Silencing the Tlr4 Gene Alleviates Methamphetamine-Induced Hepatotoxicity by Inhibiting Lipopolysaccharide-Mediated Inflammation in Mice
Abstract
:1. Introduction
2. Results
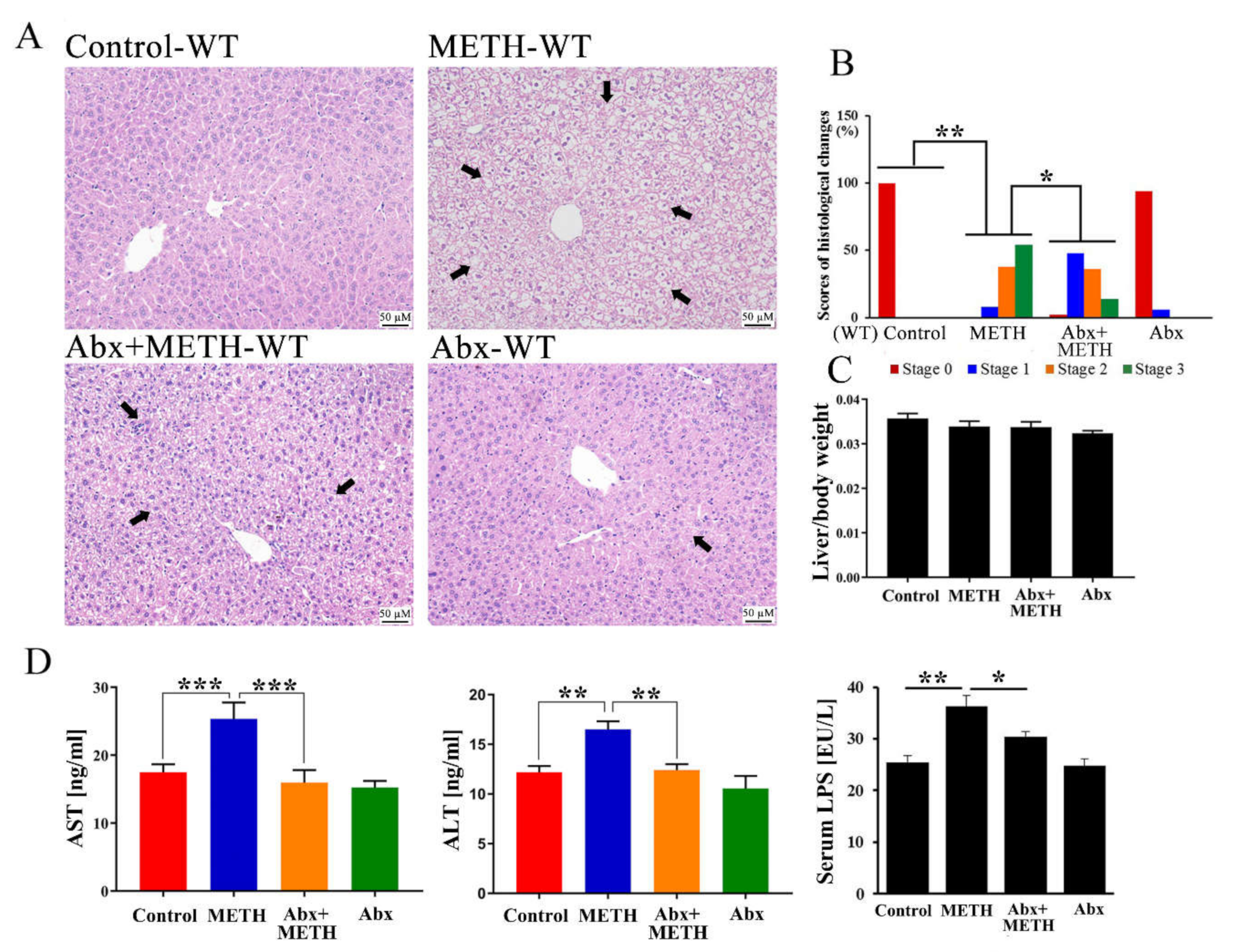
2.1. METH Induced Hepatotoxicity in Wild-Type Mice, Which Was Attenuated by Antibiotic Pretreatment
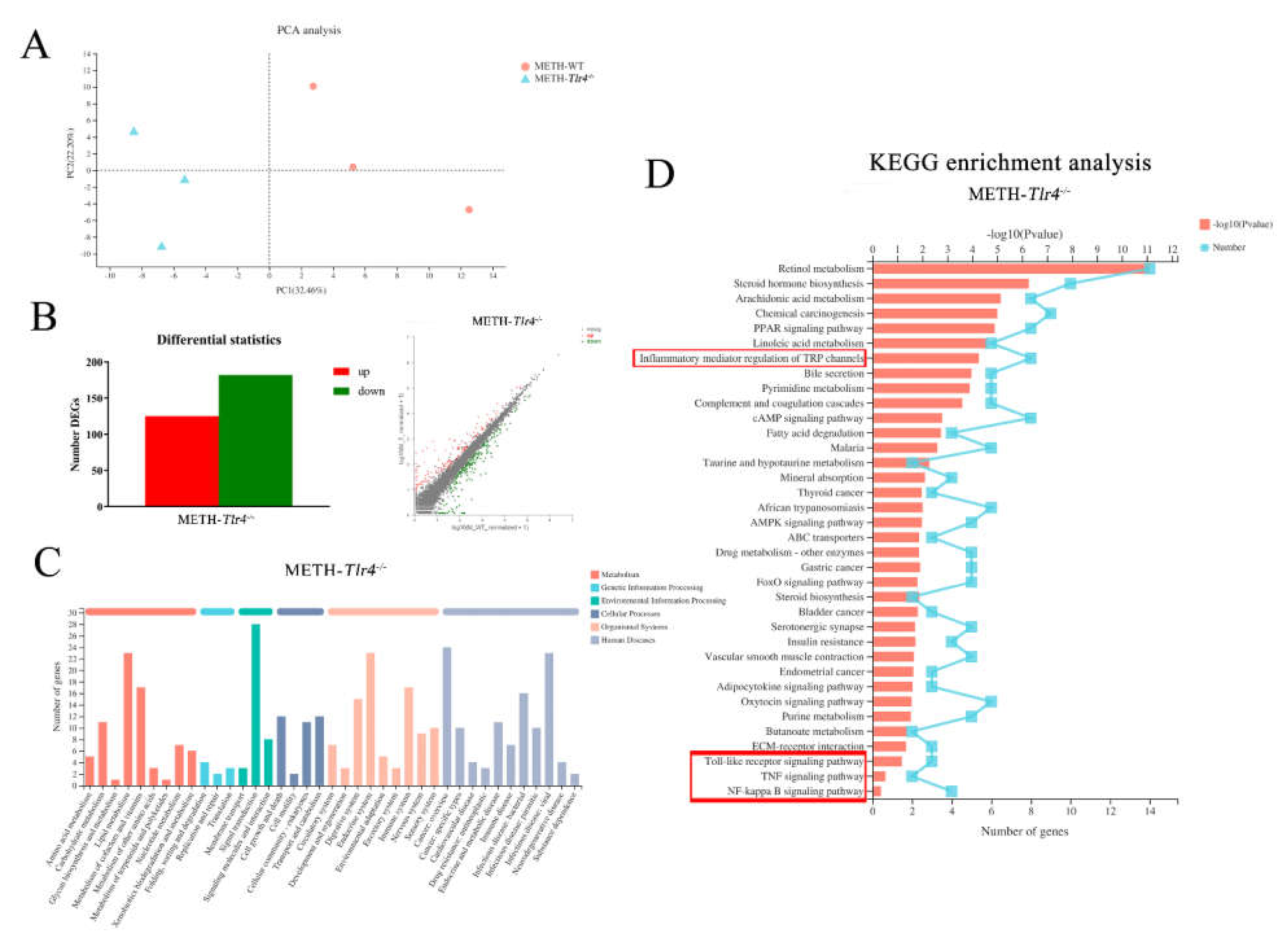
2.2. Inflammatory Pathways Were Enriched in the Livers of METH-Treated Wild-Type Mice
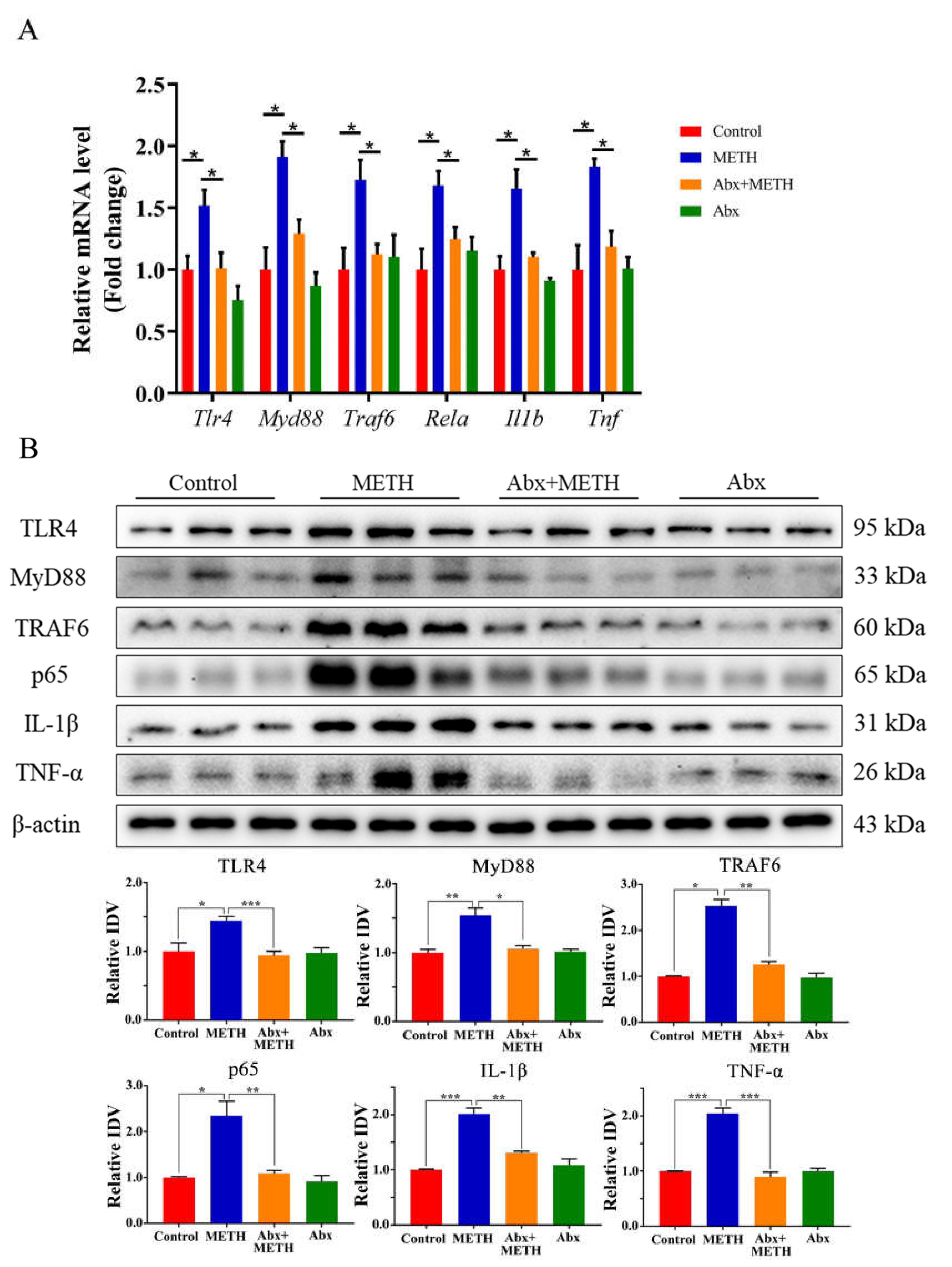
2.3. METH Treatment Aroused Liver Inflammation by Activating the TLR4 Pathway, Which Was Suppressed by Antibiotic Pretreatment
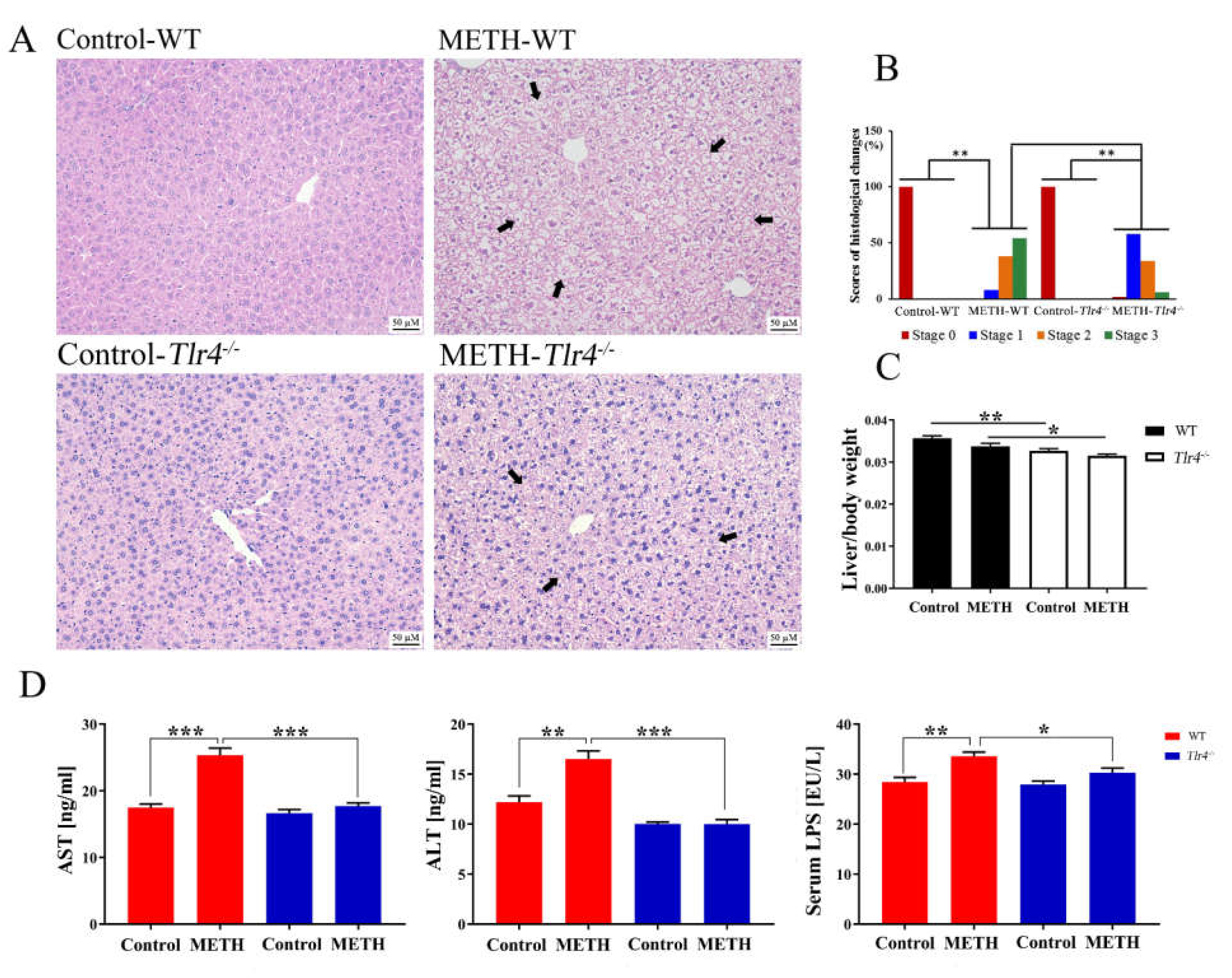
2.4. Silencing of the Tlr4 Gene in Mice Significantly Ameliorated METH-Induced Liver Injury
2.5. Enriched Inflammatory Pathways in METH-Treated Wild-Type Mice Were Regulated by Silencing Tlr4 in Mice
3. Discussion
4. Materials and Methods
4.1. Experimental Main Drugs
4.2. Animals and Treatments
4.3. Measurement of Serological Indicators
4.4. Observation of Histopathological Changes
4.5. Quantitative RT-PCR (RT-qPCR)
4.6. Western Blotting Analysis
4.7. RNA-Sequencing (RNA-Seq) and Bioinformatics Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feinberg, J. Tackle the epidemic, not the opioids. Nature 2019, 573, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, G.; Ying, L.; Ye, L.; Zhao, J.; Liu, N.; Li, J.; Liu, Y.; Zhu, M.; Wu, Y.; Xiao, B.; et al. Dopamine D1 and D2 Receptors Differentially Regulate Rac1 and Cdc42 Signaling in the Nucleus Accumbens to Modulate Behavioral and Structural Plasticity After Repeated Methamphetamine Treatment. Biol. Psychiatry 2019, 86, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Vincent, B. The multi-faceted impact of methamphetamine on Alzheimer’s disease: From a triggering role to a possible therapeutic use. Ageing Res. Rev. 2020, 60, 101062. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, R.T.; Hedlin, H.; Greuenwald, P.; Wilson, D.M.; Segal, J.I.; Jorden, M.; Kudelko, K.; Liu, J.; Hsi, A.; Rupp, A.; et al. Features and Outcomes of Methamphetamine-associated Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2018, 197, 788–800. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, L.W.; Xiao, H.Q.; Xue, Y.; Du, S.H.; Liu, Y.G.; Xie, X.L. Methamphetamine induces hepatotoxicity via inhibiting cell division, arresting cell cycle and activating apoptosis: In vivo and in vitro studies. Food Chem. Toxicol. 2017, 105, 61–72. [Google Scholar] [CrossRef]
- Halpin, L.E.; Gunning, W.T.; Yamamoto, B.K. Methamphetamine causes acute hyperthermia-dependent liver damage. Pharmacol. Res. Perspect. 2013, 1, e00008. [Google Scholar] [CrossRef]
- Soo, J.Y.; Wiese, M.D.; Dyson, R.M.; Gray, C.L.; Clarkson, A.N.; Morrison, J.L.; Berry, M.J. Methamphetamine administration increases hepatic CYP1A2 but not CYP3A activity in female guinea pigs. PLoS ONE 2020, 15, e0233010. [Google Scholar] [CrossRef]
- Zhang, K.K.; Wang, H.; Qu, D.; Chen, L.J.; Wang, L.B.; Li, J.H.; Liu, J.L.; Xu, L.L.; Yoshida, J.S.; Xu, J.T.; et al. Luteolin Alleviates Methamphetamine-Induced Hepatotoxicity by Suppressing the p53 Pathway-Mediated Apoptosis, Autophagy, and Inflammation in Rats. Front. Pharmacol. 2021, 12, 641917. [Google Scholar] [CrossRef]
- Dobosz, E.; Wadowska, M.; Kaminska, M.; Wilamowski, M.; Honarpisheh, M.; Bryzek, D.; Potempa, J.; Jura, J.; Lech, M.; Koziel, J. MCPIP-1 Restricts Inflammation via Promoting Apoptosis of Neutrophils. Front. Immunol. 2021, 12, 627922. [Google Scholar] [CrossRef]
- De Carvalho, T.G.; Garcia, V.B.; de Araújo, A.A.; da Silva Gasparotto, L.H.; Silva, H.; Guerra, G.C.B.; de Castro Miguel, E.; de Carvalho Leitão, R.F.; da Silva Costa, D.V.; Cruz, L.J.; et al. Spherical neutral gold nanoparticles improve anti-inflammatory response, oxidative stress and fibrosis in alcohol-methamphetamine-induced liver injury in rats. Int. J. Pharm. 2018, 548, 1–14. [Google Scholar] [CrossRef]
- Mashayekhi, V.; Eskandari, M.R.; Kobarfard, F.; Khajeamiri, A.; Hosseini, M.J. Induction of mitochondrial permeability transition (MPT) pore opening and ROS formation as a mechanism for methamphetamine-induced mitochondrial toxicity. Naunyn Schmiedebergs Arch Pharm. 2014, 387, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, F.; Xue, L.; Wang, B.; Li, J.; Chen, Y.; Chen, T. Methamphetamine causes neurotoxicity by promoting polarization of macrophages and inflammatory response. Hum. Exp. Toxicol. 2018, 37, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Xia, Y.; Li, X.; Chen, T.; Yan, J.; Wang, Y. Alteration of liver immunity by increasing inflammatory response during co-administration of methamphetamine and atazanavir. Immunopharmacol. Immunotoxicol. 2020, 42, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Wiest, R.; Albillos, A.; Trauner, M.; Bajaj, J.S.; Jalan, R. Targeting the gut-liver axis in liver disease. J. Hepatol. 2017, 67, 1084–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Zheng, X.; Ma, X.; Jiang, R.; Zhou, W.; Zhou, S.; Zhang, Y.; Lei, S.; Wang, S.; Kuang, J.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 2019, 10, 4971. [Google Scholar] [CrossRef] [Green Version]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef]
- Wang, W.; Zhai, S.; Xia, Y.; Wang, H.; Ruan, D.; Zhou, T.; Zhu, Y.; Zhang, H.; Zhang, M.; Ye, H.; et al. Ochratoxin A induces liver inflammation: Involvement of intestinal microbiota. Microbiome 2019, 7, 151. [Google Scholar] [CrossRef]
- Carpino, G.; Del Ben, M.; Pastori, D.; Carnevale, R.; Baratta, F.; Overi, D.; Francis, H.; Cardinale, V.; Onori, P.; Safarikia, S.; et al. Increased Liver Localization of Lipopolysaccharides in Human and Experimental NAFLD. Hepatology 2020, 72, 470–485. [Google Scholar] [CrossRef]
- Chen, L.J.; He, J.T.; Pan, M.; Liu, J.L.; Zhang, K.K.; Li, J.H.; Wang, L.B.; Xu, L.L.; Chen, Y.K.; Zhang, Q.Y.; et al. Antibiotics Attenuate Methamphetamine-Induced Hepatotoxicity by Regulating Oxidative Stress and TLR4/MyD88/Traf6 Axis. Front. Pharmacol. 2021, 12, 716703. [Google Scholar] [CrossRef]
- Yang, C.; Fu, X.; Hao, W.; Xiang, X.; Liu, T.; Yang, B.Z.; Zhang, X. Gut dysbiosis associated with the rats’ responses in methamphetamine-induced conditioned place preference. Addict. Biol. 2020, 26, e12975. [Google Scholar] [CrossRef] [PubMed]
- Ning, T.; Gong, X.; Xie, L.; Ma, B. Gut Microbiota Analysis in Rats with Methamphetamine-Induced Conditioned Place Preference. Front. Microbiol. 2017, 8, 1620. [Google Scholar] [CrossRef]
- Chen, L.J.; Zhi, X.; Zhang, K.K.; Wang, L.B.; Li, J.H.; Liu, J.L.; Xu, L.L.; Yoshida, J.S.; Xie, X.L.; Wang, Q. Escalating dose-multiple binge methamphetamine treatment elicits neurotoxicity, altering gut microbiota and fecal metabolites in mice. Food Chem. Toxicol. 2021, 148, 111946. [Google Scholar] [CrossRef]
- Cook, R.R.; Fulcher, J.A.; Tobin, N.H.; Li, F.; Lee, D.J.; Woodward, C.; Javanbakht, M.; Brookmeyer, R.; Shoptaw, S.; Bolan, R.; et al. Alterations to the Gastrointestinal Microbiome Associated with Methamphetamine Use among Young Men who have Sex with Men. Sci. Rep. 2019, 9, 14840. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, A.; Mottershead, M.; Syn, W.K.; Jones, R.; Smith, S.; Nwokolo, C.U. Ciprofloxacin suppresses bacterial overgrowth, increases fasting insulin but does not correct low acylated ghrelin concentration in non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2005, 22, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Fei, N.; Bruneau, A.; Zhang, X.; Wang, R.; Wang, J.; Rabot, S.; Gérard, P.; Zhao, L. Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. MBio 2020, 11, e03263-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.; Duan, Y.; Yang, L.; Schnabl, B. Small metabolites, possible big changes: A microbiota-centered view of non-alcoholic fatty liver disease. Gut 2019, 68, 359–370. [Google Scholar] [CrossRef]
- Wang, L.B.; Xu, L.L.; Chen, L.J.; Zhang, K.K.; Zhang, Q.Y.; Chen, Y.K.; Li, J.H.; Liu, J.L.; Wang, Q.; Xie, X.L. Methamphetamine induces intestinal injury by altering gut microbiota and promoting inflammation in mice. Toxicol. Appl. Pharmacol. 2022, 443, 116011. [Google Scholar] [CrossRef]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [Green Version]
- Shao, T.; Zhao, C.; Li, F.; Gu, Z.; Liu, L.; Zhang, L.; Wang, Y.; He, L.; Liu, Y.; Liu, Q.; et al. Intestinal HIF-1alpha deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J. Hepatol. 2018, 69, 886–895. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, Y.; Hua, J.; Yang, X.; Zhang, X.; Duan, M.; Zhu, X.; Huang, W.; Chao, J.; Zhou, R.; et al. Silencing microRNA-143 protects the integrity of the blood-brain barrier: Implications for methamphetamine abuse. Sci. Rep. 2016, 6, 35642. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Luo, H.; Wang, J.; Tang, W.; Lu, J.; Wu, S.; Xiong, Z.; Yang, G.; Chen, Z.; Lan, T.; et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp. Mol. Med. 2017, 49, e370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, J.C.; Buffa, J.A.; Org, E.; Wang, Z.; Levison, B.S.; Zhu, W.; Wagner, M.A.; Bennett, B.J.; Li, L.; DiDonato, J.A.; et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 2015, 290, 5647–5660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoolen, B.; Maronpot, R.R.; Harada, T.; Nyska, A.; Rousseaux, C.; Nolte, T.; Malarkey, D.E.; Kaufmann, W.; Küttler, K.; Deschl, U.; et al. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol. Pathol. 2010, 38, 5S–81S. [Google Scholar] [CrossRef]
- Xu, L.L.; Chen, Y.K.; Zhang, Q.Y.; Chen, L.J.; Zhang, K.K.; Li, J.H.; Liu, J.L.; Wang, Q.; Xie, X.L. Gestational exposure to GenX induces hepatic alterations by the gut-liver axis in maternal mice: A similar mechanism as PFOA. Sci. Total Environ. 2022, 820, 153281. [Google Scholar] [CrossRef]
- Coutiño-Hernández, D.; Sánchez-Tapia, M.; Leal-Vega, F.; Del Valle, M.B.; Ledezma, H.; Cervantes, R.; Pedraza-Chaverri, J.; Granados-Portillo, O.; Díaz, D.; Antunes-Ricardo, M.; et al. Modulation of gut microbiota by Mantequilla and Melipona honeys decrease low-grade inflammation caused by high fructose corn syrup or sucrose in rats. Food Res. Int. 2022, 151, 110856. [Google Scholar] [CrossRef]
- Pang, K.; Lee, J.; Kim, J.; Park, J.; Park, Y.; Hong, E.; An, H.; Ooshima, A.; Son, M.; Park, K.S.; et al. Degradation of DRAK1 by CUL3/SPOP E3 Ubiquitin ligase promotes tumor growth of paclitaxel-resistant cervical cancer cells. Cell Death Dis. 2022, 13, 169. [Google Scholar] [CrossRef]
- Zveik, O.; Fainstein, N.; Rechtman, A.; Haham, N.; Ganz, T.; Lavon, I.; Brill, L.; Vaknin-Dembinsky, A. Cerebrospinal fluid of progressive multiple sclerosis patients reduces differentiation and immune functions of oligodendrocyte progenitor cells. Glia 2022, 70, 1191–1209. [Google Scholar] [CrossRef]



| Gene Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Tlr4 | ATGGCATGGCTTACACCACC | GAGGCCAATTTTGTCTCCACA |
| Myd88 | TCATGTTCTCCATACCCTTGGT | AAACTGCGAGTGGGGTCAG |
| Traf6 | ATGCAGAGGAATCACTTGGCA | ACGGACGCAAAGCAAGGTT |
| Rela (p65) | AGGCTTCTGGGCCTTATGTG | TGCTTCTCTCGCCAGGAATAC |
| Tnf | CTGAACTTCGGGGTGATCGG | GGCTTGTCACTCGAATTTTGAGA |
| Il1b | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| Actb | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.-B.; Chen, L.-J.; Wang, Q.; Xie, X.-L. Silencing the Tlr4 Gene Alleviates Methamphetamine-Induced Hepatotoxicity by Inhibiting Lipopolysaccharide-Mediated Inflammation in Mice. Int. J. Mol. Sci. 2022, 23, 6810. https://doi.org/10.3390/ijms23126810
Wang L-B, Chen L-J, Wang Q, Xie X-L. Silencing the Tlr4 Gene Alleviates Methamphetamine-Induced Hepatotoxicity by Inhibiting Lipopolysaccharide-Mediated Inflammation in Mice. International Journal of Molecular Sciences. 2022; 23(12):6810. https://doi.org/10.3390/ijms23126810
Chicago/Turabian StyleWang, Li-Bin, Li-Jian Chen, Qi Wang, and Xiao-Li Xie. 2022. "Silencing the Tlr4 Gene Alleviates Methamphetamine-Induced Hepatotoxicity by Inhibiting Lipopolysaccharide-Mediated Inflammation in Mice" International Journal of Molecular Sciences 23, no. 12: 6810. https://doi.org/10.3390/ijms23126810






