Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) Infection in Tomato
Abstract
:1. Introduction
2. Results
2.1. Inoculation
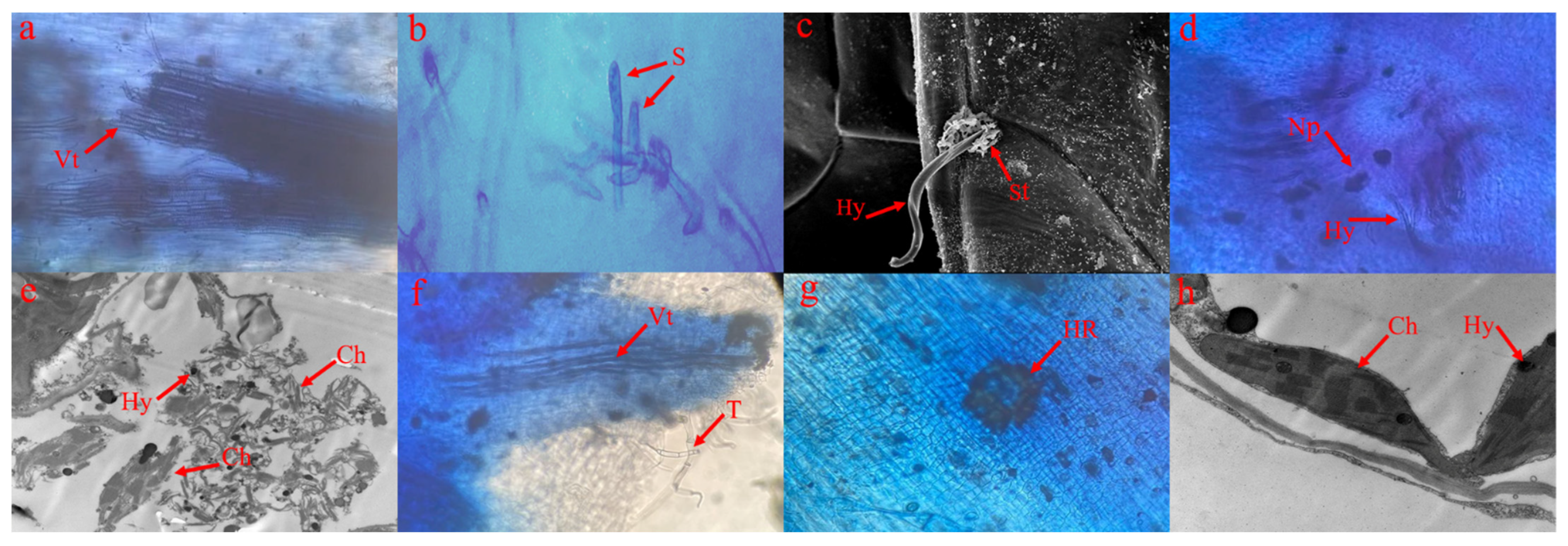
2.2. Microscopic Analysis of FORL Growth in Two Tomato Cultivars
2.3. RNA Sequencing and Identification of Transcripts
2.4. DEGs (Differentially Expressed Genes) in Response to FORL Inoculation
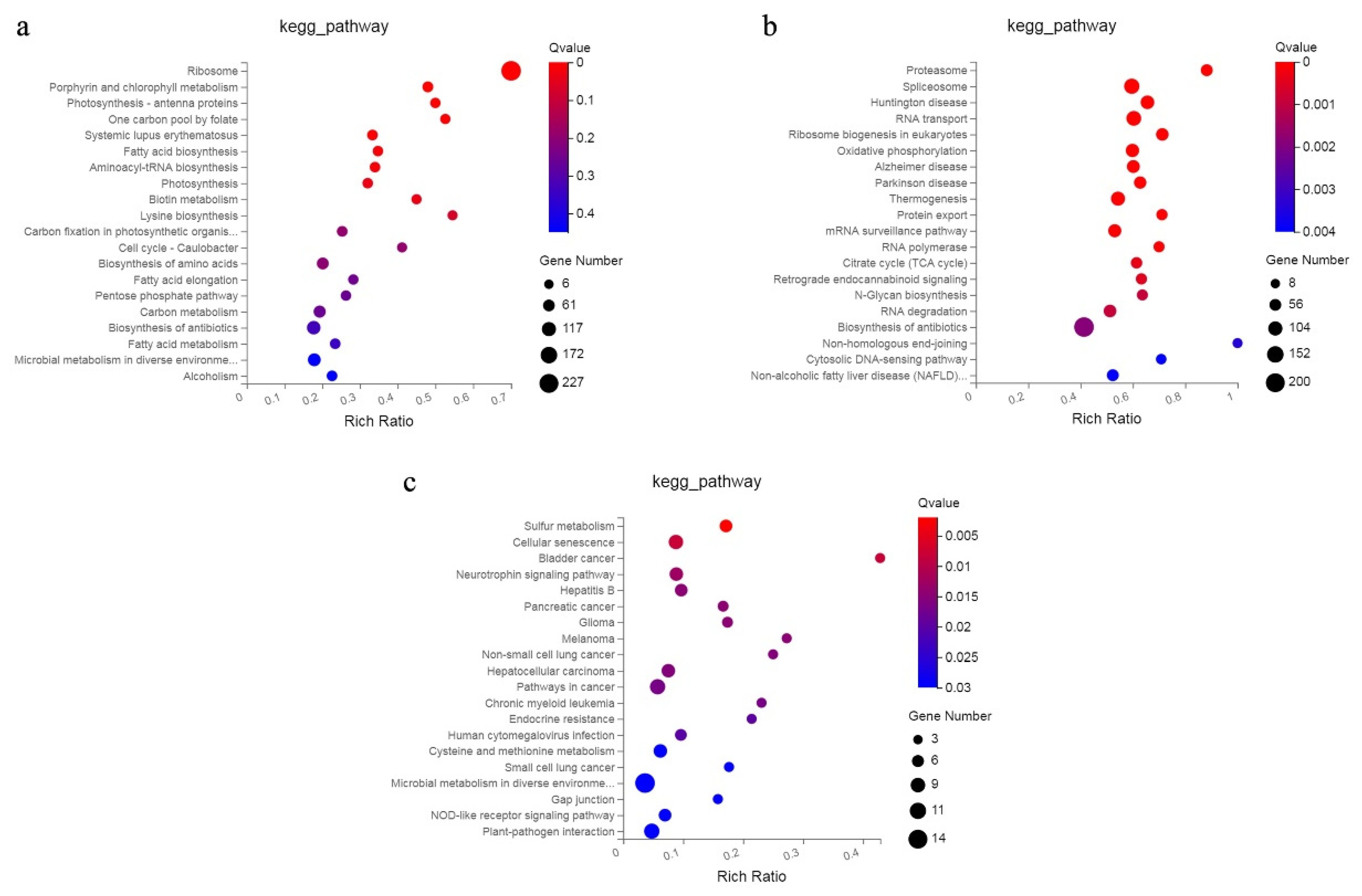
2.5. KEGG Pathway and GO Enrichment Analysis of DEGs
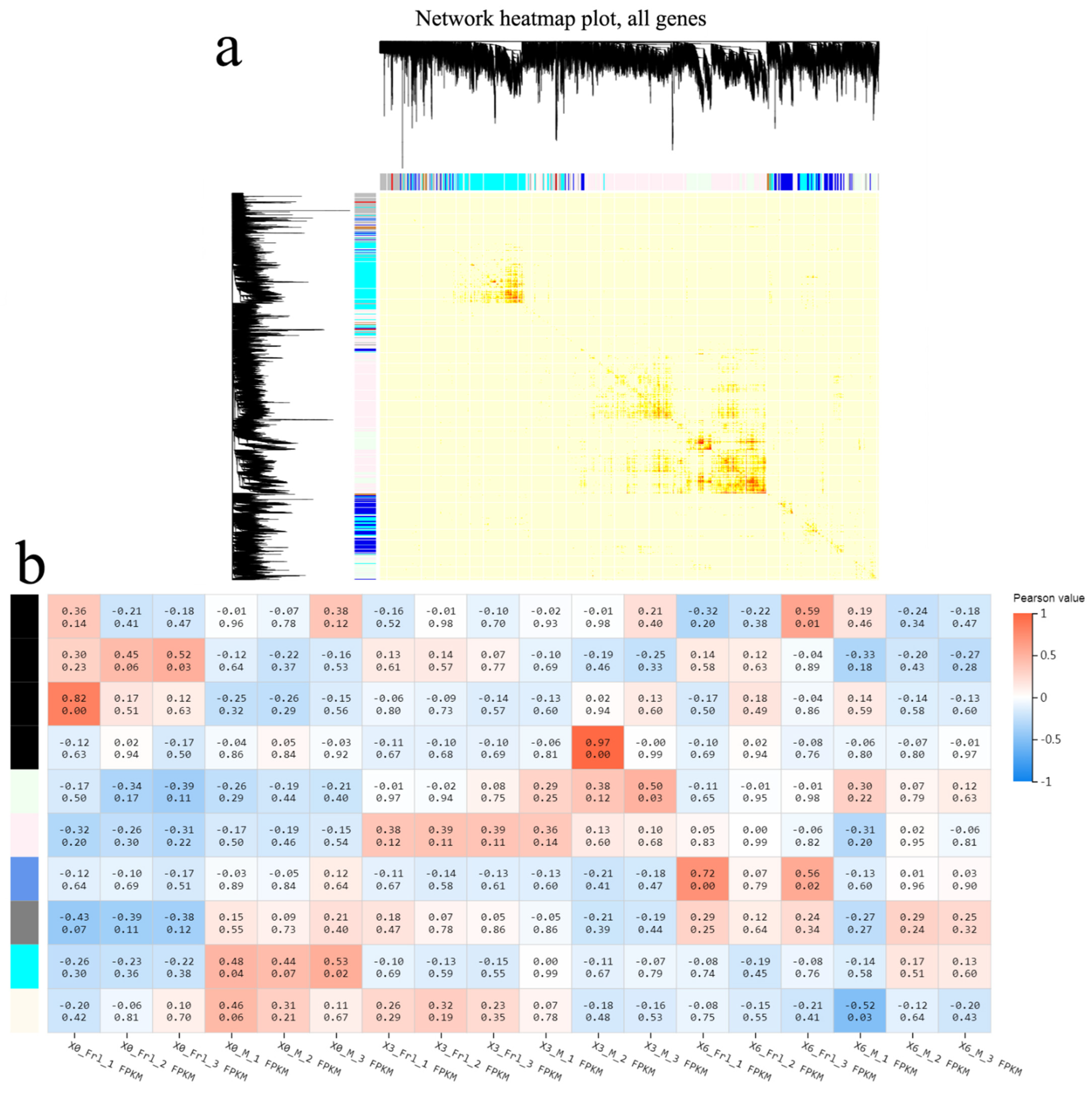
2.6. Gene Co-Expression Network Analysis
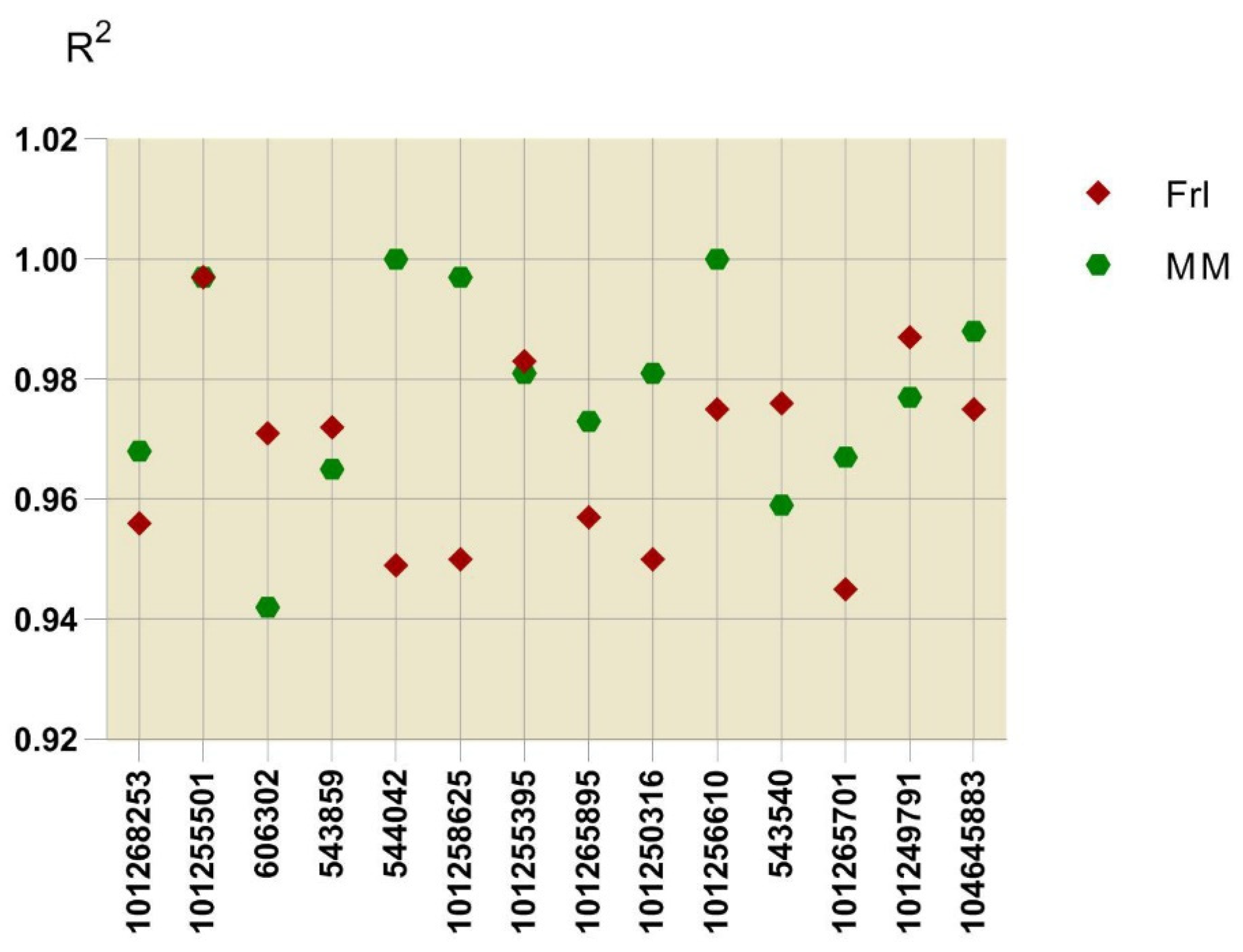
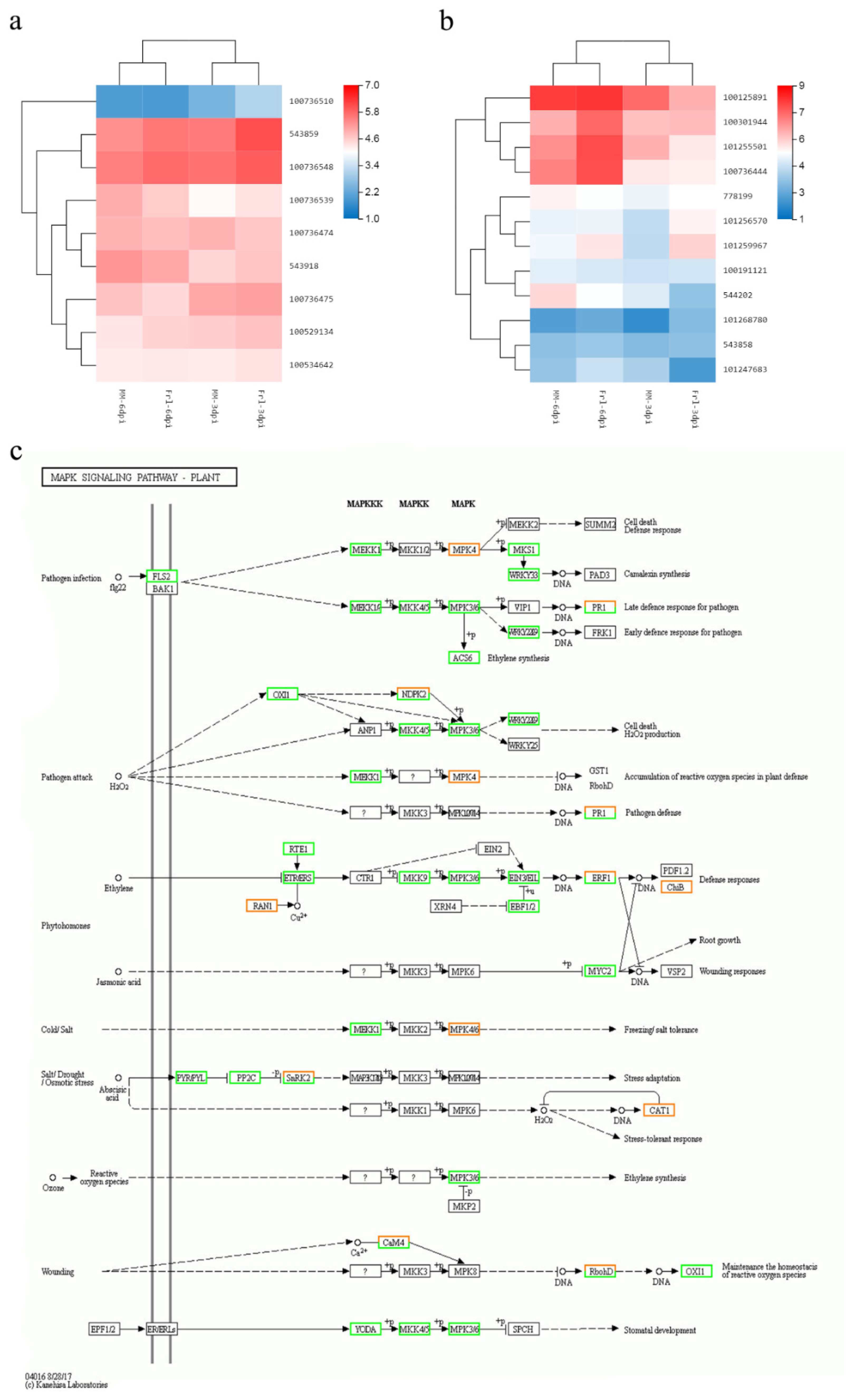
2.7. Validation of DEG Expression Patterns
2.8. Determination of Critical Genes from Vital Pathways
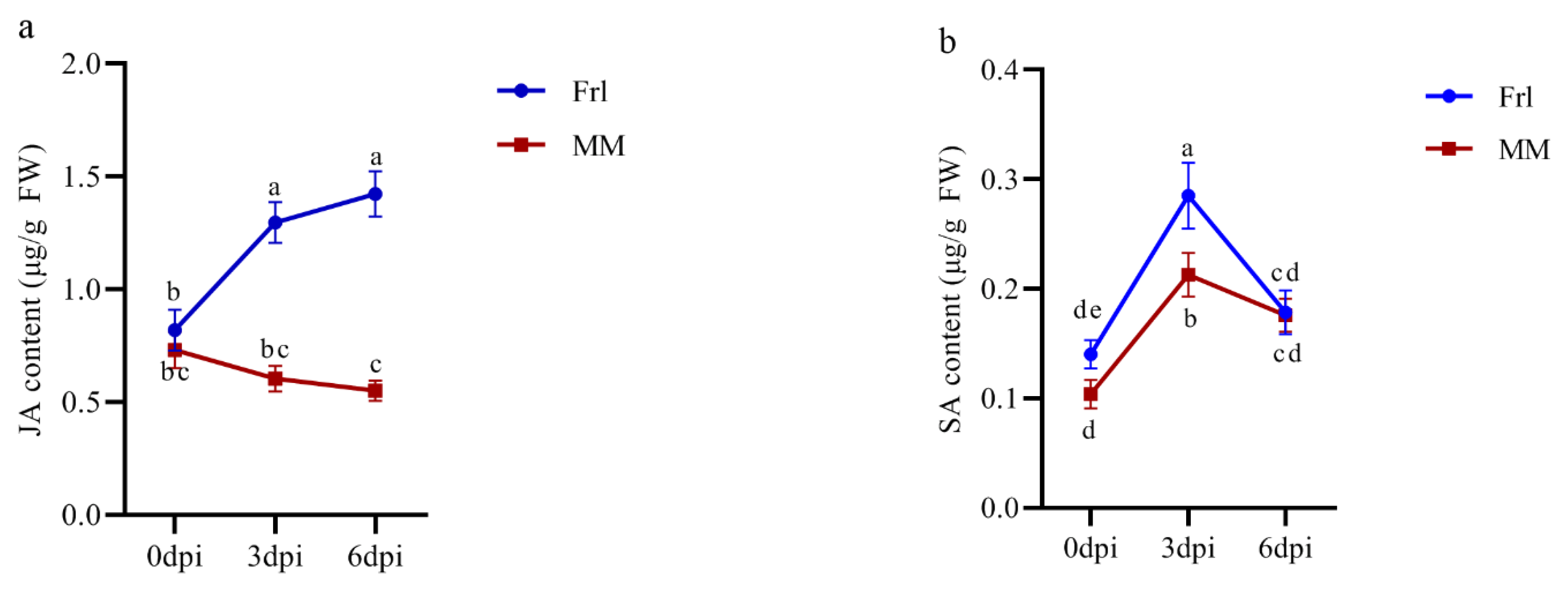
2.9. Hormone Measurements
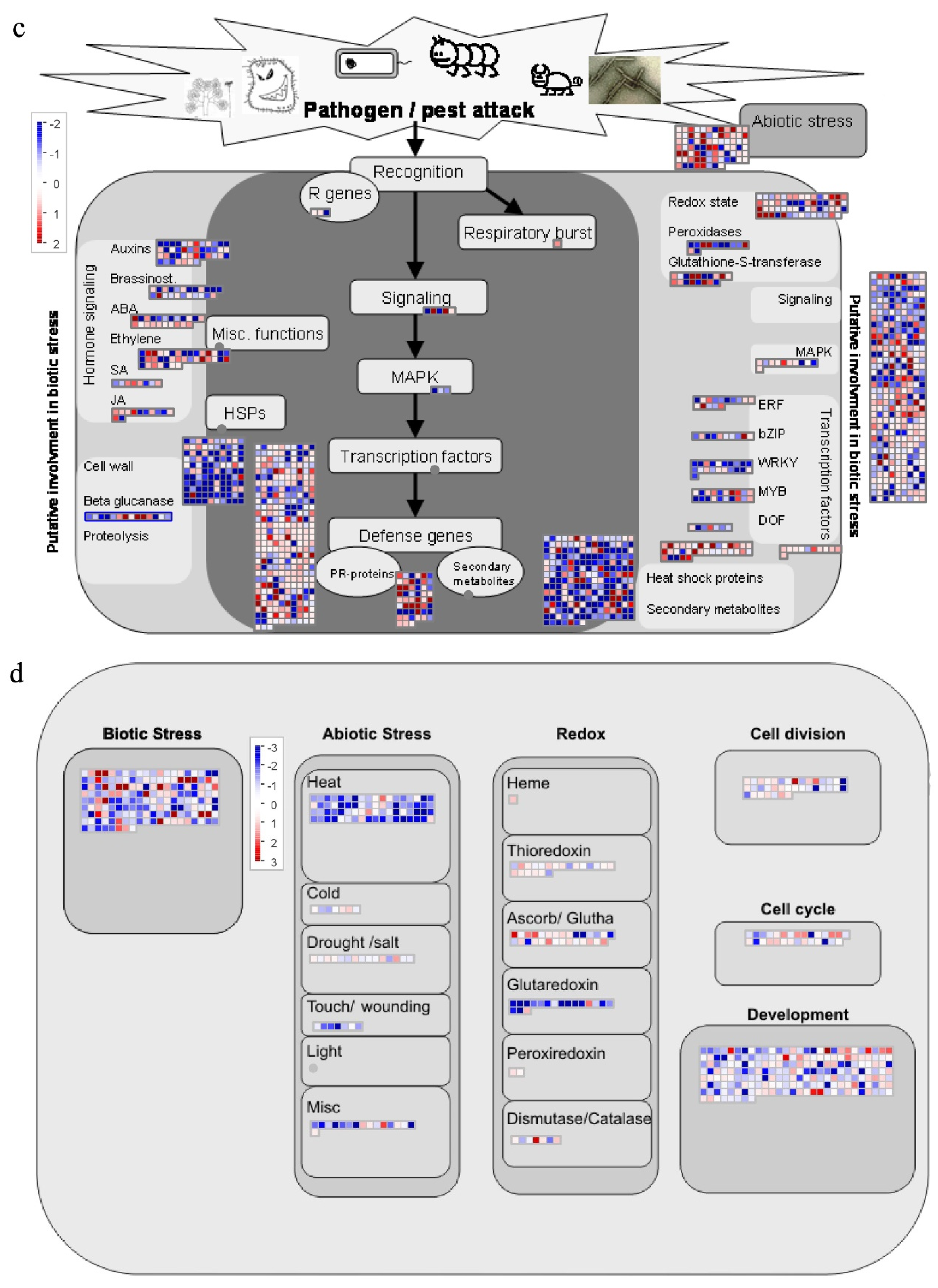
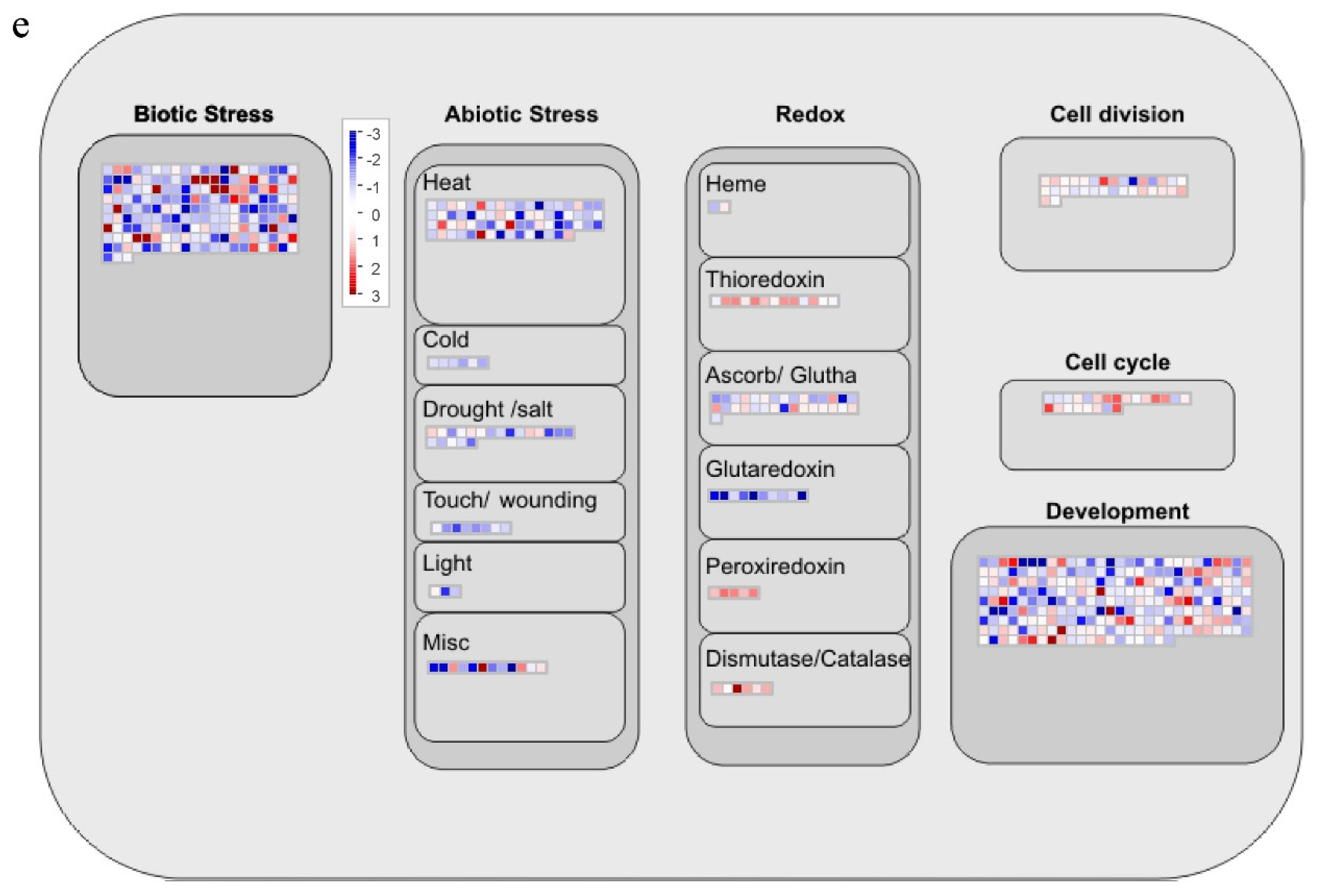
2.10. Analysis of DEGs in Response to Biotic Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and FORL Inoculation
4.2. Microscopic Observation of FORL in Frl Tomato
4.3. RNA Extraction and Transcriptome Sequencing
4.4. RNA Sequence Read Matching and Differentially Expressed Gene (DEG) Identification
4.5. Functional Annotation and Enrichment Analysis of DEGs
4.6. Determination of Endogenous Hormones
4.7. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.8. MapMan Analysis of the Biofunctions of DEGs
4.9. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamamoto, I.; Komada, H.; Kuniyasu, K.; Saito, M.; Ezuka, A. A new race of Fusarium oxysporum f. sp.: Lycopersici inducing root rot of tomato. Annu. Rep. Plant Protect. Soc. 1974, 16, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Sonoda, R. The occurrence of Fusarium root rot of tomatoes in South Florida. Plant Dis. Rep. 1976, 60, 271–274. [Google Scholar]
- Cheng, L.; Zhang, S.; Li, Y.Q.; Chen, F.D.; Cheng, F.; Zhang, X.Y.; Dong, T.; Guo, J.j. Pathogen Identification of Fusarium Crown Root Rot and Screening for Resistant Sources in Tomato. Acta Hortic. Sinica 2016, 43, 781–788. [Google Scholar]
- Liu, L.; Wang, H. Research progress of tomato Fusarium crown and root rot pathogen and resistance breeding. J. Chang. Veg. 2016, 6, 35–37. [Google Scholar]
- Zhang, X.; Liu, L.; Wang, H.; Li, W.; Wang, F. Isolation and Identification of Tomato Fusarium oxysporum in Shandong Province. Shandong Agric. Sci. 2019, 51, 78–81. [Google Scholar]
- Geng, L.; Li, C.; Chi, S.; Wang, L.; Chai, M. Identification of the pathogen causing Fusarium crown and root rot of tomato and its growth affecting factors. Acta Hortic. Sinica 2012, 42, 449–455. [Google Scholar]
- Vakalounakis, D. The genetic analysis of resistance to Fusarium crown and root rot of tomato. Plant Pathol. 1988, 37, 71–73. [Google Scholar] [CrossRef]
- Vakalounakis, D.; Laterrot, H.; Moretti, A.; Ligoxigakis, E.; Smardas, K. Linkage between Frl (Fusarium oxysporum f. sp. radicis-lycopersici) and Tm-2(tobacco mosaic virus resistance-2)loci in tomato (Lycopersicon esculentum). Ann. Appl. Biol. 1997, 130, 319–323. [Google Scholar]
- Metraux, J.P.; Signer, H.; Ryals, J.; Ward, E.; Wyss-Benz, M.; Gaudin, J.; Raschdorf, K.; Schmid, E.; Blum, W.; Inverardi, B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 1990, 250, 1004–1006. [Google Scholar] [CrossRef] [Green Version]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic Acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar] [CrossRef] [Green Version]
- Lawton, K.; Weymann, K.; Friedrich, L.; Vernooij, B.; Uknes, S.; Ryals, J. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 1995, 8, 863–870. [Google Scholar] [CrossRef]
- Summermatter, K.; Sticher, L.; Metraux, J.P. Systemic responses in Arabidopsis thaliana infected and challenged with pseudomonas syringae pv syringae. Plant Physiol. 1995, 108, 1379–1385. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Durrant, W.E.; Song, J.; Spivey, N.W.; Dong, X. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. USA 2010, 107, 22716–22721. [Google Scholar] [CrossRef] [Green Version]
- Van Verk, M.C.; Bol, J.F.; Linthorst, H.J. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006, 2, e123. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Dong, Q.; Yu, D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012, 185, 288–297. [Google Scholar] [CrossRef]
- Ren, C.M.; Zhu, Q.; Gao, B.D.; Ke, S.Y.; Yu, W.C.; Xie, D.X.; Peng, W. Transcription factor WRKY70 displays important but no indispensable roles in jasmonate and salicylic acid signaling. J. Integr. Plant Biol. 2008, 50, 630–637. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, F.; Yoshida, R.; Ichimura, K.; Mizoguchi, T.; Seo, S.; Yonezawa, M.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The mitogen-activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 2007, 19, 805–818. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Nolan, T.M.; Song, G.; Liu, S.; Xie, Z.; Chen, J.; Schnable, P.S.; Walley, J.W.; Yin, Y. FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 2018, 28, 3316–3324.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Schneider, J.D.; Lin, C.; Geng, S.; Ma, T.; Lawrence, S.R.; Dufresne, C.P.; Harmon, A.C.; Chen, S. MPK4 phosphorylation dynamics and interacting proteins in plant immunity. J. Proteome Res. 2019, 18, 826–840. [Google Scholar] [CrossRef]
- Seo, S.; Katou, S.; Seto, H.; Gomi, K.; Ohashi, Y. The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 2007, 49, 899–909. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Itoch, M. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. 2005, 4, 1128. [Google Scholar] [CrossRef]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Li, N.; Ma, X.; Gupta, V.; Zhang, D.; Li, T.; Dai, X. The Ectopic Overexpression of the Cotton Ve1 and Ve2-Homolog Sequences Leads to Resistance Response to Verticillium Wilt in Arabidopsis. Front. Plant Sci. 2017, 8, 844. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Xing, M.; Lv, H.; Ma, J.; Xu, D.; Li, H.; Yang, L.; Kang, J.; Wang, X.; Fang, Z. Transcriptome profiling of resistance to Fusarium oxysporum f. sp.conglutinansin cabbage (Brassica oleracea) roots. PLoS ONE 2016, 11, 0148048. [Google Scholar] [CrossRef]
- Tena, G.; Asai, T.; Chiu, W.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Meng, X.; Xu, J.; He, Y.; Yang, K.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 2013, 25, 1126–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Popescu, S.; Popescu, G.; Bachan, S.; Zhang, Z.; Gerstein, M.; Snyder, M.; Dinesh-Kumar, S. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Gene. Dev. 2009, 23, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Diener, K.; Tan, T.; Yao, Z. MAPKKK6, a novel mitogen-activated protein kinase kinase kinase, that associates with MAPKKK5. Biochem. Bioph. Res. Commun. 1998, 253, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Yoshihiro, T.; Nasir, K.; Ito, A.; Kanzaki, H.; Matsumura, H.; Saitoh, H.; Fujisawa, S.; Kamoun, S.; Terauchi, R. A novel MAPKK involved in cell death and defense signaling. Plant Signal. Behav. 2007, 2, 396–398. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lee, S.; OH, S.; Seo, E.; Choi, D. HSP70s enhance a phytophthora infestans effector-induced cell death via an MAPK cascade in Nicotiana benthamiana. Mol. Plant Microbe Interact. 2018, 31, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Schwessinger, B.; Zipfel, C. News from the frontline: Recent insights into PAMP triggered immunity in plants. Curr. Opin. Plant Bio. 2008, 11, 389–395. [Google Scholar] [CrossRef]
- Li, S.; Zhou, X.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 2010, 29, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Dang, F.; Liu, Z.; Wang, X.; Eulgem, T.; Lai, Y.; Yu, L.; She, J.; Shi, Y.; Lin, J.; et al. CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 2013, 14, 131–144. [Google Scholar] [CrossRef]
- Shigenaga, A.; Berens, M.; Tsuda, K.; Argueso, C. Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Bio. 2017, 38, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Bio. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coolen, S.; Proietti, S.; Hickman, R.; Davila Olivas, N.H.; Huang, P.-P.; Van Verk, M.C.; Vanpelt, J.A.; Wittenberg, A.H.J.; De Vos, M.; Prins, M.; et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016, 86, 249–267. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T. Fine Mapping of Tomato Leaf Mould Resistance Gene Cf-19 and Screening of Resistance Response Genes. Ph.D. Thesis, Northeast Agricultural University Harbin, Harbin, China, 2016. [Google Scholar]
- Huang, C.; Cui, Y.; Weng, C.; Zabel, P.; Lindhout, P. Development of diagnostic PCR markers closely linked to the tomato powdery mildew resistance gene Ol-1 on chromosome 6 of tomato. Theor. Appl. Genet. 2000, 101, 918–924. [Google Scholar] [CrossRef]
- Faoro, F.; Maffi, D.; Cantu, D.; Iriti, M. Chemical-induced resistance against powdery mildew in barley: The effects of chitosan and benzothiadiazole. Biocontrol 2008, 53, 387–401. [Google Scholar] [CrossRef]
- Kumro, F.; O’Neil, E.; Ciernia, L.; Moraes, J.; Spencer, T.; Lucy, M. Scanning electron microscopy of the surface epithelium of the bovine endometrium. J. Dairy Sci. 2020, 103, 12083–12090. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.; Park, J.; Kim, J.; Kim, J.; Kim, Y.; Kim, J.; Kim, S. Comparison of the MGISEQ-2000 and Illumina HiSeq 4000 sequencing platforms for RNA sequencing. Genom. Inform. 2019, 17, 32. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Song, K.; Li, L.; Zhang, G. Bias and Correction in RNA-seq Data for Marine Species. Mar. Biotechnol. 2017, 19, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.F.; Xu, S.Y.; Long, J.P.; Wang, F.; Chen, Y.D. RNA-Sequence Analysis Reveals Differentially Expressed Genes (DEGs) in Patients Exhibiting Different Risks of Tumor Metastasis. Med. Sci. Monitor. 2017, 23, 2842–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdi, H. The Bonferonni and Sidák Corrections for Multiple Comparisons. Encycl. Meas. Stat. 2007, 3, 103–107. [Google Scholar]
- Adie, B.; Pérez-Pérez, J.; Pérez-Pérez, M.; Godoy, M.; Sánchez-Serrano, J.; Schmelz, E.; Solano, R. ABA is an essential signal for plant resistance topathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007, 19, 1665–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Llugany, M.; Martin, S.; Barceló, J.; Poschenrieder, C. Endogenous jasmonic and salicylic acids levels in the cd-hyperaccumulator Noccaea (Thlaspi) praecox exposed to fungal infection and/or mechanical stress. Plant Cell Rep. 2013, 32, 1243–1249. [Google Scholar] [CrossRef]
- Liu, G.; Liu, J.; Zhang, C.; You, X.; Zhao, T. Physiological and RNA-seq analyses provide insights into the response mechanism of the Cf-10-mediated resistance to Cladosporium fulvum infection in tomato. Plant Mol. Biol. 2018, 19, 15. [Google Scholar] [CrossRef]
- Thimm, O.; Blaesing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Muller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Rotenberg, D.; Thompson, T.; German, T. Willis DK. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 2006, 138, 49–59. [Google Scholar] [CrossRef]

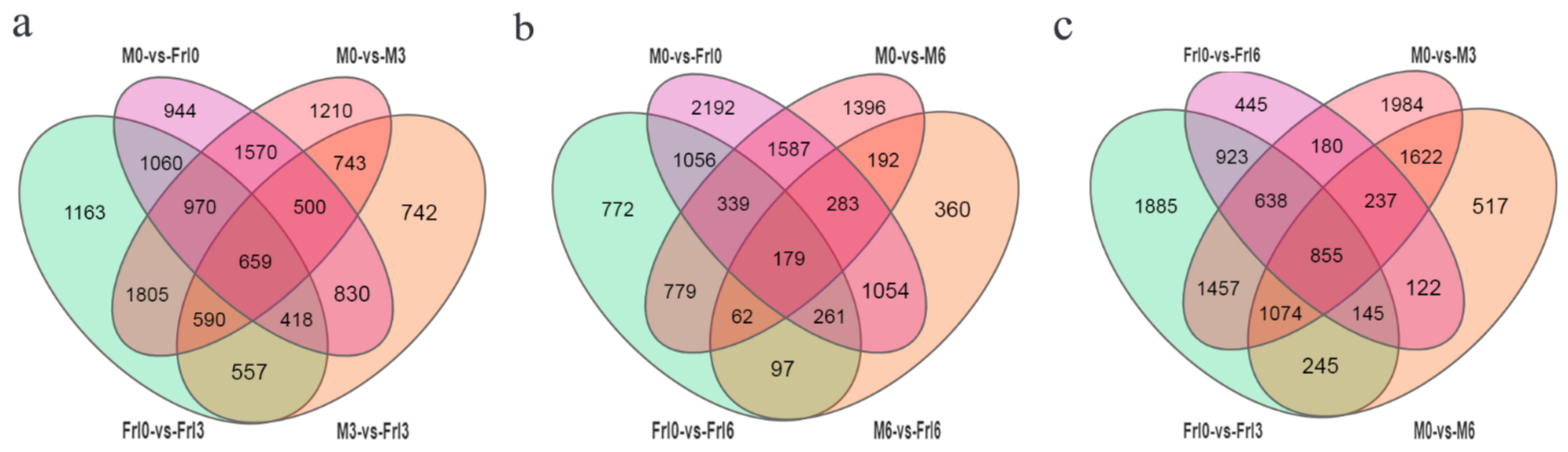


| DEG Set | Total DEGs | Upregulated | Downregulated |
|---|---|---|---|
| Frl0 vs. Frl3 | 7222 | 3374 | 3848 |
| Frl0 vs. Frl6 | 3545 | 1567 | 1978 |
| Frl3 vs. Frl6 | 4291 | 2398 | 1893 |
| M0 vs. M3 | 8047 | 3972 | 4075 |
| M0 vs. M6 | 4817 | 2316 | 2501 |
| M3 vs. M6 | 2944 | 1473 | 1471 |
| Frl0 vs. M0 | 6951 | 3487 | 3464 |
| Frl3 vs. M3 | 5039 | 2358 | 2681 |
| Frl6 vs. M6 | 2488 | 1222 | 1266 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Yang, H.; Li, J. Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) Infection in Tomato. Int. J. Mol. Sci. 2022, 23, 7078. https://doi.org/10.3390/ijms23137078
Sun Y, Yang H, Li J. Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) Infection in Tomato. International Journal of Molecular Sciences. 2022; 23(13):7078. https://doi.org/10.3390/ijms23137078
Chicago/Turabian StyleSun, Yuqing, Huanhuan Yang, and Jingfu Li. 2022. "Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) Infection in Tomato" International Journal of Molecular Sciences 23, no. 13: 7078. https://doi.org/10.3390/ijms23137078
APA StyleSun, Y., Yang, H., & Li, J. (2022). Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) Infection in Tomato. International Journal of Molecular Sciences, 23(13), 7078. https://doi.org/10.3390/ijms23137078





