Phosphomimicry on STAU1 Serine 20 Impairs STAU1 Posttranscriptional Functions and Induces Apoptosis in Human Transformed Cells
Abstract
:1. Introduction
2. Results
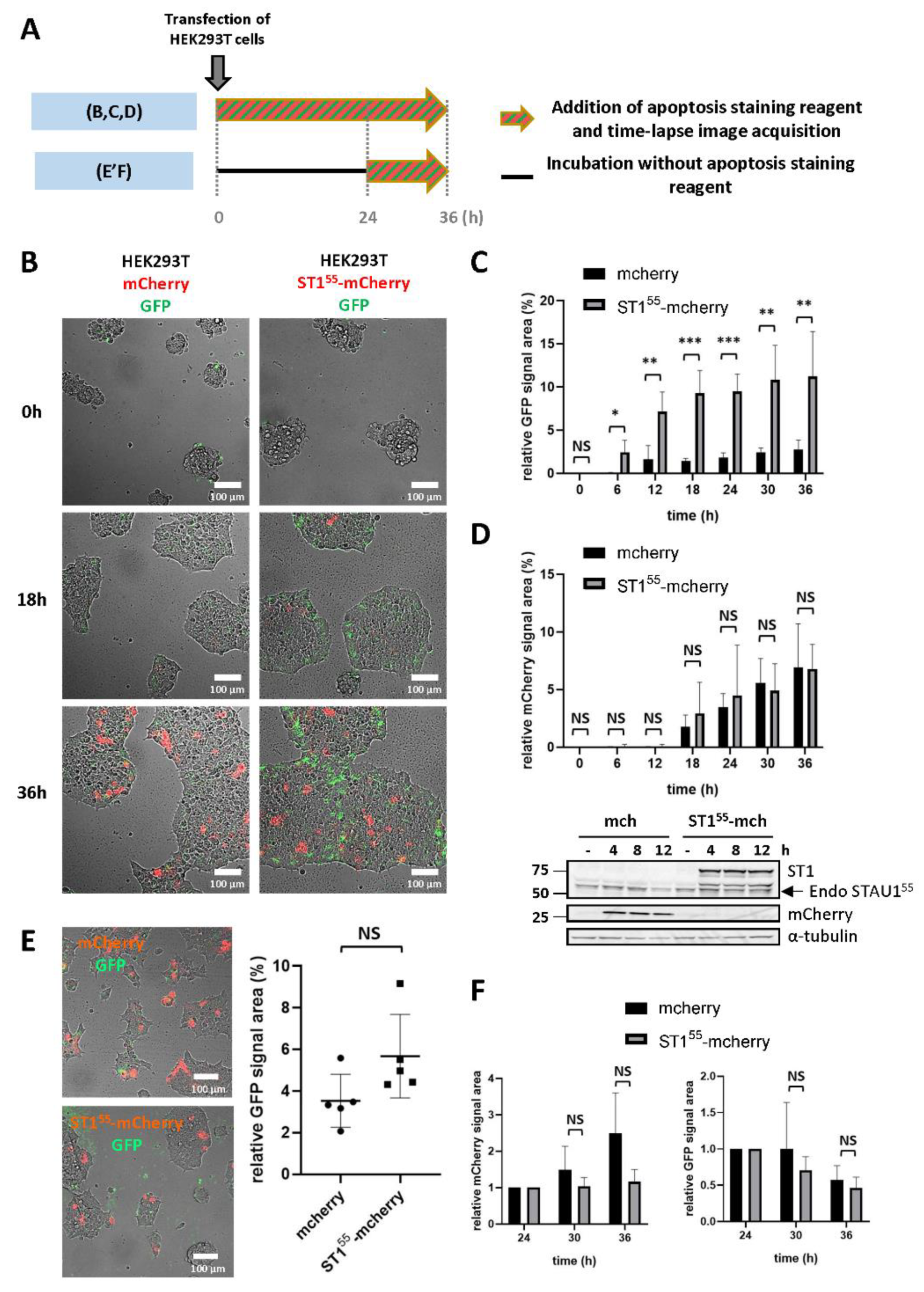
2.1. STAU155 Overexpression Causes a Fast-Acting Apoptosis Response in Transformed Cells
2.2. Amino Acids 18–25 Carry the Molecular Determinant That Impairs Cell Proliferation
2.3. Phosphomimicry on S20 and T21 Controls Cell Proliferation
2.4. S20 Phosphomimicry Controls STAU155-Mediated Post-Transcriptional Regulation
2.5. RBD2 Expression Is Sufficient to Impair Cell Proliferation and to Induce Apoptosis
2.6. RBD2 Interferes with Endogenous STAU155 to Impair SMD
3. Discussion
3.1. STAU155 Overexpression Impairs Cell Proliferation and Triggers Apoptosis of Transformed Cells
3.2. STAU155 Regulates Cell Proliferation through Modifications of Serine 20/Threonine 21
3.3. Expression of RBD2 Alone Impairs Cell Proliferation and Triggers Apoptosis
3.4. Serine 20 Regulates STAU155 Posttranscriptional Functions
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Plasmids and Cloning Strategies
4.3. Time Lapse Microscopy and Image Analysis
4.4. Antibodies and Reagents
4.5. Western Blot Analysis
4.6. RNA Isolation and RT-qPCR
4.7. Gene Expression Assays
4.8. Growth Curve Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gookin, S.; Min, M.; Phadke, H.; Chung, M.; Moser, J.; Miller, I.; Carter, D.; Spencer, S.L. A map of protein dynamics during cell-cycle progression and cell-cycle exit. PLoS Biol. 2017, 15, e2003268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visconti, R.; Della Monica, R.; Grieco, D. Cell cycle checkpoint in cancer: A therapeutically targetable double-edged sword. J. Exp. Clin. Cancer Res. 2016, 35, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harper, J.W.; Bennett, E.J. Proteome complexity and the forces that drive proteome imbalance. Nature 2016, 537, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbett, A.H. Post-transcriptional regulation of gene expression and human disease. Curr. Opin. Cell Biol. 2018, 52, 96–104. [Google Scholar] [CrossRef] [PubMed]
- El Hiani, Y.; Egom, E.E.; Dong, X.P. mTOR signalling: Jack-of-all-trades. Biochem Cell Biol 2019, 97, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Kontomanolis, E.N.; Koutras, A.; Syllaios, A.; Schizas, D.; Mastoraki, A.; Garmpis, N.; Diakosavvas, M.; Angelou, K.; Tsatsaris, G.; Pagkalos, A.; et al. Role of Oncogenes and Tumor-suppressor Genes in Carcinogenesis: A Review. Anticancer Res. 2020, 40, 6009–6015. [Google Scholar] [CrossRef]
- Liu, X.L.; Ding, J.; Meng, L.H. Oncogene-induced senescence: A double edged sword in cancer. Acta Pharmacol. Sin. 2018, 39, 1553–1558. [Google Scholar] [CrossRef]
- Jan, R.; Chaudhry, G.E. Understanding Apoptosis and Apoptotic Pathways Targeted Cancer Therapeutics. Adv. Pharm. Bull. 2019, 9, 205–218. [Google Scholar] [CrossRef] [Green Version]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 2016, 8, 603–619. [Google Scholar] [CrossRef] [Green Version]
- Wickham, L.; Duchaine, T.; Luo, M.; Nabi, I.R.; DesGroseillers, L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999, 19, 2220–2230. [Google Scholar] [CrossRef] [Green Version]
- Marion, R.M.; Fortes, P.; Beloso, A.; Dotti, C.; Ortin, J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999, 19, 2212–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duchaine, T.; Wang, H.J.; Luo, M.; Steinberg, S.V.; Nabi, I.R.; DesGroseillers, L. A novel murine Staufen isoform modulates the RNA content of Staufen complexes. Mol. Cell. Biol. 2000, 20, 5592–5601. [Google Scholar] [CrossRef] [Green Version]
- Bonnet-Magnaval, F.; DesGroseillers, L. The Staufen1-dependent cell cycle regulon or how a misregulated RNA-binding protein leads to cancer. Biol. Rev. Camb. Philos Soc. 2021, 96, 2192–2208. [Google Scholar] [CrossRef] [PubMed]
- Furic, L.; Maher-Laporte, M.; DesGroseillers, L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 2008, 14, 324–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, E.P.; Kucukural, A.; Cenik, C.; Mercier, B.C.; Singh, G.; Heyer, E.E.; Ashar-Patel, A.; Peng, L.; Moore, M.J. Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol. 2014, 21, 26–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Lucas, S.; Oliveros, J.C.; Chagoyen, M.; Ortin, J. Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Res. 2014, 42, 4516–4526. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef] [Green Version]
- Zheng, D.; Cho, H.; Wang, W.; Rambout, X.; Tian, B.; Maquat, L.E. 3′READS + RIP defines differential Staufen1 binding to alternative 3′UTR isoforms and reveals structures and sequence motifs influencing binding and polysome association. RNA 2020, 26, 1621–1636. [Google Scholar] [CrossRef]
- Sossin, W.S.; DesGroseillers, L. Intracellular trafficking of RNA in neurons. Traffic 2006, 7, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Kohrmann, M.; Luo, M.; Kaether, C.; DesGroseillers, L.; Dotti, C.G.; Kiebler, M.A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 1999, 10, 2945–2953. [Google Scholar] [CrossRef]
- Ravel-Chapuis, A.; Belanger, G.; Yadava, R.S.; Mahadevan, M.S.; DesGroseillers, L.; Cote, J.; Jasmin, B.J. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012, 196, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Dugre-Brisson, S.; Elvira, G.; Boulay, K.; Chatel-Chaix, L.; Mouland, A.J.; DesGroseillers, L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005, 33, 4797–4812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.K.; Furic, L.; Desgroseillers, L.; Maquat, L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 2005, 120, 195–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, M.; Duchaine, T.F.; DesGroseillers, L. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem. J. 2002, 365, 817–824. [Google Scholar] [CrossRef] [Green Version]
- Ramos, H.; Monette, A.; Niu, M.; Barrera, A.; Lopez-Ulloa, B.; Fuentes, Y.; Guizar, P.; Pino, K.; DesGroseillers, L.; Mouland, A.J.; et al. The double-stranded RNA-binding protein, Staufen1, is an IRES-transacting factor regulating HIV-1 cap-independent translation initiation. Nucleic Acids Res. 2021, 50, 411–429. [Google Scholar] [CrossRef]
- Sanchez-Carbente, M.; DesGroseillers, L. Understanding the importance of mRNA transport in memory. Prog. Brain Res. 2008, 169, 41–58. [Google Scholar]
- Heraud-Farlow, J.E.; Sharangdhar, T.; Li, X.; Pfeifer, P.; Tauber, S.; Orozco, D.; Hoermann, A.; Thomas, S.; Bakosova, A.; Farlow, A.R.; et al. Staufen2 Regulates Neuronal Target RNAs. Cell Rep. 2013, 5, 1511–1518. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Gleghorn, M.L.; Maquat, L.E. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. USA 2013, 110, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Almasi, S.; Jasmin, B.J. The multifunctional RNA-binding protein Staufen1: An emerging regulator of oncogenesis through its various roles in key cellular events. Cell Mol. Life Sci. 2021, 78, 7145–7160. [Google Scholar] [CrossRef]
- Luo, W.; Slebos, R.J.; Hill, S.; Li, M.; Brabek, J.; Amanchy, R.; Chaerkady, R.; Pandey, A.; Ham, A.J.; Hanks, S.K. Global impact of oncogenic Src on a phosphotyrosine proteome. J. Proteome Res. 2008, 7, 3447–3460. [Google Scholar] [CrossRef] [Green Version]
- Ghram, M.; Bonnet-Magnaval, F.; Hotea, D.I.; Doran, B.; Ly, S.; DesGroseillers, L. Staufen1 is Essential for Cell-Cycle Transitions and Cell Proliferation Via the Control of E2F1 Expression. J. Mol. Biol. 2020, 432, 3881–3897. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Liu, X.; Zheng, J.; Song, J.; Liu, Y.; Ruan, X.; Shen, S.; Shao, L.; Yang, C.; Wang, D.; et al. Lin28A promotes IRF6-regulated aerobic glycolysis in glioma cells by stabilizing SNHG14. Cell Death Dis 2020, 11, 447. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.; Zheng, J.; Liu, X.; Liu, Y.; Liu, L.; Ma, J.; He, Q.; Yang, C.; Wang, D.; Cai, H.; et al. lncRNA LINC00665 Stabilized by TAF15 Impeded the Malignant Biological Behaviors of Glioma Cells via STAU1-Mediated mRNA Degradation. Mol. Ther. Nucleic Acids 2020, 20, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Marcellus, K.A.; Crawford Parks, T.E.; Almasi, S.; Jasmin, B.J. Distinct roles for the RNA-binding protein Staufen1 in prostate cancer. BMC Cancer 2021, 21, 120. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [Green Version]
- Jing, F.; Ruan, X.; Liu, X.; Yang, C.; Wang, D.; Zheng, J.; Xue, Y.; Shen, S.; Shao, L.; Yang, Y.; et al. The PABPC5/HCG15/ZNF331 Feedback Loop Regulates Vasculogenic Mimicry of Glioma via STAU1-Mediated mRNA Decay. Mol. Ther. Oncolytics 2020, 17, 216–231. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Wang, J.; Chen, R.; Li, J. A novel 12-gene signature as independent prognostic model in stage IA and IB lung squamous cell carcinoma patients. Clin. Transl. Oncol. 2021, 23, 2368–2381. [Google Scholar] [CrossRef]
- Gyorffy, B.; Surowiak, P.; Budczies, J.; Lanczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [Green Version]
- Nagy, A.; Lanczky, A.; Menyhart, O.; Gyorffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef]
- Bonnet-Magnaval, F.; Diallo, L.H.; Brunchault, V.; Laugero, N.; Morfoisse, F.; David, F.; Roussel, E.; Nougue, M.; Zamora, A.; Marchaud, E.; et al. High Level of Staufen1 Expression Confers Longer Recurrence Free Survival to Non-Small Cell Lung Cancer Patients by Promoting THBS1 mRNA Degradation. Int. J. Mol. Sci. 2021, 23, 215. [Google Scholar] [CrossRef]
- Boulay, K.; Ghram, M.; Viranaicken, W.; Trepanier, V.; Mollet, S.; Frechina, C.; DesGroseillers, L. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res. 2014, 42, 7867–7883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damas, N.D.; Marcatti, M.; Come, C.; Christensen, L.L.; Nielsen, M.M.; Baumgartner, R.; Gylling, H.M.; Maglieri, G.; Rundsten, C.F.; Seemann, S.E.; et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat. Commun. 2016, 7, 13875. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Shiromoto, Y.; Ota, H.; Song, C.Z.; Kossenkov, A.V.; Wickramasinghe, J.; Showe, L.C.; Skordalakes, E.; Tang, H.Y.; Speicher, D.W.; et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017, 24, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Gandelman, M.; Dansithong, W.; Figueroa, K.P.; Paul, S.; Scoles, D.R.; Pulst, S.M. Staufen 1 amplifies proapoptotic activation of the unfolded protein response. Cell Death Differ. 2020, 27, 2942–2951. [Google Scholar] [CrossRef]
- Su, R.; Ma, J.; Zheng, J.; Liu, X.; Liu, Y.; Ruan, X.; Shen, S.; Yang, C.; Wang, D.; Cai, H.; et al. PABPC1-induced stabilization of BDNF-AS inhibits malignant progression of glioblastoma cells through STAU1-mediated decay. Cell Death Dis. 2020, 11, 81. [Google Scholar] [CrossRef]
- Hassine, S.; Bonnet-Magnaval, F.; Benoit Bouvrette, L.P.; Doran, B.; Ghram, M.; Bouthillette, M.; Lecuyer, E.; DesGroseillers, L. Staufen1 localizes to the mitotic spindle and controls the localization of RNA populations to the spindle. J. Cell Sci. 2020, 133, jcs247155. [Google Scholar] [CrossRef]
- Martel, C.; Dugre-Brisson, S.; Boulay, K.; Breton, B.; Lapointe, G.; Armando, S.; Trepanier, V.; Duchaine, T.; Bouvier, M.; Desgroseillers, L. Multimerization of Staufen1 in live cells. RNA 2010, 16, 585–597. [Google Scholar] [CrossRef] [Green Version]
- Gleghorn, M.L.; Gong, C.; Kielkopf, C.L.; Maquat, L.E. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat. Struct. Mol. Biol. 2013, 20, 515–524. [Google Scholar] [CrossRef] [Green Version]
- Cicenas, J.; Zalyte, E.; Bairoch, A.; Gaudet, P. Kinases and Cancer. Cancers 2018, 10, 63. [Google Scholar] [CrossRef] [Green Version]
- Bhullar, K.S.; Lagaron, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Rao, S.; Hassine, S.; Monette, A.; Amorim, R.; DesGroseillers, L.; Mouland, A.J. HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles. RNA 2019, 25, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Dansithong, W.; Figueroa, K.P.; Scoles, D.R.; Pulst, S.M. Staufen1 links RNA stress granules and autophagy in a model of neurodegeneration. Nat. Commun. 2018, 9, 3648. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.G.; Tosar, L.J.; Desbats, M.A.; Leishman, C.C.; Boccaccio, G.L. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 2009, 122, 563–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez Quesada, Y.; Bonnet-Magnaval, F.; DesGroseillers, L. Phosphomimicry on STAU1 Serine 20 Impairs STAU1 Posttranscriptional Functions and Induces Apoptosis in Human Transformed Cells. Int. J. Mol. Sci. 2022, 23, 7344. https://doi.org/10.3390/ijms23137344
Gonzalez Quesada Y, Bonnet-Magnaval F, DesGroseillers L. Phosphomimicry on STAU1 Serine 20 Impairs STAU1 Posttranscriptional Functions and Induces Apoptosis in Human Transformed Cells. International Journal of Molecular Sciences. 2022; 23(13):7344. https://doi.org/10.3390/ijms23137344
Chicago/Turabian StyleGonzalez Quesada, Yulemi, Florence Bonnet-Magnaval, and Luc DesGroseillers. 2022. "Phosphomimicry on STAU1 Serine 20 Impairs STAU1 Posttranscriptional Functions and Induces Apoptosis in Human Transformed Cells" International Journal of Molecular Sciences 23, no. 13: 7344. https://doi.org/10.3390/ijms23137344
APA StyleGonzalez Quesada, Y., Bonnet-Magnaval, F., & DesGroseillers, L. (2022). Phosphomimicry on STAU1 Serine 20 Impairs STAU1 Posttranscriptional Functions and Induces Apoptosis in Human Transformed Cells. International Journal of Molecular Sciences, 23(13), 7344. https://doi.org/10.3390/ijms23137344





