Construction and Analysis of Disuse Atrophy Model of the Gastrocnemius Muscle in Chicken
Abstract
:1. Introduction
2. Results
2.1. Atrophy Model Leads to Loss of Muscle Mass
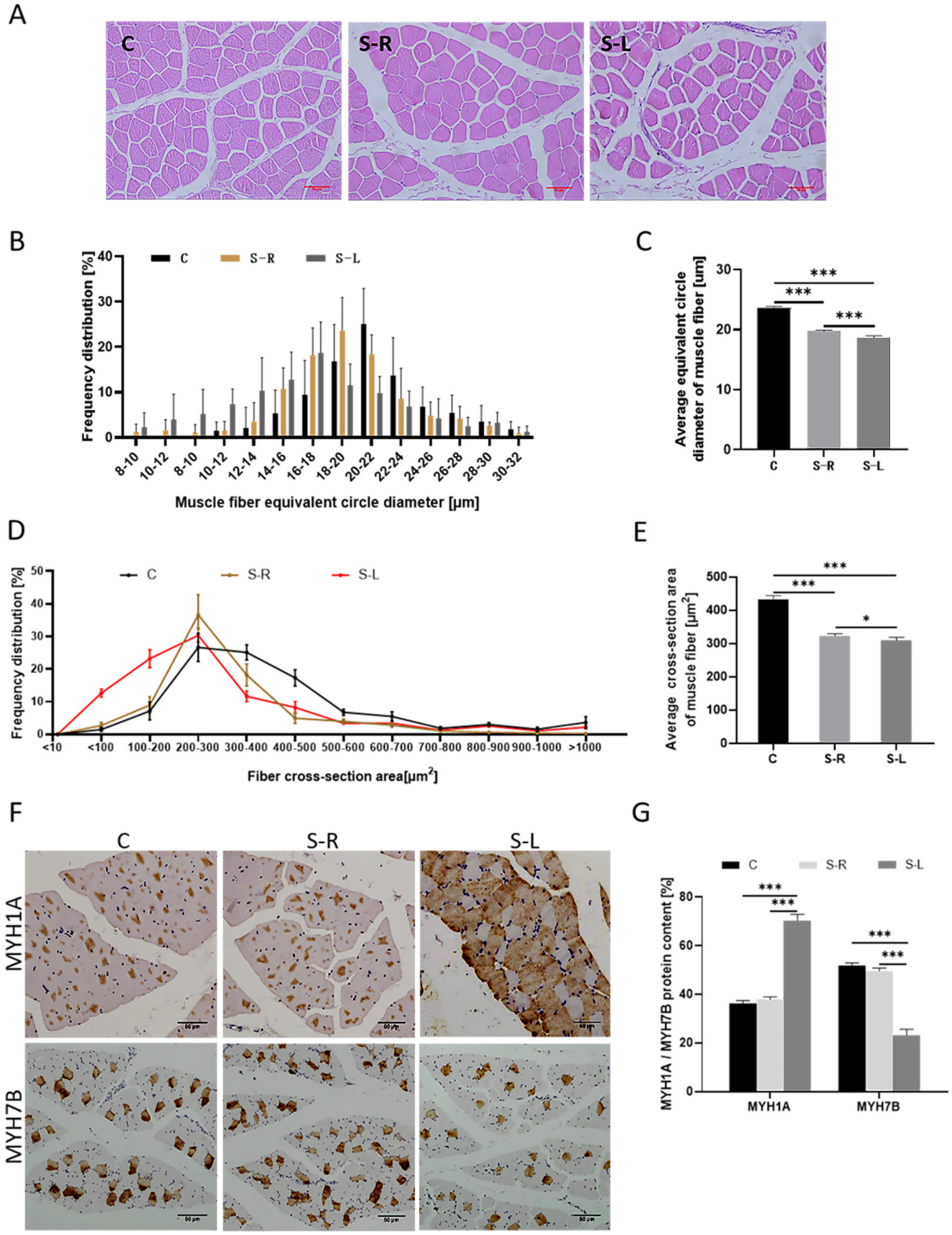
2.2. Atrophy Model Results in a Reduction of Muscle Fiber Diameter and Altered Muscle Fiber Composition Type
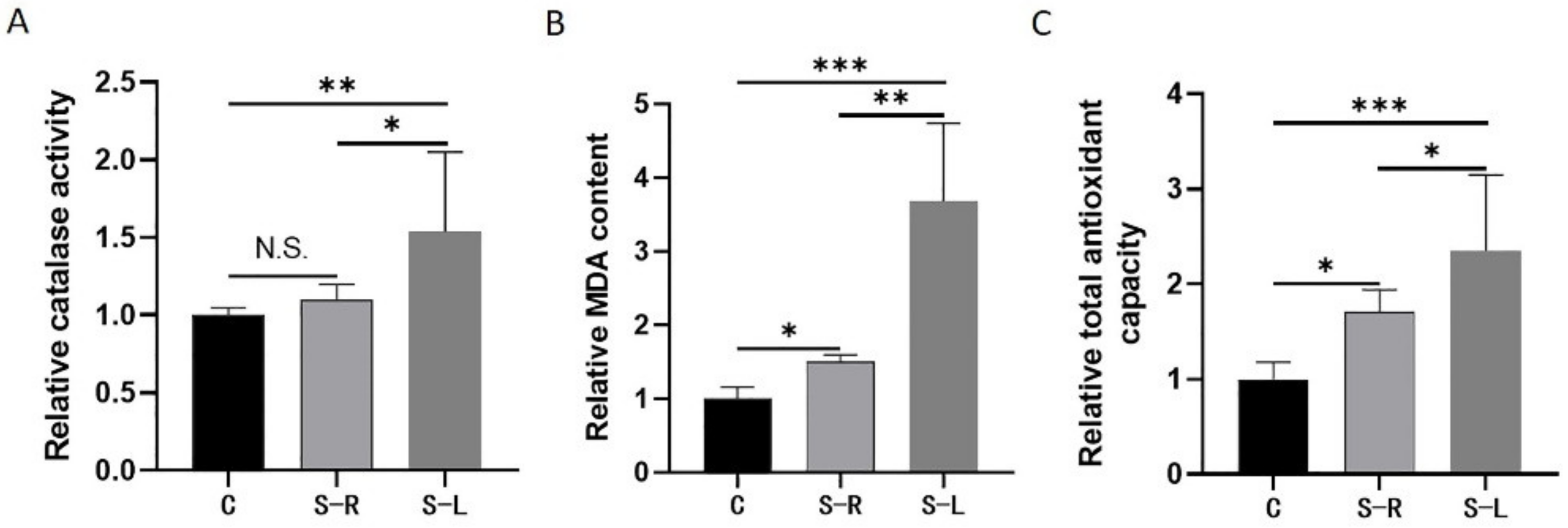
2.3. Increased Levels of Oxidative Stress in Atrophy Model
2.4. Transcriptome Sequencing Analysis
2.5. DEGs among Three Groups (C Group, S-R Group, and S-L Group) of Gastrocnemius Tissues
2.6. GO and KEGG Pathway Analysis for DEGs
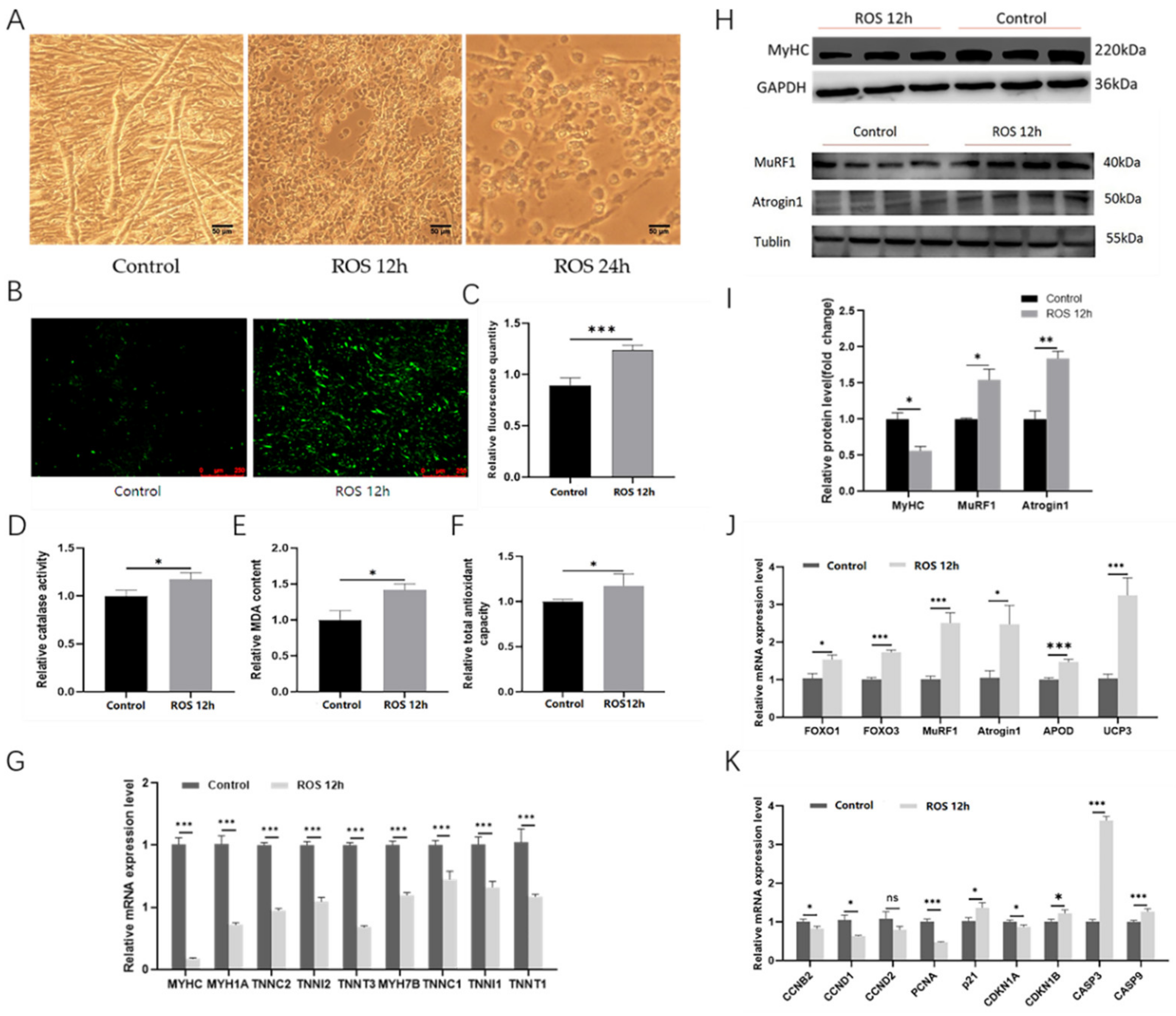
2.7. The ROS Can Induce Myotube Atrophy
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Animals
4.3. Total RNA Extraction and RNA Sequencing
4.4. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (qRT-PCR)
4.5. Cell Culture
4.6. Hematoxylin and Eosin Staining (HE)
4.7. Immunohistochemical Analysis
4.8. Western Blotting Assay
4.9. Catalase (CAT) Activity Assay
4.10. Malondialdehyde (MDA) Content Assay
4.11. Total Antioxidant Capacity (T-AOC) Assay
4.12. Reactive Oxygen Species (ROS) Assay
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stump, C.S.; Henriksen, E.J.; Wei, Y.; Sowers, J.R. The metabolic syndrome: Role of skeletal muscle metablism. Ann. Med. 2006, 38, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2015, 50, 500–509. [Google Scholar] [CrossRef]
- Schiaffino, S.; Sandri, M.; Murgia, M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology 2007, 22, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, K. Relationship of skeletal muscle atrophy to functional status: A systematic research review. Biol. Res. Nurs. 2000, 2, 117–131. [Google Scholar] [CrossRef]
- Xi, S.; Yue, S. Research progress of disuse muscle atrophy. Chin. J. Clin. Rehabil. 2003, 7, 710–714. [Google Scholar]
- Kenji, H.; Yuichi, U.; Hajime, O. Disuse muscle atrophy of lower limbs in hemiplegic patients. Arch. Phys. Med. Rehabil. 1997, 78, 197–206. [Google Scholar]
- Frimel, T.N.; Kapadia, F.; Gaidosh, G.S.; Li, Y.; Walter, G.A.; Vandenborne, K. A model of muscle atrophy using cast immobilization in mice. Med. Sci. Sports Exerc. 2005, 32, 197–206. [Google Scholar] [CrossRef]
- Bloomfield, S.A. Changes in musculoskeletal structure and function with prolonged bed rest. Med. Sci. Sports Exerc. 1997, 29, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Seifried, H.E.; Anderson, D.E.; Fisher, E.I.; Milner, J.A. A review of the interaction among dietary antioxidants and reactive oxygen species. J. Nutr. Biochem. 2007, 18, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Winyard, P.G.; Moody, C.J.; Jacob, C. Oxidative activation of antioxidant defence. Trends Biochem. Sci. 2005, 30, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Lawler, J.M.; Song, W.; Demaree, S.R. Hindlimb unloading increases oxidatve stress and disrupts antioxidant capacity in skeletal muscle. Free Radic. Biol. Med. 2003, 35, 9–16. [Google Scholar] [CrossRef]
- Matsushima, Y.; Nanri, H.; Nara, S.; Okufuji, T.; Ohta, M.; Hachisuka, K.; Ikeda, M. Hindlimb unloading decreases thioredoxin-related antioxidant proteins and increases thioredoxin-binding protein-2 in rat skeletal muscle. Free Radic. Res. 2006, 40, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.; Song, W.; Jang, Y.C.; Liu, Y.; Sabia, M.; Richardson, A.; Van Remmen, H. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, 1159–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, S.K.; Kavazis, A.N.; DeRuisseau, K.C. Mechanisms of disuse muscle atrophy: Role of oxidative stress. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyler, B.; Maret, G.T. From animal to human: Evidence linking oxidative stress as a causative factor in muscle atrophy. J. Physiol. 2007, 583, 421–422. [Google Scholar]
- Andrianjafiniony, T.; Dupré-Aucouturier, S.; Letexier, D.; Couchoux, H.; Desplanches, D. Oxidative stress, apoptosis, and proteolysis in skeletal muscle repair after unloading. Am. J. Physiol. Cell Physiol. 2010, 299, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Booth, F.W. Effect of limb immobilization on skeletal muscle. J. Appl. Physiol. 1982, 52, 1113–1118. [Google Scholar] [CrossRef]
- Ni, G.X.; Cheng, H.P.; Su, L.; Tang, J.K. Effects of immobilization on morphology of soleus and gastrocnemius muscles. China J. Rehabl. Med. 2000, 15, 137–139. [Google Scholar]
- Itai, Y.; Kariya, Y.; Hoshino, Y. Morphological changes in rat hindlimb muscle fibres during recovery from disuse atrophy. Acta Physiol. Scand. 2004, 181, 217–224. [Google Scholar] [CrossRef]
- Ferreira, R.; Vitorino, R.; Neuparth, M.J.; Appell, H.J.; Amado, F.; Duarte, J.A. Cellular patterns of the atrophic response in murinesoleus andgastrocnemiusmuscles submitted to simulated weightlessness. Eur. J. Appl. Physiol. 2007, 101, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.R.; Haddad, F.; Baldwin, K.M. Iereaction of creatine depletion and muscle unloading: Effects on postural and locomotor muscles. J. Appl. Physiol. 1994, 77, 1198–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavazis, A.N.; Talbert, E.E.; Smuder, A.J.; Hudson, M.B.; Nelson, W.B.; Powers, S.K. Mechanical ventilation induces diaphragmatic mitochondrial dysfunction and increased oxidant production. Free Radic. Biol. Med. 2009, 46, 842–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowaltowski, A.J.; Vercesi, A.E. Mitochondrial damage induced by conditions of oxidative stress. Free Radic. Biol. Med. 1999, 26, 463–471. [Google Scholar] [CrossRef]
- Guo, C.; Sun, L.; Chen, X.; Zhang, D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural. Regen. Res. 2013, 8, 2003–2014. [Google Scholar]
- Lee, S.H.; Joo, S.T.; Ryu, Y.C. Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality. Meat Sci. 2010, 86, 166–170. [Google Scholar] [CrossRef]
- Xu, H.Y. Functional Research of FHL3 Regulating Muscle Growth and Muscle Fiber Type Transformation; Huazhong Agricultural University: Wuhan, China, 2018. [Google Scholar]
- Senthil, K.; Aranganathan, S.; Nalini, N. Evidence of oxidative stress in the circulation of ovarian cancer patients. Clin. Chim. Acta 2004, 339, 27–32. [Google Scholar] [CrossRef]
- Fernández, M.I.G.; Gheduzzi, D.; Boraldi, F.; Paolinelli, C.D.; Sánchez, P.S.; Valdivielso, P.; Morilla, M.J.; Quaglino, D.; Guerra, D.; Casolari, S.; et al. Parameters of oxidative stress are present in the circulation of PXE patients. Biochim. Biophys. Acta 2008, 1782, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Billings, F.T., IV; Ball, S.K.; Roberts, L.J., II; Pretorius, M. Postoperative acute kidney injury is associated with hemoglobinemia and an enhanced oxidative stress response. Free Radic. Biol. Med. 2011, 50, 1480–1487. [Google Scholar] [CrossRef] [Green Version]
- Powers, S.K.; Smuder, A.J.; Criswell, D.S. Mechanistic links between oxidative stress and disuse muscle atrophy. Antioxid. Redox Signal. 2011, 15, 2519–2528. [Google Scholar] [CrossRef] [Green Version]
- Pellegrino, M.A.; Desaphy, J.F.; Brocca, L.; Pierno, S.; Camerino, D.C.; Bottinelli, R. Redox homeostasis, oxidative stress and disuse muscleatrophy. Am. J. Physiol. 2011, 589, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Calvani, R.; Joseph, A.-M.; Adhihetty, P.J.; Miccheli, A.; Bossola, M.; Leeuwenburgh, C.; Bernabei, R.; Marzetti, E. Mitochondrial pathways in sarcopenia of aging and disuse muscle atrophy. Biol. Chem. 2013, 394, 393–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, S.L.; Gagnon, B.J.; Senf, S.M.; Hain, B.A.; Judge, A.R. Ros-mediated activation of NF-κB and Foxo during muscle disuse. Muscle Nerve 2010, 41, 110–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Lecker, S.H.; Jagoe, R.T.; Gilbert, A.; Gomes, M.; Baracos, V.; Bailey, J.; Price, S.R.; Mitch, W.E.; Goldberg, A.L. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004, 18, 39–51. [Google Scholar] [CrossRef]
- Foletta, V.C.; White, L.J.; Larsen, A.E.; Léger, B.; Russell, A.P. The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflug. Arch. Eur. J. Physiol. 2011, 461, 325–335. [Google Scholar] [CrossRef]
- Clarke, B.A.; Drujan, D.; Willis, M.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 coordinately activates protein deg-radation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef] [Green Version]
- Siems, W.; Grune, T. Intracellular metabolism of 4-hydroxynonenal. Mol. Asp. Med. 2003, 24, 167–175. [Google Scholar] [CrossRef]
- Furuno, K.; Goldberg, A.L. The activation of protein degradation in muscle by Ca2+ or muscle injury does not involve a lysosomal mechanism. Biochem. J. 1986, 237, 859–864. [Google Scholar] [CrossRef] [Green Version]
- Goll, D.E.; Thompson, V.F.; Li, H.; Wei, W.; Cong, J. The calpain system. Physiol. Rev. 2003, 83, 731–801. [Google Scholar] [CrossRef] [PubMed]
- Purintrapiban, J.; Wang, M.C.; Forsberg, N.E. Degradation of sarcomeric and cytoskeletal proteins in cultured skeletal muscle cells. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 136, 393–401. [Google Scholar] [CrossRef]
- Tidball, J.G.; Spencer, M.J. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J. Physiol. 2002, 545, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Qaisar, R.; Bhaskaran, S.; Ranjit, R.; Sataranatarajan, K.; Premkumar, P.; Huseman, K.; Van Remmen, H. Restoration of SERCA ATPase prevents oxidative stress-related muscle atrophy and weakness. Redox Biol. 2019, 20, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Ganfornina, M.D.; Do Carmo, S.; Lora, J.M.; Torres-Schumann, S.; Vogel, M.; Allhorn, M.; González, C.; Bastiani, M.J.; Rassart, E.; Sanchez, D. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 2008, 7, 506–515. [Google Scholar] [CrossRef] [Green Version]
- Bajo-Grañeras, R.; Ganfornina, M.D.; Martín-Tejedor, E.; Sanchez, D. Apolipoprotein D mediates autocrine protection of astrocytes and controls their reactivity level, contributing to the functional maintenance of paraquat-challenged dopaminergic systems. GLIA 2011, 59, 1551–1566. [Google Scholar] [CrossRef] [Green Version]
- Brand, M.D.; Pamplona, R.; Portero-Otin, M.; Requena, J.R.; Roebuck, S.J.; Buckingham, J.A.; Clapham, J.C.; Cadenas, S. Oxidative damage and phospholipid fatty acyl composition in skeletal muscle mitochondria from mice underexpressing or overexpressing uncoupling protein 3. Biochem. J. 2002, 368, 597–603. [Google Scholar] [CrossRef]
- Rousset, S.; Alves-Guerra, M.-C.; Mozo, J.; Miroux, B.; Cassard-Doulcier, A.-M.; Bouillaud, F.; Ricquier, D. The biology of mitochondrial uncoupling proteins. Diabetes 2004, 53, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Clapham, J.C.; Arch, J.R.S.; Chapman, H.; Haynes, A.; Lister, C.; Moore, G.B.T.; Piercy, V.; Carter, S.A.; Lehner, I.; Smith, S.A.; et al. Mice overexpressing human uncoupling protein-3 in skeletal muscle are hyperphagic and lean. Nature 2000, 406, 415–418. [Google Scholar] [CrossRef]
- Moore, G.B.; Himms-Hagen, J.; Harper, M.-E.; Clapham, J.C. Overexpression of UCP-3 in skeletal muscle of mice results in increased expression of mitochondrial thioesterase mRNA. Biochem. Biophys. Res. Commun. 2001, 283, 785–790. [Google Scholar] [CrossRef]
- Vidal-Puig, A.; Grujic, D.; Zhang, C.-Y.; Hagen, T.; Boss, O.; Ido, Y.; Szczepanik, A.; Wade, J.; Mootha, V.; Cortright, R.; et al. Energy metabolism in uncoupling protein 3 gene knockout mice. J. Biol. Chem. 2000, 275, 16258–16266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echtay, K.S.; Esteves, T.C.; Pakay, J.L.; Jekabsons, M.B.; Lambert, A.J.; Portero-Otín, M.; Pamplona, R.; Vidal-Puig, A.J.; Wang, S.; Roebuck, S.J.; et al. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003, 22, 4103–4110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirement. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]




| Control Group (C Group) | Treatment Group (S Group) | ||
|---|---|---|---|
| Left lower limb | Right lower limb | Left lower limb | Right lower limb |
| (C-L group) | (C-R group) | (S-L group) | (S-R group) |
| No treatment | No treatment | Immobilization | No treatment |
| Pathways | −log (p Value) | Rich Ratio | Focus Molecules |
|---|---|---|---|
| TGF-beta signaling pathway | 3.67 | 0.13 | ID1, ID2, ID3, SMAD6, SMAD7, BMP4, BMP5, FMOD, RBX1 |
| FOXO signaling pathway | 2.33 | 0.09 | FBXO32, FOXO3, PLK2, PRKAB1, SGK1, HOMER3, GABARAPL1 |
| Thyroid cancer | 2.21 | 0.15 | GADD45B, GADD45G, TPM3, CDKN1A, CCND1 |
| Breast cancer | 2.01 | 0.08 | GADD45B, GADD45G, DLL4, CDKN1A, CCND1, NOTCH1 |
| Glioma | 2.00 | 0.10 | GADD45B, GADD45G, CCND1, CDKN1A, PDGFB |
| Endometrial cancer | 1.98 | 0.12 | GADD45B, GADD45G, CCND1, CDKN1A, AXIN2 |
| Small cell lung cancer | 1.89 | 0.09 | GADD45B, GADD45G, CCND1, CDKN1A, XIAP |
| Cardiac muscle contraction | 1.86 | 0.11 | MYH7B, MYH7, TNNC1, MYL3, MYH1B, ACTC1, TPM3 |
| Notch signaling pathway | 1.69 | 0.11 | HES4, NOTCH1, LFNG, JAG1, DLL4 |
| Calcium signaling pathway | 1.66 | 0.07 | ATP2A2, ATP2A3, ATP2B2, CAMK1D, CAMK4, GRIN2C |
| Nitrogen metabolism | 1.66 | 0.11 | HES4, NOTCH1, LFNG, JAG1, DLL4 |
| Apelin signaling pathway | 1.60 | 0.18 | PRKAB1, APLNR, MYLK3 |
| p53 signaling pathway | 1.54 | 0.09 | CDKN1A, CCND1, GADD45B, GADD45G, SESN1, AIFM2 |
| Ribosome biogenesis in eukaryotes | 1.51 | 0.09 | RPP38, RPP30, HEATR1, NOP56, NAT10, UTP4 |
| Pathways in cancer | 1.44 | 0.05 | GADD45B, GADD45G, XIAP, CDKN1A, CCND1, RBX1, ETS1 |
| Other types of O-glycan biosynthesis | 1.42 | 0.14 | LFNG, ST6GAL2, EOGT |
| Hippo signaling pathway | 1.41 | 0.07 | WWC1, CCND1, FRMD6 |
| Retinol metabolism | 1.40 | 0.11 | BCO1, ALDH1A2, CYP26A1, AOX1 |
| cAMP signaling pathway | 1.34 | 0.06 | EDNRA, RELA, SOX9, PDE3A |
| PI3K-Akt signaling pathway | 1.28 | 0.06 | ANGPT4, PDGFB, F2R, KITLG, KDR, FLT1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, J.; Wang, Z.; Liu, Q.; Li, Z.; Nie, Q. Construction and Analysis of Disuse Atrophy Model of the Gastrocnemius Muscle in Chicken. Int. J. Mol. Sci. 2022, 23, 6892. https://doi.org/10.3390/ijms23136892
Mo J, Wang Z, Liu Q, Li Z, Nie Q. Construction and Analysis of Disuse Atrophy Model of the Gastrocnemius Muscle in Chicken. International Journal of Molecular Sciences. 2022; 23(13):6892. https://doi.org/10.3390/ijms23136892
Chicago/Turabian StyleMo, Jiawei, Zhijun Wang, Qingchun Liu, Zhenhui Li, and Qinghua Nie. 2022. "Construction and Analysis of Disuse Atrophy Model of the Gastrocnemius Muscle in Chicken" International Journal of Molecular Sciences 23, no. 13: 6892. https://doi.org/10.3390/ijms23136892
APA StyleMo, J., Wang, Z., Liu, Q., Li, Z., & Nie, Q. (2022). Construction and Analysis of Disuse Atrophy Model of the Gastrocnemius Muscle in Chicken. International Journal of Molecular Sciences, 23(13), 6892. https://doi.org/10.3390/ijms23136892








