Degradation Mechanism of Autophagy-Related Proteins and Research Progress
Abstract
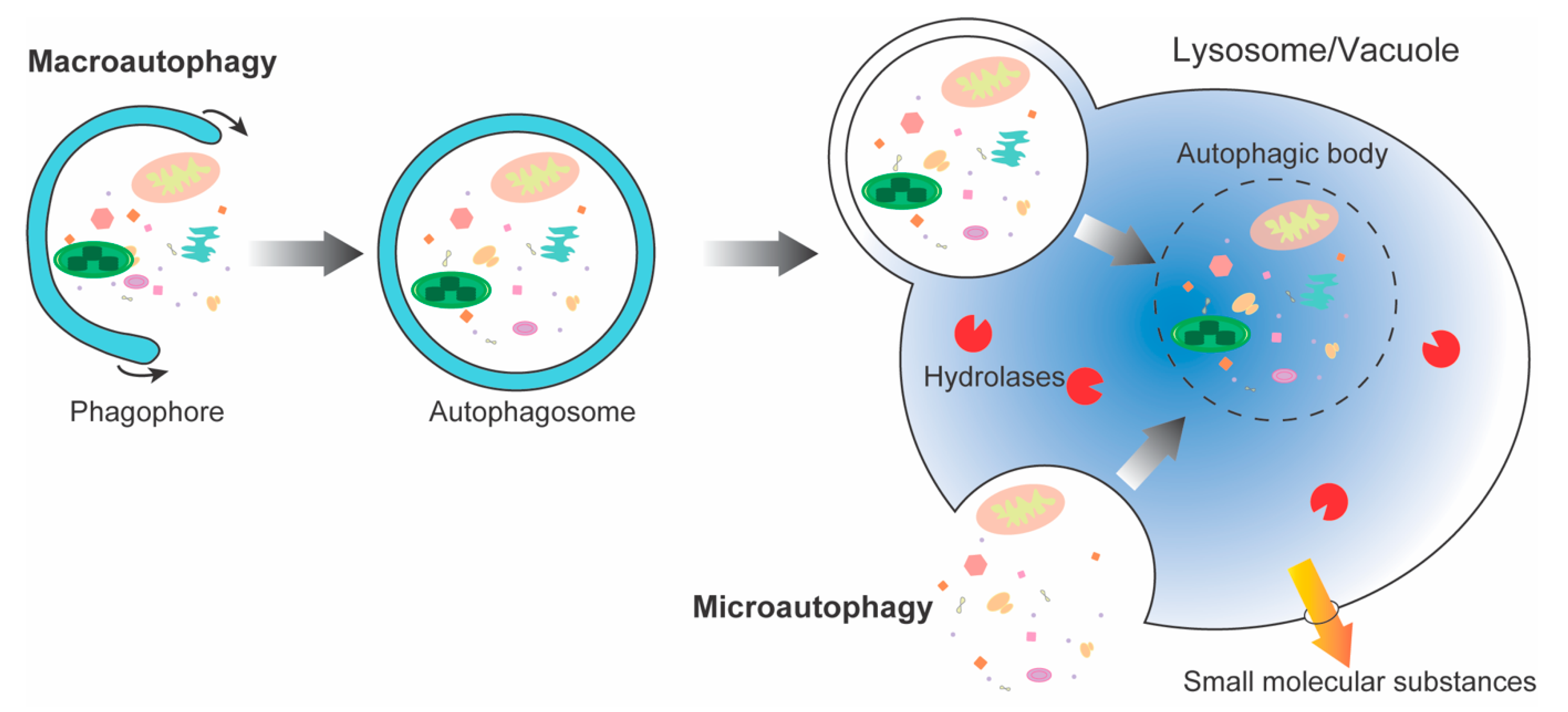
:1. Introduction
2. ATG Complexes in Plants and Animals
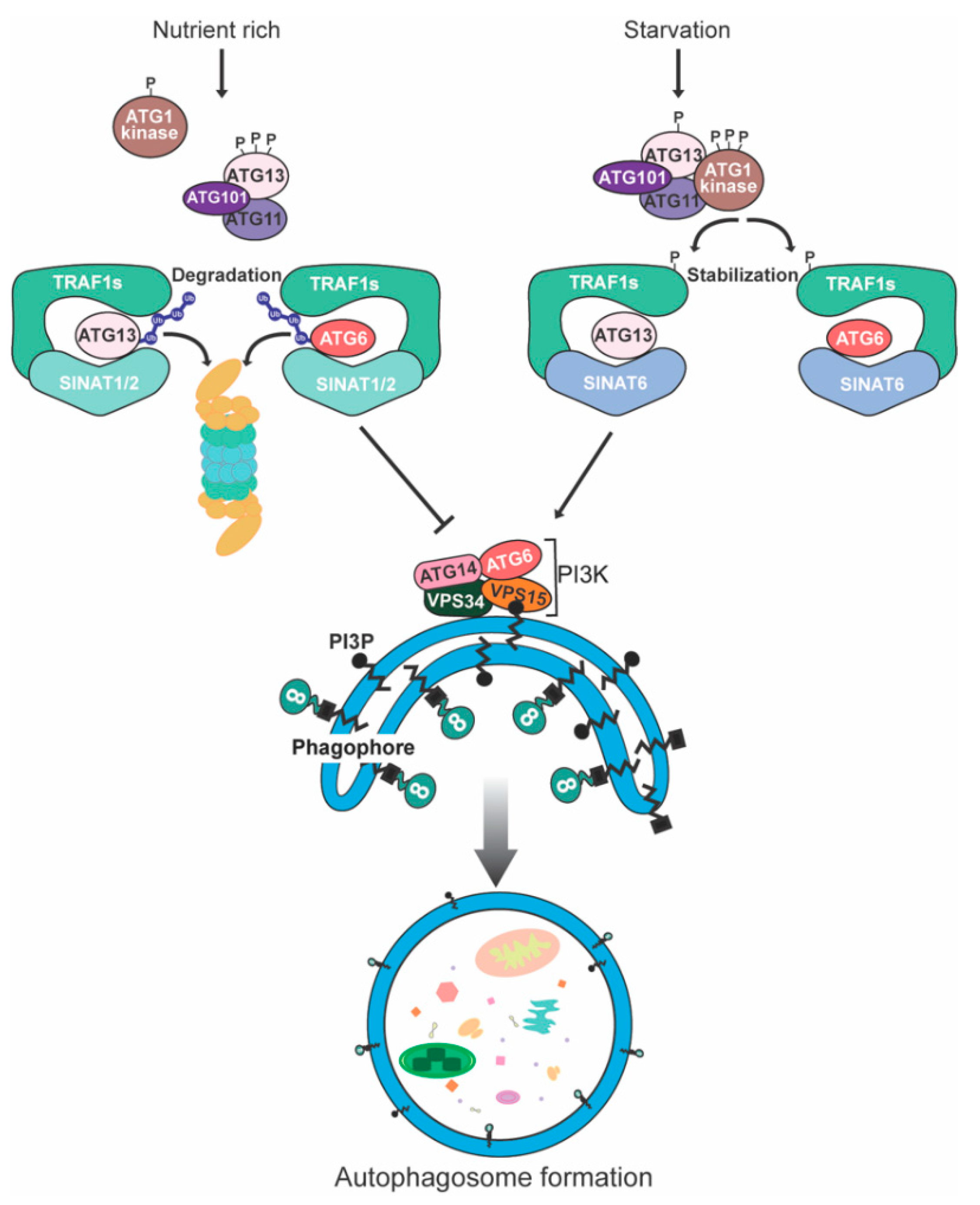
2.1. ATG1/ATG13 Protein Kinase Complex
2.2. ATG9/2/18 Transmembrane Complex
2.3. Phosphatidylinositol-3-Kinase (PI3K) Complex
2.4. ATG5/ATG12 and ATG8-Phosphatidylethanolamine (PE) Conjugation Systems
3. Protein Degradation Pathways in Eukaryotic Cells
4. Degradation of Yeast Autophagy-Related Proteins
5. Degradation of Autophagy-Related Proteins in Metazoa
5.1. Degradation of ULK Complex Components
5.2. Degradation of ATG9 Complex Components
5.3. Degradation of PI3K Complex Components
5.4. Degradation of the ATG12-Conjugation System Components
5.5. Degradation of the LC3-Conjugation System Components
6. Degradation of Plant Autophagy-Related Proteins
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarika, C.; Sreedevi, P.; Mishra, S.; Singh, S.N. Multifaceted Housekeeping Functions of Autophagy. J. Indian Inst. Sci. 2017, 97, 79–94. [Google Scholar]
- Huang, H.; Ayaz, A.; Zheng, M.; Yang, X.; Zaman, W.; Zhao, H.; Lü, S. Arabidopsis KCS5 and KCS6 Play Redundant Roles in Wax Synthesis. Int. J. Mol. Sci. 2022, 23, 4450. [Google Scholar] [CrossRef]
- van Doorn, W.G.; Papini, A. Ultrastructure of autophagy in plant cells: A review. Autophagy 2013, 9, 1922–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolliet, V.; Labonne, J.; Olazcuaga, L.; Panserat, S.; Seiliez, I. Modeling of autophagy-related gene expression dynamics during long term fasting in European eel (Anguilla anguilla). Sci. Rep. 2017, 7, 17896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamark, T.; Johansen, T. Mechanisms of selective autophagy. Annu. Rev. Cell Dev. Biol. 2021, 37, 143–169. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Vierstra, R.D. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014, 26, 788–807. [Google Scholar] [CrossRef] [Green Version]
- Marion, J.; Le Bars, R.; Besse, L.; Batoko, H.; Satiat-Jeunemaitre, B. Multiscale and Multimodal Approaches to Study Autophagy in Model Plants. Cells 2018, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef]
- Sieńko, K.; Poormassalehgoo, A.; Yamada, K.; Goto-Yamada, S. Microautophagy in Plants: Consideration of Its Molecular Mechanism. Cells 2020, 9, 887. [Google Scholar] [CrossRef] [Green Version]
- Liaquat, F.; Munis, M.F.H.; Arif, S.; Haroon, U.; Shi, J.; Saqib, S.; Zaman, W.; Che, S.; Liu, Q. PacBio single-molecule long-read sequencing reveals genes tolerating manganese stress in Schima superba saplings. Front. Genet. 2021, 12, 635043. [Google Scholar] [CrossRef]
- Ayaz, A.; Huang, H.; Zheng, M.; Zaman, W.; Li, D.; Saqib, S.; Zhao, H.; Lü, S. Molecular cloning and functional analysis of GmLACS2-3 reveals its involvement in cutin and suberin biosynthesis along with abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) role in plant development and coping with different stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Hussain, A.; Ullah, A.; Gul, S.; Shaban, M.; Khan, A.H.; Ali, M.; Sani, S.G.A.S.; Chaudhary, H.J.; Munis, M.F.H. Expression analysis of defense related genes in wheat and maize against Bipolaris sorokiniana. Physiol. Mol. Plant Pathol. 2018, 103, 36–46. [Google Scholar] [CrossRef]
- Ali, Q.; Yu, C.; Hussain, A.; Ali, M.; Ahmar, S.; Sohail, M.A.; Riaz, M.; Ashraf, M.F.; Abdalmegeed, D.; Wang, X. Genome engineering technology for durable disease resistance: Recent progress and future outlooks for sustainable agriculture. Front. Plant Sci. 2022, 13, 860281. [Google Scholar] [CrossRef]
- Ali, Q.; Zheng, H.; Rao, M.J.; Ali, M.; Hussain, A.; Saleem, M.H.; Nehela, Y.; Sohail, M.A.; Ahmed, A.M.; Kubar, K.A. Advances, limitations, and prospects of biosensing technology for detecting phytopathogenic bacteria. Chemosphere 2022, 296, 133773. [Google Scholar] [CrossRef]
- Liaquat, F.; Qunlu, L.; Arif, S.; Haroon, U.; Saqib, S.; Zaman, W.; Jianxin, S.; Shengquan, C.; Li, L.X.; Akbar, M. Isolation and characterization of pathogen causing brown rot in lemon and its control by using ecofriendly botanicals. Physiol. Mol. Plant Pathol. 2021, 114, 101639. [Google Scholar] [CrossRef]
- Senft, D.; Ronai, Z.A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 2015, 40, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Manghwar, H.; Li, J. Endoplasmic Reticulum Stress and Unfolded Protein Response Signaling in Plants. Int. J. Mol. Sci. 2022, 23, 828. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A. Mechanism of tobacco osmotin gene in plant responses to biotic and abiotic stress tolerance: A brief history. Biocell 2022, 46, 623. [Google Scholar] [CrossRef]
- Yim, W.W.-Y.; Mizushima, N. Lysosome biology in autophagy. Cell Discov. 2020, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Bu, F.; Huang, W.; Chen, L. Multiple Regulatory Levels Shape Autophagy Activity in Plants. Front. Plant Sci. 2019, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Luo, M.; Zhuang, X.; Li, F.; Gao, C. Transcriptional and epigenetic regulation of autophagy in plants. Trends Genet. 2020, 36, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Yao, Z.; Klionsky, D.J. How to control self-digestion: Transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015, 25, 354–363. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Wang, W.; Sun, X.; Xu, D.; Wang, C.; Zhang, Q.; Wang, H.; Luo, W.; Chen, Y.; Chen, H.; et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy 2016, 12, 1447–1459. [Google Scholar] [CrossRef] [Green Version]
- Laureano-Marín, A.M.; Aroca, Á.; Pérez-Pérez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic Acid-Triggered Persulfidation of the Cys Protease ATG4 Mediates Regulation of Autophagy by Sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade-an interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555. [Google Scholar] [CrossRef]
- Nanji, T.; Liu, X.; Chew, L.H.; Li, F.K.; Biswas, M.; Yu, Z.Q.; Lu, S.; Dong, M.Q.; Du, L.L.; Klionsky, D.J.; et al. Conserved and unique features of the fission yeast core Atg1 complex. Autophagy 2017, 13, 2018–2027. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The Master of Bulk and Selective Recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Rehman, N.U.; Zeng, P.; Mo, Z.; Guo, S.; Liu, Y.; Huang, Y.; Xie, Q. Conserved and Diversified Mechanism of Autophagy between Plants and Animals upon Various Stresses. Antioxidants 2021, 10, 1736. [Google Scholar] [CrossRef] [PubMed]
- Budovskaya, Y.V.; Stephan, J.S.; Reggiori, F.; Klionsky, D.J.; Herman, P.K. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20663–20671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.; Gao, C.; Kang, B.H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E426–E435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, U.; Cao, Y.; Xie, Z.; Klionsky, D.J. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 2010, 285, 11476–11488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Hu, W.; Li, F.; Marshall, R.S.; Zarza, X.; Munnik, T.; Vierstra, R.D. AUTOPHAGY-RELATED14 and Its Associated Phosphatidylinositol 3-Kinase Complex Promote Autophagy in Arabidopsis. Plant Cell 2020, 32, 3939–3960. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. The ATG4 protease integrates redox and stress signals to regulate autophagy. J. Exp. Bot. 2021, 72, 3340–3351. [Google Scholar] [CrossRef]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010, 62, 483–493. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef]
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786. [Google Scholar] [CrossRef]
- Nedelsky, N.B.; Todd, P.K.; Taylor, J.P. Autophagy and the ubiquitin-proteasome system: Collaborators in neuroprotection. Biochim. Biophys. Acta 2008, 1782, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Komander, D. Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 2010, 54, 69–87. [Google Scholar] [PubMed]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Xia, F.N.; Xie, L.J.; Yu, L.J.; Chen, Q.F.; Zhuang, X.H.; Wang, Q.; Li, F.; Jiang, L.; Xie, Q.; et al. TRAF Family Proteins Regulate Autophagy Dynamics by Modulating AUTOPHAGY PROTEIN6 Stability in Arabidopsis. Plant Cell 2017, 29, 890–911. [Google Scholar] [CrossRef] [Green Version]
- Su, T.; Yang, M.; Wang, P.; Zhao, Y.; Ma, C. Interplay between the Ubiquitin Proteasome System and Ubiquitin-Mediated Autophagy in Plants. Cells 2020, 9, 2219. [Google Scholar] [CrossRef]
- Jin, M.; He, D.; Backues, S.K.; Freeberg, M.A.; Liu, X.; Kim, J.K.; Klionsky, D.J. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr. Biol. 2014, 24, 1314–1322. [Google Scholar] [CrossRef] [Green Version]
- Smothers, D.B.; Kozubowski, L.; Dixon, C.; Goebl, M.G.; Mathias, N. The abundance of Met30p limits SCF(Met30p) complex activity and is regulated by methionine availability. Mol. Cell Biol. 2000, 20, 7845–7852. [Google Scholar] [CrossRef]
- Feng, Y.; Ariosa, A.R.; Yang, Y.; Hu, Z.; Dengjel, J.; Klionsky, D.J. Downregulation of autophagy by Met30-mediated Atg9 ubiquitination. Proc. Natl. Acad. Sci. USA 2021, 118, e2005539118. [Google Scholar] [CrossRef]
- Hu, G.; Rios, L.; Yan, Z.; Jasper, A.M.; Luera, D.; Luo, S.; Rao, H. Autophagy regulator Atg9 is degraded by the proteasome. Biochem. Biophys. Res. Commun. 2020, 522, 254–258. [Google Scholar] [CrossRef]
- Kanki, T.; Wang, K.; Cao, Y.; Baba, M.; Klionsky, D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 2009, 17, 98–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012, 14, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Tian, W.; Hu, Z.; Chen, G.; Huang, L.; Li, W.; Zhang, X.; Xue, P.; Zhou, C.; Liu, L.; et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014, 15, 566–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levchenko, M.; Lorenzi, I.; Dudek, J. The Degradation Pathway of the Mitophagy Receptor Atg32 Is Re-Routed by a Posttranslational Modification. PLoS ONE 2016, 11, e0168518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camougrand, N.; Vigié, P.; Gonzalez, C.; Manon, S.; Bhatia-Kiššová, I. The yeast mitophagy receptor Atg32 is ubiquitinated and degraded by the proteasome. PLoS ONE 2020, 15, e0241576. [Google Scholar] [CrossRef]
- Liu, C.C.; Lin, Y.C.; Chen, Y.H.; Chen, C.M.; Pang, L.Y.; Chen, H.A.; Wu, P.R.; Lin, M.Y.; Jiang, S.T.; Tsai, T.F.; et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol. Cell 2016, 61, 84–97. [Google Scholar] [CrossRef]
- Deng, R.; Zhang, H.L.; Huang, J.H.; Cai, R.Z.; Wang, Y.; Chen, Y.H.; Hu, B.X.; Ye, Z.P.; Li, Z.L.; Mai, J.; et al. MAPK1/3 kinase-dependent ULK1 degradation attenuates mitophagy and promotes breast cancer bone metastasis. Autophagy 2021, 17, 3011–3029. [Google Scholar] [CrossRef]
- Shi, J.H.; Ling, C.; Wang, T.T.; Zhang, L.N.; Liu, W.W.; Qin, Y.; Tan, Y.H.; Cui, N.P.; Ni, Z.Y. TRK-fused gene (TFG) regulates ULK1 stability via TRAF3-mediated ubiquitination and protects macrophages from LPS-induced pyroptosis. Cell Death Dis. 2022, 13, 93. [Google Scholar] [CrossRef]
- Nazio, F.; Carinci, M.; Valacca, C.; Bielli, P.; Strappazzon, F.; Antonioli, M.; Ciccosanti, F.; Rodolfo, C.; Campello, S.; Fimia, G.M.; et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 2016, 215, 841–856. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, J.; Shin, J.; Kang, Y.; Choi, J.; Cheong, H. ATG101 Degradation by HUWE1-Mediated Ubiquitination Impairs Autophagy and Reduces Survival in Cancer Cells. Int. J. Mol. Sci. 2021, 22, 9182. [Google Scholar] [CrossRef]
- Wan, W.; You, Z.; Zhou, L.; Xu, Y.; Peng, C.; Zhou, T.; Yi, C.; Shi, Y.; Liu, W. mTORC1-Regulated and HUWE1-Mediated WIPI2 Degradation Controls Autophagy Flux. Mol. Cell 2018, 72, 303–315.e6. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Dong, K.; Liang, W.; Xu, D.; Xia, H.; Geng, J.; Najafov, A.; Liu, M.; Li, Y.; Han, X.; et al. G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. eLife 2015, 4, e06734. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Feng, K.; Zhao, X.; Huang, S.; Cheng, Y.; Qian, L.; Wang, Y.; Sun, H.; Jin, M.; Chuang, T.H.; et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy 2014, 10, 2239–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Yang, K.B.; Chen, W.; Mai, J.; Wu, X.Q.; Sun, T.; Wu, R.Y.; Jiao, L.; Li, D.D.; Ji, J.; et al. CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy 2021, 17, 4323–4340. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.S.; Kehrl, J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010, 3, ra42. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Huang, T.Y.; Lin, Y.T.; Lin, S.Y.; Li, W.H.; Hsiao, H.J.; Yan, R.L.; Tang, H.W.; Shen, Z.Q.; Chen, G.C.; et al. VPS34 K29/K48 branched ubiquitination governed by UBE3C and TRABID regulates autophagy, proteostasis and liver metabolism. Nat. Commun. 2021, 12, 1322. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhang, T.; Xu, D.; Wang, H.; Cai, Y.; Jin, T.; Liu, M.; Jin, M.; Wu, K.; Yuan, J. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015, 29, 184–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scrivo, A.; Codogno, P.; Bomont, P. Gigaxonin E3 ligase governs ATG16L1 turnover to control autophagosome production. Nat. Commun. 2019, 10, 780. [Google Scholar] [CrossRef]
- Jia, R.; Bonifacino, J.S. Negative regulation of autophagy by UBA6-BIRC6-mediated ubiquitination of LC3. eLife 2019, 8, e50034. [Google Scholar] [CrossRef]
- Ma, K.; Fu, W.; Tang, M.; Zhang, C.; Hou, T.; Li, R.; Lu, X.; Wang, Y.; Zhou, J.; Li, X.; et al. PTK2-mediated degradation of ATG3 impedes cancer cells susceptible to DNA damage treatment. Autophagy 2017, 13, 579–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, H.; Li, J.; Xia, F.N.; Chen, J.Y.; Lei, X.; Han, M.Q.; Xie, L.J.; Zhou, Q.M.; Xiao, S. Arabidopsis SINAT Proteins Control Autophagy by Mediating Ubiquitylation and Degradation of ATG13. Plant Cell 2020, 32, 263–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, J.X.; Raffeiner, M.; Spinti, D.; Langin, G.; Franz-Wachtel, M.; Guzman, A.R.; Kim, J.G.; Pandey, P.; Minina, A.E.; Macek, B.; et al. A bacterial effector counteracts host autophagy by promoting degradation of an autophagy component. EMBO J. 2022, e110352. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; White, E.; Rubinsztein, D.C. Breakthroughs and bottlenecks in autophagy research. Trends Mol. Med. 2021, 27, 835–838. [Google Scholar] [CrossRef]
- Kim, J.H.; Seo, D.; Kim, S.J.; Choi, D.W.; Park, J.S.; Ha, J.; Choi, J.; Lee, J.H.; Jung, S.M.; Seo, K.W.; et al. The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation. EMBO Rep. 2018, 19, e44378. [Google Scholar] [CrossRef]
- Hosokawa, N.; Sasaki, T.; Iemura, S.; Natsume, T.; Hara, T.; Mizushima, N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009, 5, 973–979. [Google Scholar] [CrossRef] [Green Version]
- Mercer, C.A.; Kaliappan, A.; Dennis, P.B. A novel, hu...uman Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009, 5, 649–662. [Google Scholar] [CrossRef] [Green Version]
- Kao, S.H.; Wu, H.T.; Wu, K.J. Ubiquitination by HUWE1 in tumorigenesis and beyond. J. Biomed. Sci. 2018, 25, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.G.; Chen, Y.; Miao, G.; Zhao, H.; Qu, W.; Li, D.; Wang, Z.; Liu, N.; Li, L.; Chen, S.; et al. The ER-Localized Transmembrane Protein EPG-3/VMP1 Regulates SERCA Activity to Control ER-Isolation Membrane Contacts for Autophagosome Formation. Mol. Cell 2017, 67, 974–989.e6. [Google Scholar] [CrossRef] [Green Version]
- Thurston, T.L.; Boyle, K.B.; Allen, M.; Ravenhill, B.J.; Karpiyevich, M.; Bloor, S.; Kaul, A.; Noad, J.; Foeglein, A.; Matthews, S.A.; et al. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. EMBO J. 2016, 35, 1779–1792. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Shizukuishi, S.; Ogawa, M.; Ryo, A.; Ohnishi, M. Streptococcus pneumoniae promotes its own survival via choline-binding protein CbpC-mediated degradation of ATG14. Autophagy 2020, 16, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef]
- Funderburk, S.F.; Wang, Q.J.; Yue, Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Han, T.; Guo, M.; Gan, M.; Yu, B.; Tian, X.; Wang, J.B. TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy 2018, 14, 2035–2048. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Tian, S.; Chen, Y.; Zhang, C.; Xie, W.; Xia, X.; Cui, J.; Wang, R.F. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016, 35, 866–880. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Ma, Y.; He, H.W.; Wang, J.P.; Jiang, J.D.; Shao, R.G. SLC9A3R1 stimulates autophagy via BECN1 stabilization in breast cancer cells. Autophagy 2015, 11, 2323–2334. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Young, L.N. Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Jin, S.; Wu, Y.; Xian, H.; Tian, S.; Liu, D.A.; Guo, Z.; Cui, J. Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. Cell Rep. 2020, 30, 2807–2819.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, M.; Li, L.; Chen, S.; Wang, X. Ubiquitination of the PI3-kinase VPS-34 promotes VPS-34 stability and phagosome maturation. J. Cell Biol. 2018, 217, 347–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef]
- Tang, J.; Sun, F.; Deng, X.J.; Ma, Y.L.; Li, K.Y.; Tang, Y.; Chen, G.J. Ubiquitination status does not affect Vps34 degradation. Genes Dis. 2020, 7, 401–407. [Google Scholar] [CrossRef]
- van der Veen, A.G.; Ploegh, H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012, 81, 323–357. [Google Scholar] [CrossRef]
- Haller, M.; Hock, A.K.; Giampazolias, E.; Oberst, A.; Green, D.R.; Debnath, J.; Ryan, K.M.; Vousden, K.H.; Tait, S.W. Ubiquitination and proteasomal degradation of ATG12 regulates its proapoptotic activity. Autophagy 2014, 10, 2269–2278. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, Y.; Lu, H.; Xiang, Y.; Huang, X.; Hu, R.; Chen, Z.; Yuan, W.; Peng, R.; Peng, J.; et al. Spodoptera litura autophagy-related protein 1 interacts with autophagy-related protein 5 and enhances its degradation. Insect Mol. Biol. 2017, 26, 190–203. [Google Scholar] [CrossRef]
- Xie, X.; Bi, H.L.; Lai, S.; Zhang, Y.L.; Li, N.; Cao, H.J.; Han, L.; Wang, H.X.; Li, H.H. The immunoproteasome catalytic β5i subunit regulates cardiac hypertrophy by targeting the autophagy protein ATG5 for degradation. Sci. Adv. 2019, 5, eaau0495. [Google Scholar] [CrossRef] [Green Version]
- Mallik, A.; Yammani, R.R. Saturated fatty acid palmitate negatively regulates autophagy by promoting ATG5 protein degradation in meniscus cells. Biochem. Biophys. Res. Commun. 2018, 502, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117 Pt 13, 2805–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanida, I.; Sou, Y.S.; Minematsu-Ikeguchi, N.; Ueno, T.; Kominami, E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006, 273, 2553–2562. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.X.; Zou, J.B.; Zhu, Q.Q.; Liu, C.H.; Wang, G.F.; Du, T.T.; Luo, Z.Y.; Guo, F.; Zhou, L.M.; Liu, J.J.; et al. SIP/CacyBP promotes autophagy by regulating levels of BRUCE/Apollon, which stimulates LC3-I degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 13404–13413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Levine, B. Autosis and autophagic cell death: The dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhang, J.; Li, Y.Q.; Liu, Q.X.; Zhou, D.; Deng, X.F.; Qiu, Y.; Chen, Q.; Li, M.Y.; Liu, X.Q.; et al. Plasmodium Circumsporozoite Protein Enhances the Efficacy of Gefitinib in Lung Adenocarcinoma Cells by Inhibiting Autophagy via Proteasomal Degradation of LC3B. Front. Cell Dev. Biol. 2022, 10, 830046. [Google Scholar] [CrossRef]
- Besteiro, S.; Brooks, C.F.; Striepen, B.; Dubremetz, J.F. Autophagy protein Atg3 is essential for maintaining mitochondrial integrity and for normal intracellular development of Toxoplasma gondii tachyzoites. PLoS Pathog. 2011, 7, e1002416. [Google Scholar] [CrossRef] [Green Version]
- Metlagel, Z.; Otomo, C.; Takaesu, G.; Otomo, T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc. Natl. Acad. Sci. USA 2013, 110, 18844–18849. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [Green Version]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Wang, C.; Jiang, C.; Pan, J.; Zhang, G.; Liu, H.; Zhang, H. The tumor necrosis factor receptor-associated factor (TRAF)-like family protein SEVEN IN ABSENTIA 2 (SINA2) promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytol. 2014, 202, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Gao, W.; Chen, Q.F.; Chan, S.W.; Zheng, S.X.; Ma, J.; Wang, M.; Welti, R.; Chye, M.L. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 2010, 22, 1463–1482. [Google Scholar] [CrossRef] [Green Version]
- Leary, A.Y.; Sanguankiattichai, N.; Duggan, C.; Tumtas, Y.; Pandey, P.; Segretin, M.E.; Linares, J.S.; Savage, Z.D.; Yow, R.J.; Bozkurt, T.O. Modulation of plant autophagy during pathogen attack. J. Exp. Bot. 2018, 69, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Seto, D.; Subramaniam, R.; Desveaux, D. Oh, the places they’ll go! A survey of phytopathogen effectors and their host targets. Plant J. 2018, 93, 651–663. [Google Scholar] [CrossRef]
- Zhuang, X.; Jiang, L. Autophagosome biogenesis in plants: Roles of SH3P2. Autophagy 2014, 10, 704–705. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Wang, T.; Han, J.; Li, M.; Zhao, Y.; Su, T.; Ma, C. Plant Autophagy: An Intricate Process Controlled by Various Signaling Pathways. Front. Plant Sci. 2021, 12, 754982. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef] [Green Version]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Sedaghatmehr, M.; Thirumalaikumar, V.P.; Kamranfar, I.; Marmagne, A.; Masclaux-Daubresse, C.; Balazadeh, S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019, 42, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [Green Version]
- Deretic, V. Autophagy as an innate immunity paradigm: Expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 2012, 24, 21–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Eukaryotes | ATGs | E3 Ligases | Ubiquitin Chain Types | References |
|---|---|---|---|---|
| Yeast | Atg9 | Met30 | Unknown | [49] |
| Atg32 | Rsp5 | Unknown | [55] | |
| Mammalian | ULK1 | CUL3-KLHL20 | K48 | [56] |
| BTRC | K48 | [57] | ||
| TRAF3 | K48 | [58] | ||
| NEDD4L | K27 and K29 | [59] | ||
| ATG101 | HUWE1 | K48 | [60] | |
| WIPI2 | HUWE1 | Unknown | [61] | |
| ATG14L | ZBTB16-CUL3-Roc1 | Unknown | [62] | |
| BECN1 | NEDD4 | K11 | [63] | |
| RNF216 | K48 | [64] | ||
| CUL3-KLHL20 | Unknown | [65] | ||
| CUL3-KLHL38 | K48 | [65] | ||
| TRAF6 | K63 | [66] | ||
| VPS34 | UBE3C | K29/K48 branched | [67] | |
| FBXL20-CUL1-SKP1 | Unknown | [68] | ||
| ATG16L1 | Gigaxonin | K48 | [69] | |
| LC3B | BIRC6 | Single ubiquitin | [70] | |
| ATG3 | PTK2 | Unknown | [71] | |
| Plants | ATG6 | SINAT1/2 | Unknown | [45] |
| ATG13 | SINAT1/2 | K48 | [72] | |
| SH3P2 | XopL | Unknown | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Manghwar, H.; Hu, W.; Liu, F. Degradation Mechanism of Autophagy-Related Proteins and Research Progress. Int. J. Mol. Sci. 2022, 23, 7301. https://doi.org/10.3390/ijms23137301
Zhou Y, Manghwar H, Hu W, Liu F. Degradation Mechanism of Autophagy-Related Proteins and Research Progress. International Journal of Molecular Sciences. 2022; 23(13):7301. https://doi.org/10.3390/ijms23137301
Chicago/Turabian StyleZhou, Yanhui, Hakim Manghwar, Weiming Hu, and Fen Liu. 2022. "Degradation Mechanism of Autophagy-Related Proteins and Research Progress" International Journal of Molecular Sciences 23, no. 13: 7301. https://doi.org/10.3390/ijms23137301
APA StyleZhou, Y., Manghwar, H., Hu, W., & Liu, F. (2022). Degradation Mechanism of Autophagy-Related Proteins and Research Progress. International Journal of Molecular Sciences, 23(13), 7301. https://doi.org/10.3390/ijms23137301








