A regulates transport of cell surface receptors modulating multinucleation and lysosome-related organelles in osteoclasts. Sci. Rep. 2015, 5, 9620. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Sakai, E.; Okamoto, K.; Kajiya, H.; Okabe, K.; Naito, M.; Kadowaki, T.; Tsukuba, T. Rab44, a novel large Rab GTPase, negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation. Cell. Mol. Life Sci. 2018, 75, 33–48. [Google Scholar] [CrossRef]
- Tsukuba, T.; Yamaguchi, Y.; Kadowaki, T. Large Rab GTPases: Novel Membrane Trafficking Regulators with a Calcium Sensor and Functional Domains. Int. J. Mol. Sci. 2021, 22, 7691. [Google Scholar] [CrossRef]
- Chan, K.T.; Creed, S.J.; Bear, J.E. Unraveling the enigma: Progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 2011, 21, 481–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Shang, Y.; Ren, G.; Zhou, L.; Feng, B.; Li, K.; Deng, L.; Liang, J.; Lu, Y.; Wang, X. Coronin3 regulates gastric cancer invasion and metastasis by interacting with Arp2. Cancer Biol. Ther. 2014, 15, 1163–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, R.C.; Cowell, C.A.M.; Reville, T.; Roper, J.A.; Rendall, T.C.S.; Bass, M.D. Coronin-1C Protein and Caveolin Protein Provide Constitutive and Inducible Mechanisms of Rac1 Protein Trafficking. J. Biol. Chem. 2015, 290, 15437–15449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.T.; Roadcap, D.W.; Holoweckyj, N.; Bear, J.E. Coronin 1C harbours a second actin-binding site that confers co-operative binding to F-actin. Biochem. J. 2012, 444, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Ren, G.; Tian, Q.; An, Y.; Feng, B.; Lu, Y.; Liang, J.; Li, K.; Shang, Y.; Nie, Y.; Wang, X.; et al. Coronin 3 promotes gastric cancer metastasis via the up-regulation of MMP-9 and cathepsin K. Mol. Cancer 2012, 11, 67. [Google Scholar] [CrossRef] [Green Version]
- Thal, D.; Xavier, C.P.; Rosentreter, A.; Linder, S.; Friedrichs, B.; Waha, A.; Pietsch, T.; Stumpf, M.; Noegel, A.; Clemen, C. Expression of coronin-3 (coronin-1C) in diffuse gliomas is related to malignancy. J. Pathol. 2008, 214, 415–424. [Google Scholar] [CrossRef]
- Koch, D.; Rai, A.; Ali, I.; Bleimling, N.; Friese, T.; Brockmeyer, A.; Janning, P.; Goud, B.; Itzen, A.; Müller, M.P.; et al. A pull-down procedure for the identification of unknown GEFs for small GTPases. Small GTPases 2016, 7, 93–106. [Google Scholar] [CrossRef] [Green Version]
- Kadowaki, T.; Yamaguchi, Y.; Ogawa, K.; Tokuhisa, M.; Okamoto, K.; Tsukuba, T. Rab44 isoforms similarly promote lysosomal exocytosis, but exhibit differential localization in mast cells. FEBS Open Bio 2021, 11, 1165–1185. [Google Scholar] [CrossRef]
- Kimura, T.; Kaneko, Y.; Yamada, S.; Ishihara, H.; Senda, T.; Iwamatsu, A.; Niki, I. The GDP-dependent Rab27a effector coronin 3 controls endocytosis of secretory membrane in insulin-secreting cell lines. J. Cell Sci. 2008, 121, 3092–3098. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, M.; Ishizaki, T.; Kimura, T. GTP- and GDP-Dependent Rab27a Effectors in Pancreatic Beta-Cells. Biol. Pharm. Bull. 2015, 38, 663–668. [Google Scholar] [CrossRef] [Green Version]
- Kimura, T.; Yamaoka, M.; Taniguchi, S.; Okamoto, M.; Takei, M.; Ando, T.; Iwamatsu, A.; Watanabe, T.; Kaibuchi, K.; Ishizaki, T.; et al. Activated Cdc42-bound IQGAP1 determines the cellular endocytic site. Mol. Cell. Biol. 2013, 33, 4834–4843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, K.; Ramalho, J.S.; Izumi, T. Exophilin8 transiently clusters insulin granules at the actin-rich cell cortex prior to exocytosis. Mol. Biol. Cell 2011, 22, 1716–1726. [Google Scholar] [CrossRef] [PubMed]
- Brozzi, F.; Diraison, F.; Lajus, S.; Rajatileka, S.; Philips, T.; Regazzi, R.; Fukuda, M.; Verkade, P.; Molnár, E.; Váradi, A. Molecular mechanism of myosin Va recruitment to dense core secretory granules. Traffic 2012, 13, 54–69. [Google Scholar] [CrossRef]
- Wang, H.; Ishizaki, R.; Xu, J.; Kasai, K.; Kobayashi, E.; Gomi, H.; Izumi, T. The Rab27a effector exophilin7 promotes fusion of secretory granules that have not been docked to the plasma membrane. Mol. Biol. Cell 2013, 24, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Ohmae, S.; Noma, N.; Toyomoto, M.; Shinohara, M.; Takeiri, M.; Fuji, H.; Takemoto, K.; Iwaisako, K.; Fujita, T.; Takeda, N.; et al. Actin-binding protein coronin 1A controls osteoclastic bone resorption by regulating lysosomal secretion of cathepsin K. Sci. Rep. 2017, 7, 41710. [Google Scholar] [CrossRef] [Green Version]
- Behrens, J.; Solga, R.; Ziemann, A.; Rastetter, R.H.; Berwanger, C.; Herrmann, H.; Noegel, A.A.; Clemen, C.S. Coronin 1C-free primary mouse fibroblasts exhibit robust rearrangements in the orientation of actin filaments, microtubules and intermediate filaments. Eur. J. Cell Biol. 2016, 95, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Wang, X.; Wu, Z.; Tan, S.; Zhu, T.; Ding, K. CORO1C expression is associated with poor survival rates in gastric cancer and promotes metastasis in vitro. FEBS Open Bio 2019, 9, 1097–1108. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Jia, L.; Sun, Y.; Li, C.; Zhang, L.; Wang, X.; Chen, H. CORO1C is Associated With Poor Prognosis and Promotes Metastasis Through PI3K/AKT Pathway in Colorectal Cancer. Front. Mol. Biosci. 2021, 8, 682594. [Google Scholar] [CrossRef]
- Tagliatela, A.C.; Hempstead, S.C.; Hibshman, P.S.; Hockenberry, M.A.; Brighton, H.E.; Pecot, C.V.; Bear, J.E. Coronin 1C inhibits melanoma metastasis through regulation of MT1-MMP-containing extracellular vesicle secretion. Sci. Rep. 2020, 10, 11958. [Google Scholar] [CrossRef]
- Lim, J.P.; Shyamasundar, S.; Gunaratne, J.; Scully, O.J.; Matsumoto, K.; Bay, B.H. YBX1 gene silencing inhibits migratory and invasive potential via CORO1C in breast cancer in vitro. BMC Cancer 2017, 17, 201. [Google Scholar] [CrossRef] [Green Version]
- Shao, J.; Zhang, H.; Wang, Z. Coronin 1c and F-actin Promote Metastasis of Breast Cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 5980–5987. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; Jia, M.K.; Cao, H.; Bian, P.; Fang, X.D. Knockdown of Coronin-1C disrupts Rac1 activation and impairs tumorigenic potential in hepatocellular carcinoma cells. Oncol. Rep. 2013, 29, 1066–1072. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Li, Q.; Yu, S.; Zhang, Z.; Qiu, P.; Zhang, Y.; Yang, W.; Xu, G.; Xu, T. Coronin 3 Promotes the Development of Oncogenic Properties in Glioma Through the Wnt/β-Catenin Signaling Pathway. Onco Targets Ther. 2020, 13, 6661–6673. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Yamaguchi, Y.; Kido, M.A.; Abe, T.; Ogawa, K.; Tokuhisa, M.; Gao, W.; Okamoto, K.; Kiyonari, H.; Tsukuba, T. The large GTPase Rab44 regulates granule exocytosis in mast cells and IgE-mediated anaphylaxis. Cell. Mol. Immunol. 2020, 17, 1287–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narahara, H.; Sakai, E.; Yamaguchi, Y.; Narahara, S.; Iwatake, M.; Okamoto, K.; Yoshida, N.; Tsukuba, T. Actin binding LIM 1 (abLIM1) negatively controls osteoclastogenesis by regulating cell migration and fusion. J. Cell. Physiol. 2018, 234, 486–499. [Google Scholar] [CrossRef] [Green Version]
- Kukita, T.; Wada, N.; Kukita, A.; Kakimoto, T.; Sandra, F.; Toh, K.; Nagata, K.; Iijima, T.; Horiuchi, M.; Matsusaki, H.; et al. RANKL-induced DC-STAMP is essential for osteoclastogenesis. J. Exp. Med. 2004, 200, 941–946. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Kukita, T.; Kukita, A.; Wada, N.; Toh, K.; Nagata, K.; Nomiyama, H.; Iijima, T. Direct stimulation of osteoclastogenesis by MIP-1alpha: Evidence obtained from studies using RAW264 cell clone highly responsive to RANKL. J. Endocrinol. 2004, 180, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Sakai, E.; Aoki, Y.; Yoshimatsu, M.; Nishishita, K.; Iwatake, M.; Fukuma, Y.; Okamoto, K.; Tanaka, T.; Tsukuba, T. Sanguiin H-6, a constituent of Rubus parvifolius L., inhibits receptor activator of nuclear factor-κB ligand-induced osteoclastogenesis and bone resorption in vitro and prevents tumor necrosis factor-α-induced osteoclast formation in vivo. Phytomedicine Int. J. Phytother. Phytopharm. 2016, 23, 828–837. [Google Scholar] [CrossRef]
- Tokuhisa, M.; Kadowaki, T.; Ogawa, K.; Yamaguchi, Y.; Kido, M.A.; Gao, W.; Umeda, M.; Tsukuba, T. Expression and localisation of Rab44 in immune-related cells change during cell differentiation and stimulation. Sci. Rep. 2020, 10, 10728. [Google Scholar] [CrossRef]
- Ogawa, K.; Kadowaki, T.; Tokuhisa, M.; Yamaguchi, Y.; Umeda, M.; Tsukuba, T. Role of the EF-hand and coiled-coil domains of human Rab44 in localisation and organelle formation. Sci. Rep. 2020, 10, 19149. [Google Scholar] [CrossRef]
- Narahara, S.; Sakai, E.; Kadowaki, T.; Yamaguchi, Y.; Narahara, H.; Okamoto, K.; Asahina, I.; Tsukuba, T. KBTBD11, a novel BTB-Kelch protein, is a negative regulator of osteoclastogenesis through controlling Cullin3-mediated ubiquitination of NFATc1. Sci. Rep. 2019, 9, 3523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagawa, M.; Tsukuba, T.; Nishioku, T.; Okamoto, Y.; Okamoto, K.; Takii, R.; Terada, Y.; Nakayama, K.I.; Kadowaki, T.; Yamamoto, K. Cathepsin E deficiency induces a novel form of lysosomal storage disorder showing the accumulation of lysosomal membrane sialoglycoproteins and the elevation of lysosomal pH in macrophages. J. Biol. Chem. 2007, 282, 1851–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamaguchi, Y.; Kadowaki, T.; Aibara, N.; Ohyama, K.; Okamoto, K.; Sakai, E.; Tsukuba, T. Coronin1C Is a GDP-Specific Rab44 Effector That Controls Osteoclast Formation by Regulating Cell Motility in Macrophages. Int. J. Mol. Sci. 2022, 23, 6619. https://doi.org/10.3390/ijms23126619
Yamaguchi Y, Kadowaki T, Aibara N, Ohyama K, Okamoto K, Sakai E, Tsukuba T. Coronin1C Is a GDP-Specific Rab44 Effector That Controls Osteoclast Formation by Regulating Cell Motility in Macrophages. International Journal of Molecular Sciences. 2022; 23(12):6619. https://doi.org/10.3390/ijms23126619
Chicago/Turabian StyleYamaguchi, Yu, Tomoko Kadowaki, Nozomi Aibara, Kaname Ohyama, Kuniaki Okamoto, Eiko Sakai, and Takayuki Tsukuba. 2022. "Coronin1C Is a GDP-Specific Rab44 Effector That Controls Osteoclast Formation by Regulating Cell Motility in Macrophages" International Journal of Molecular Sciences 23, no. 12: 6619. https://doi.org/10.3390/ijms23126619
APA StyleYamaguchi, Y., Kadowaki, T., Aibara, N., Ohyama, K., Okamoto, K., Sakai, E., & Tsukuba, T. (2022). Coronin1C Is a GDP-Specific Rab44 Effector That Controls Osteoclast Formation by Regulating Cell Motility in Macrophages. International Journal of Molecular Sciences, 23(12), 6619. https://doi.org/10.3390/ijms23126619






