Overexpression of Neurogenin 1 Negatively Regulates Osteoclast and Osteoblast Differentiation
Abstract
1. Introduction
2. Results
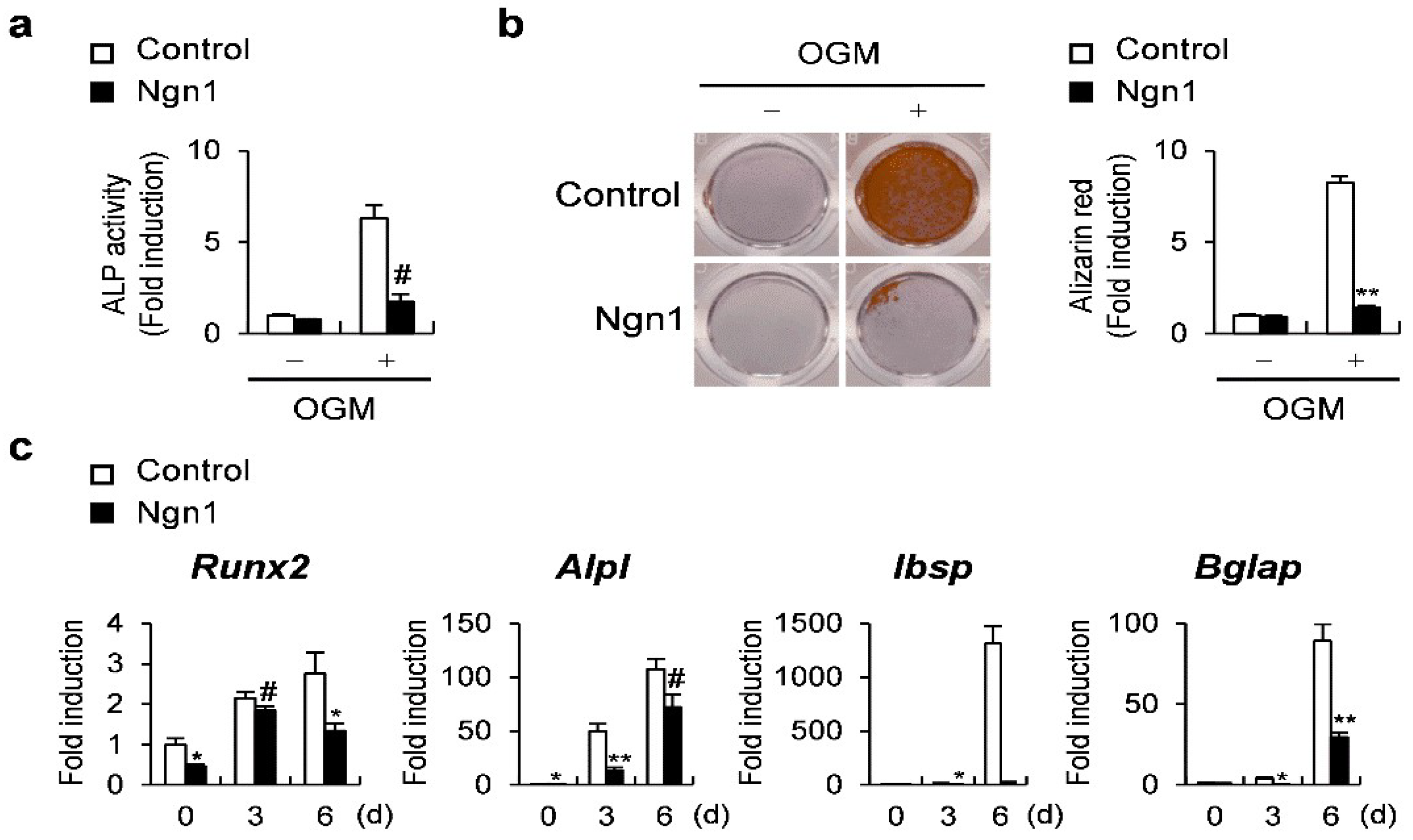
2.1. Ngn1 Inhibited Osteoclast and Osteoblast Differentiation
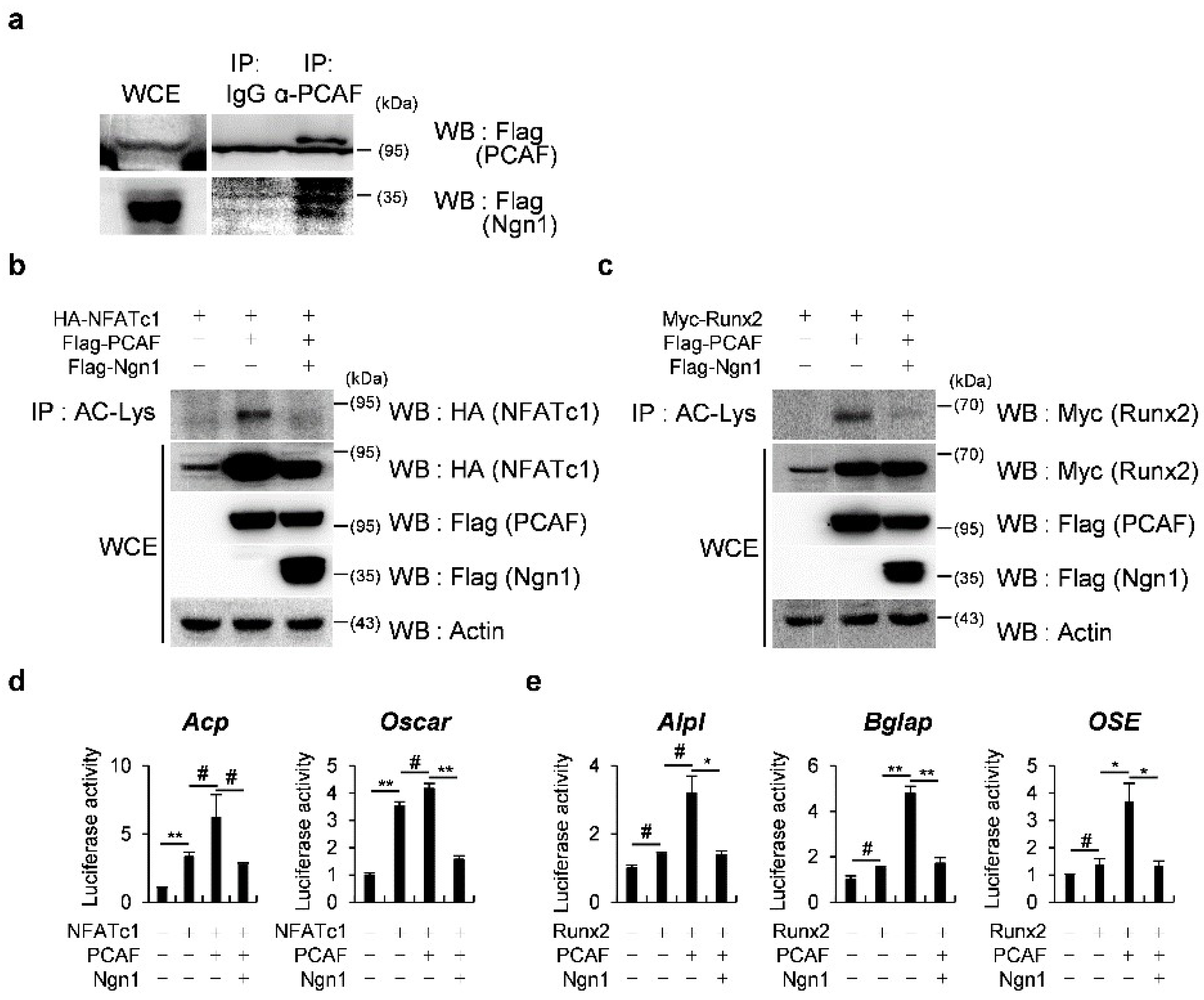
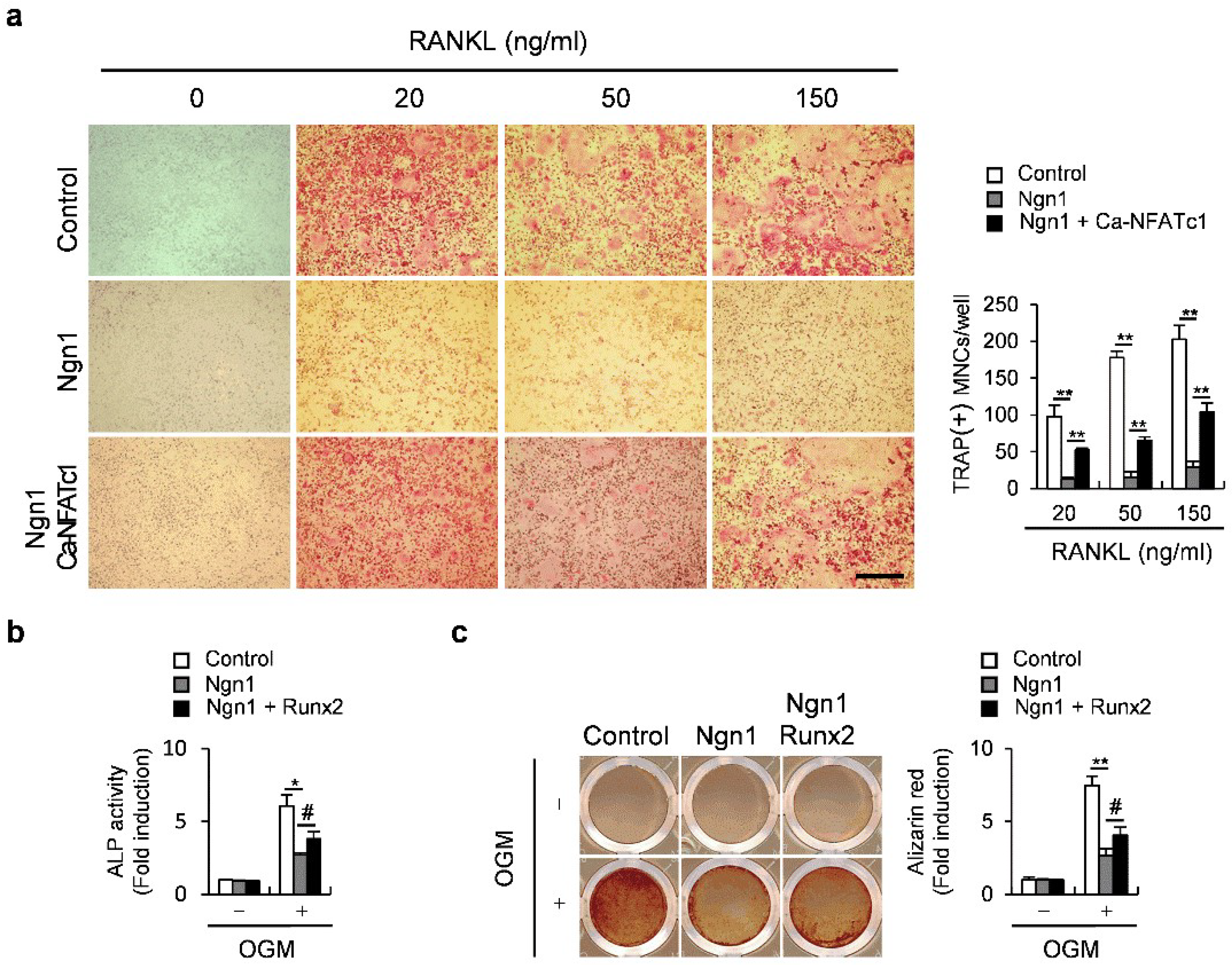
2.2. Ngn1 Inhibited NFATc1 and Runx2 Transcriptional Activities through Interaction with PCAF
2.3. Ngn1 Inhibited the Osteoclast and Osteoblast Differentiation Mediated by Inflammatory Cytokines
3. Discussion
4. Materials and Methods
4.1. Osteoclast Differentiation
4.2. Osteoblast Differentiation
4.3. Retroviral Transduction
4.4. Quantitative Reverse-Transcription PCR (qRT-PCR)
4.5. Luciferase Reporter Assay
4.6. Immunoprecipitation
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Inoue, K.; Ng, C.; Xia, Y.; Zhao, B. Regulation of Osteoclastogenesis and Bone Resorption by miRNAs. Front. Cell Dev. Biol. 2021, 9, 651161. [Google Scholar] [CrossRef] [PubMed]
- Schett, G.; Gravallese, E. Bone erosion in rheumatoid arthritis: Mechanisms, diagnosis and treatment. Nat. Rev. Rheumatol. 2012, 8, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Goldring, S.R.; Purdue, P.E.; Crotti, T.N.; Shen, Z.; Flannery, M.R.; Binder, N.B.; Ross, F.P.; McHugh, K.P. Bone remodelling in inflammatory arthritis. Ann. Rheum. Dis. 2013, 72 (Suppl. S2), ii52–ii55. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, N. Bone Cell Communication Factors Provide a New Therapeutic Strategy for Osteoporosis. Chonnam. Med. J. 2020, 56, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Koh, J.M. Coupling factors involved in preserving bone balance. Cell Mol. Life Sci. 2019, 76, 1243–1253. [Google Scholar] [CrossRef]
- Kong, Y.Y.; Yoshida, H.; Sarosi, I.; Tan, H.L.; Timms, E.; Capparelli, C.; Morony, S.; Oliveira-dos-Santos, A.J.; Van, G.; Itie, A.; et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999, 397, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Onal, M.; Jilka, R.L.; Weinstein, R.S.; Manolagas, S.C.; O’Brien, C.A. Matrix-embedded cells control osteoclast formation. Nat. Med. 2011, 17, 1235–1241. [Google Scholar] [CrossRef]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Kim, W.J.; Shin, H.L.; Kim, B.S.; Kim, H.J.; Ryoo, H.M. RUNX2-modifying enzymes: Therapeutic targets for bone diseases. Exp. Mol. Med. 2020, 52, 1178–1184. [Google Scholar] [CrossRef]
- Narayanan, A.; Srinaath, N.; Rohini, M.; Selvamurugan, N. Regulation of Runx2 by MicroRNAs in osteoblast differentiation. Life Sci. 2019, 232, 116676. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, M.; Hamade, E.; Badran, B.; Buchet, R.; Magne, D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J. Stem. Cells 2013, 5, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.H.; O’Shea, K.S.; Wys, N.L.; Velkey, J.M.; Prieskorn, D.M.; Wesolowski, K.; Miller, J.M.; Altschuler, R.A. Glutamatergic neuronal differentiation of mouse embryonic stem cells after transient expression of neurogenin 1 and treatment with BDNF and GDNF: In vitro and in vivo studies. J. Neurosci. 2008, 28, 12622–12631. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.J.; Liu, W.; Fritzsch, B.; Bianchi, L.M.; Reichardt, L.F.; Xiang, M. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development 2001, 128, 2421–2432. [Google Scholar] [CrossRef]
- Karis, A.; Pata, I.; van Doorninck, J.H.; Grosveld, F.; de Zeeuw, C.I.; de Caprona, D.; Fritzsch, B. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J. Comp. Neurol. 2001, 429, 615–630. [Google Scholar] [CrossRef]
- Fritzsch, B. Development of inner ear afferent connections: Forming primary neurons and connecting them to the developing sensory epithelia. Brain Res. Bull. 2003, 60, 423–433. [Google Scholar] [CrossRef][Green Version]
- Abeynayake, N.; Arthur, A.; Gronthos, S. Crosstalk between skeletal and neural tissues is critical for skeletal health. Bone 2021, 142, 115645. [Google Scholar] [CrossRef]
- Liu, Q.; Lei, L.; Yu, T.; Jiang, T.; Kang, Y. Effect of Brain-Derived Neurotrophic Factor on the Neurogenesis and Osteogenesis in Bone Engineering. Tissue Eng. Part A 2018, 24, 1283–1292. [Google Scholar] [CrossRef]
- Ramnemark, A.; Nyberg, L.; Lorentzon, R.; Englund, U.; Gustafson, Y. Progressive hemiosteoporosis on the paretic side and increased bone mineral density in the nonparetic arm the first year after severe stroke. Osteoporos. Int. 1999, 9, 269–275. [Google Scholar] [CrossRef]
- Hayashi, M.; Nakashima, T.; Taniguchi, M.; Kodama, T.; Kumanogoh, A.; Takayanagi, H. Osteoprotection by semaphorin 3A. Nature 2012, 485, 69–74. [Google Scholar] [CrossRef]
- Unger, R.E.; Dohle, E.; Kirkpatrick, C.J. Improving vascularization of engineered bone through the generation of pro-angiogenic effects in co-culture systems. Adv. Drug Deliv. Rev. 2015, 94, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Luxey, M.; Jungas, T.; Laussu, J.; Audouard, C.; Garces, A.; Davy, A. Eph:ephrin-B1 forward signaling controls fasciculation of sensory and motor axons. Dev. Biol. 2013, 383, 264–274. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eberhart, J.; Barr, J.; O’Connell, S.; Flagg, A.; Swartz, M.E.; Cramer, K.S.; Tosney, K.W.; Pasquale, E.B.; Krull, C.E. Ephrin-A5 exerts positive or inhibitory effects on distinct subsets of EphA4-positive motor neurons. J. Neurosci. 2004, 24, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kokabu, S.; Enoki, Y.; Hayashi, N.; Matsumoto, M.; Nakahira, M.; Sugasawa, M.; Yoda, T. Functional Roles of Netrin-1 in Osteoblast Differentiation. In Vivo 2017, 31, 321–328. [Google Scholar] [CrossRef]
- Zhu, S.; Zhu, J.; Zhen, G.; Hu, Y.; An, S.; Li, Y.; Zheng, Q.; Chen, Z.; Yang, Y.; Wan, M.; et al. Subchondral bone osteoclasts induce sensory innervation and osteoarthritis pain. J. Clin. Investig. 2019, 129, 1076–1093. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Koppel, A.M.; Luo, Y.; Raper, J.A. A role for collapsin-1 in olfactory and cranial sensory axon guidance. J. Neurosci. 1997, 17, 8339–8352. [Google Scholar] [CrossRef]
- Xu, R.; Yallowitz, A.; Qin, A.; Wu, Z.; Shin, D.Y.; Kim, J.M.; Debnath, S.; Ji, G.; Bostrom, M.P.; Yang, X.; et al. Targeting skeletal endothelium to ameliorate bone loss. Nat. Med. 2018, 24, 823–833. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Jacobs, J.R.; Goodman, C.S.; Artavanis-Tsakonas, S. Slit: An extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev. 1990, 4, 2169–2187. [Google Scholar] [CrossRef]
- Zhong, J.; Wang, Z.; Yuan, W.; Shen, Y.; Chen, L. Interleukin-17 promotes osteoclastogenesis and periodontal damage via autophagy in vitro and in vivo. Int. Immunopharmacol. 2022, 107, 108631. [Google Scholar] [CrossRef]
- Yao, Z.; Getting, S.J.; Locke, I.C. Regulation of TNF-Induced Osteoclast Differentiation. Cells 2021, 11, 132. [Google Scholar] [CrossRef]
- Yi, S.J.; Lee, H.; Lee, J.; Lee, K.; Kim, J.; Kim, Y.; Park, J.I.; Kim, K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. Int. J. Mol. Sci. 2019, 20, 3147. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yao, W.; Shao, Y.; Zheng, R.; Huang, F. PCAF fine-tunes hepatic metabolic syndrome, inflammatory disease, and cancer. J. Cell Mol. Med. 2018, 22, 5787–5800. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H. RANKL as the master regulator of osteoclast differentiation. J. Bone Miner. Metab. 2021, 39, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Khotib, J.; Gani, M.A.; Budiatin, A.S.; Lestari, M.; Rahadiansyah, E.; Ardianto, C. Signaling Pathway and Transcriptional Regulation in Osteoblasts during Bone Healing: Direct Involvement of Hydroxyapatite as a Biomaterial. Pharmaceuticals 2021, 14, 615. [Google Scholar] [CrossRef]
- Gomathi, K.; Akshaya, N.; Srinaath, N.; Moorthi, A.; Selvamurugan, N. Regulation of Runx2 by post-translational modifications in osteoblast differentiation. Life Sci. 2020, 245, 117389. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, N. Regulation of NFATc1 in Osteoclast Differentiation. J. Bone Metab. 2014, 21, 233–241. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Youn, B.U.; Jin, H.M.; Kim, J.Y.; Moon, J.B.; Ko, A.; Seo, S.B.; Lee, K.Y.; Kim, N. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem. J. 2011, 436, 253–262. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Jin, H.M.; Song, I.; Youn, B.U.; Lee, S.H.; Choi, Y.; Kim, N. Negative feedback control of osteoclast formation through ubiquitin-mediated down-regulation of NFATc1. J. Biol. Chem. 2010, 285, 5224–5231. [Google Scholar] [CrossRef]
- Wang, C.Y.; Yang, S.F.; Wang, Z.; Tan, J.M.; Xing, S.M.; Chen, D.C.; Xu, S.M.; Yuan, W. PCAF acetylates Runx2 and promotes osteoblast differentiation. J. Bone Miner. Metab. 2013, 31, 381–389. [Google Scholar] [CrossRef]
- Sun, Y.; Nadal-Vicens, M.; Misono, S.; Lin, M.Z.; Zubiaga, A.; Hua, X.; Fan, G.; Greenberg, M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 2001, 104, 365–376. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, Y.; Zhang, S.; Xu, J.; Zou, J. Osteoimmunological insights into the pathogenesis of ankylosing spondylitis. J. Cell Physiol. 2021, 236, 6090–6100. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Sieper, J. Ankylosing spondylitis. Lancet 2007, 369, 1379–1390. [Google Scholar] [CrossRef]
- Penttinen, M.A.; Heiskanen, K.M.; Mohapatra, R.; DeLay, M.L.; Colbert, R.A.; Sistonen, L.; Granfors, K. Enhanced intracellular replication of Salmonella enteritidis in HLA-B27-expressing human monocytic cells: Dependency on glutamic acid at position 45 in the B pocket of HLA-B27. Arthritis Rheum. 2004, 50, 2255–2263. [Google Scholar] [CrossRef] [PubMed]
- Alnaeeli, M.; Penninger, J.M.; Teng, Y.T. Immune interactions with CD4+ T cells promote the development of functional osteoclasts from murine CD11c+ dendritic cells. J. Immunol. 2006, 177, 3314–3326. [Google Scholar] [CrossRef] [PubMed]
- Dakwar, E.; Reddy, J.; Vale, F.L.; Uribe, J.S. A review of the pathogenesis of ankylosing spondylitis. Neurosurg. Focus 2008, 24, e2. [Google Scholar] [CrossRef]
- Klingberg, E.; Lorentzon, M.; Göthlin, J.; Mellström, D.; Geijer, M.; Ohlsson, C.; Atkinson, E.J.; Khosla, S.; Carlsten, H.; Forsblad-d’Elia, H. Bone microarchitecture in ankylosing spondylitis and the association with bone mineral density, fractures, and syndesmophytes. Arthritis Res. Ther. 2013, 15, R179. [Google Scholar] [CrossRef]
- Wang, C.M.; Tsai, S.C.; Lin, J.C.; Wu, Y.J.; Wu, J.; Chen, J.Y. Association of Genetic Variants of RANK, RANKL, and OPG with Ankylosing Spondylitis Clinical Features in Taiwanese. Mediat. Inflamm. 2019, 2019, 8029863. [Google Scholar] [CrossRef]
- Sieper, J.; Appel, H.; Braun, J.; Rudwaleit, M. Critical appraisal of assessment of structural damage in ankylosing spondylitis: Implications for treatment outcomes. Arthritis Rheum. 2008, 58, 649–656. [Google Scholar] [CrossRef]
- Zheng, G.; Xie, Z.; Wang, P.; Li, J.; Li, M.; Cen, S.; Tang, S.; Liu, W.; Ye, G.; Li, Y.; et al. Enhanced osteogenic differentiation of mesenchymal stem cells in ankylosing spondylitis: A study based on a three-dimensional biomimetic environment. Cell Death Dis. 2019, 10, 350. [Google Scholar] [CrossRef]
- Jo, S.; Wang, S.E.; Lee, Y.L.; Kang, S.; Lee, B.; Han, J.; Sung, I.H.; Park, Y.S.; Bae, S.C.; Kim, T.H. IL-17A induces osteoblast differentiation by activating JAK2/STAT3 in ankylosing spondylitis. Arthritis Res. Ther. 2018, 20, 115. [Google Scholar] [CrossRef]
- Van der Heijde, D.; Landewé, R.; Einstein, S.; Ory, P.; Vosse, D.; Ni, L.; Lin, S.L.; Tsuji, W.; Davis, J.C., Jr. Radiographic progression of ankylosing spondylitis after up to two years of treatment with etanercept. Arthritis Rheum. 2008, 58, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Molnar, C.; Scherer, A.; Baraliakos, X.; de Hooge, M.; Micheroli, R.; Exer, P.; Kissling, R.O.; Tamborrini, G.; Wildi, L.M.; Nissen, M.J.; et al. TNF blockers inhibit spinal radiographic progression in ankylosing spondylitis by reducing disease activity: Results from the Swiss Clinical Quality Management cohort. Ann. Rheum. Dis. 2018, 77, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Baeten, D.; Baraliakos, X.; Braun, J.; Sieper, J.; Emery, P.; van der Heijde, D.; McInnes, I.; van Laar, J.M.; Landewé, R.; Wordsworth, P.; et al. Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: A randomised, double-blind, placebo-controlled trial. Lancet 2013, 382, 1705–1713. [Google Scholar] [CrossRef]
- Braun, J.; Baraliakos, X.; Deodhar, A.; Baeten, D.; Sieper, J.; Emery, P.; Readie, A.; Martin, R.; Mpofu, S.; Richards, H.B. Effect of secukinumab on clinical and radiographic outcomes in ankylosing spondylitis: 2-year results from the randomised phase III MEASURE 1 study. Ann. Rheum. Dis. 2017, 76, 1070–1077. [Google Scholar] [CrossRef]
- Yan, R.T.; He, L.; Zhan, W.; Wang, S.Z. Induction of ectopic retina-like tissue by transgenic expression of neurogenin. PLoS ONE 2015, 10, e0116171. [Google Scholar] [CrossRef][Green Version]
- Song, Z.; Jadali, A.; Fritzsch, B.; Kwan, K.Y. NEUROG1 Regulates CDK2 to Promote Proliferation in Otic Progenitors. Stem. Cell Rep. 2017, 9, 1516–1529. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Kim, K.; Kim, I.; Seong, S.; Koh, J.-T.; Kim, N. Overexpression of Neurogenin 1 Negatively Regulates Osteoclast and Osteoblast Differentiation. Int. J. Mol. Sci. 2022, 23, 6708. https://doi.org/10.3390/ijms23126708
Kim JH, Kim K, Kim I, Seong S, Koh J-T, Kim N. Overexpression of Neurogenin 1 Negatively Regulates Osteoclast and Osteoblast Differentiation. International Journal of Molecular Sciences. 2022; 23(12):6708. https://doi.org/10.3390/ijms23126708
Chicago/Turabian StyleKim, Jung Ha, Kabsun Kim, Inyoung Kim, Semun Seong, Jeong-Tae Koh, and Nacksung Kim. 2022. "Overexpression of Neurogenin 1 Negatively Regulates Osteoclast and Osteoblast Differentiation" International Journal of Molecular Sciences 23, no. 12: 6708. https://doi.org/10.3390/ijms23126708
APA StyleKim, J. H., Kim, K., Kim, I., Seong, S., Koh, J.-T., & Kim, N. (2022). Overexpression of Neurogenin 1 Negatively Regulates Osteoclast and Osteoblast Differentiation. International Journal of Molecular Sciences, 23(12), 6708. https://doi.org/10.3390/ijms23126708





