Analyzing BMP2, FGFR, and TGF Beta Expressions in High-Grade Osteosarcoma Untreated and Treated Autografts Using Proteomic Analysis
Abstract
:1. Introduction
2. Results
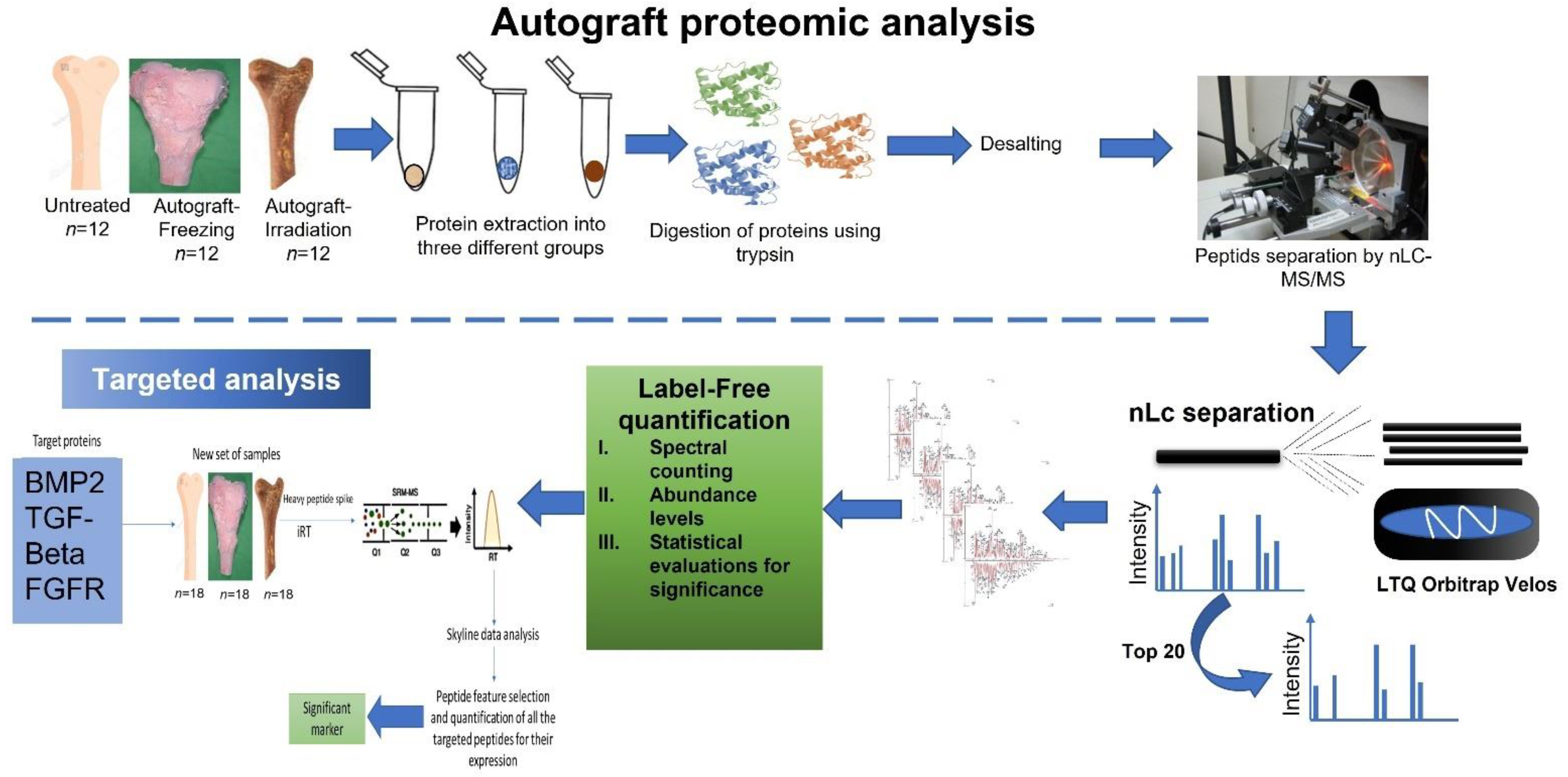
2.1. Sample Processing Information
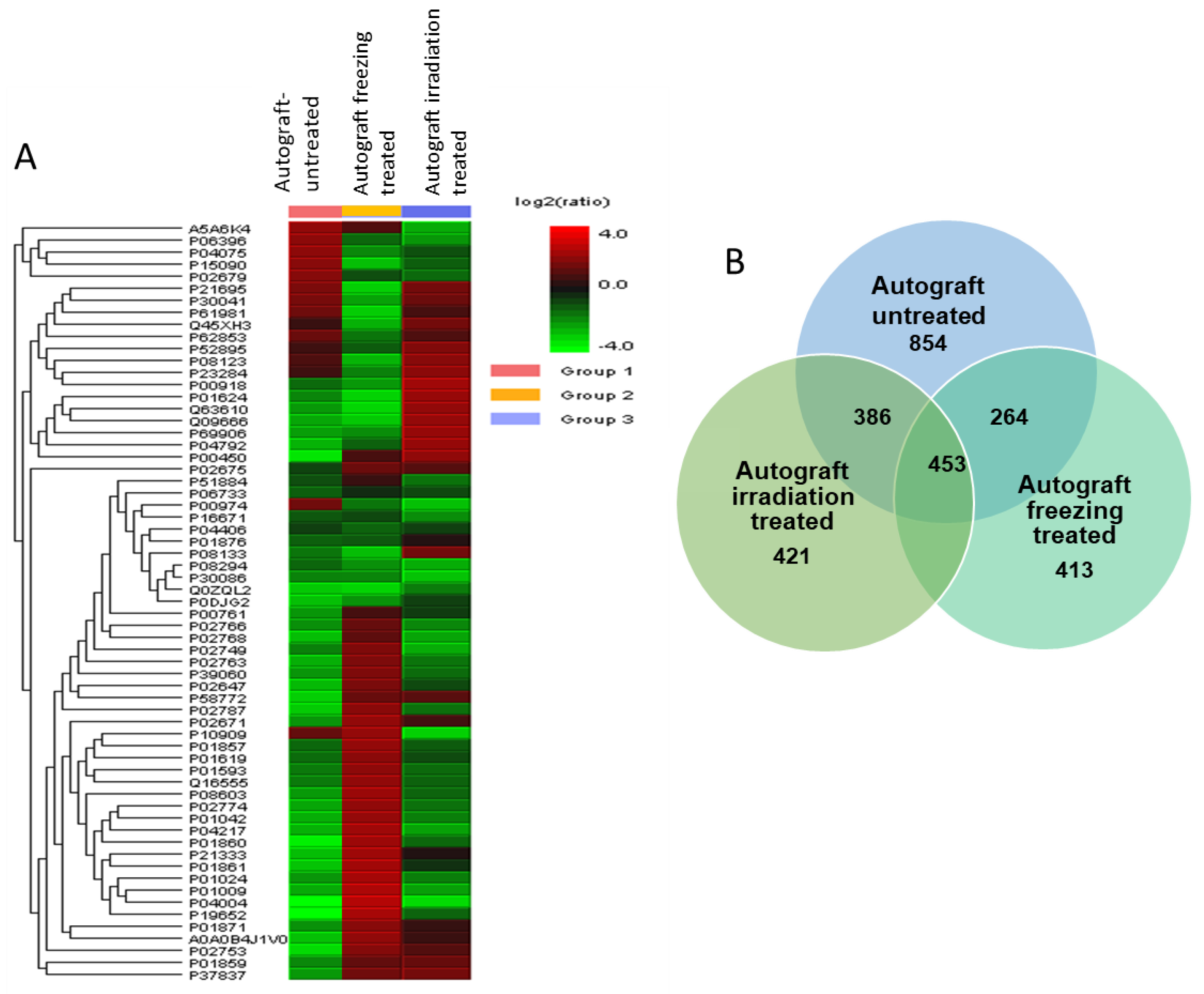
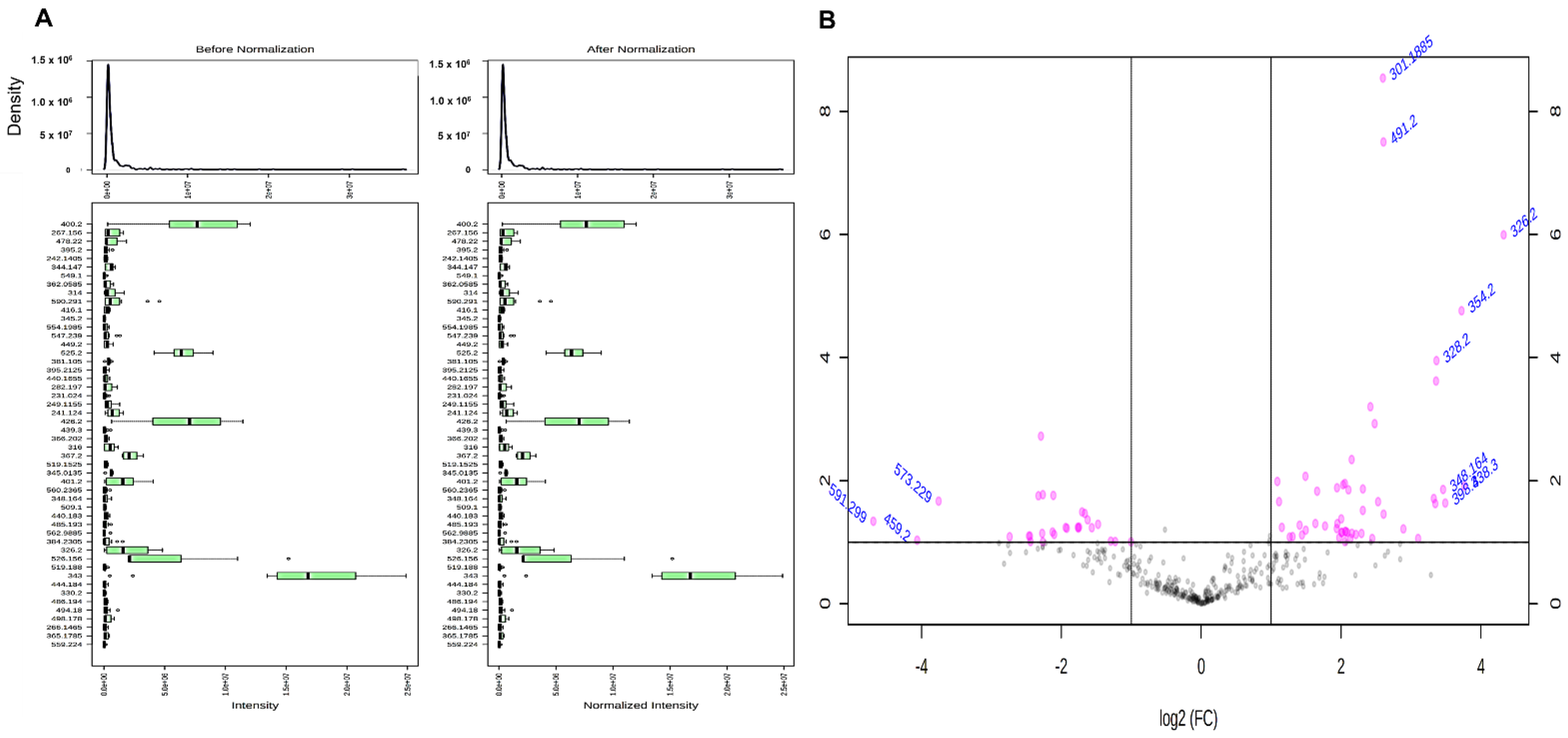
2.2. Differential Protein Expressions among LN-Freezing Treated and Irradiation-Treated Autografts with Untreated Autografts
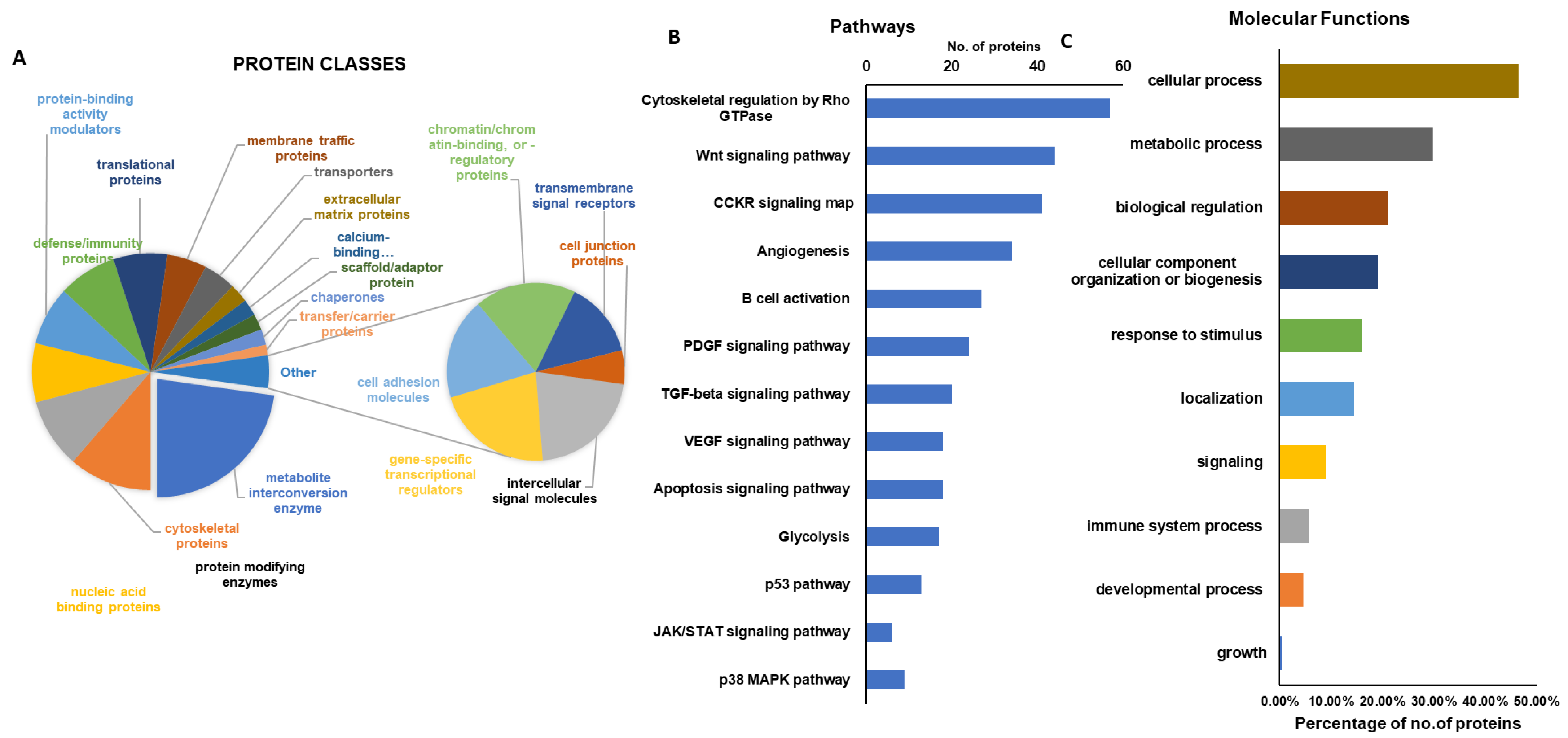
2.3. Gene Ontology Analysis and KEGG Pathway
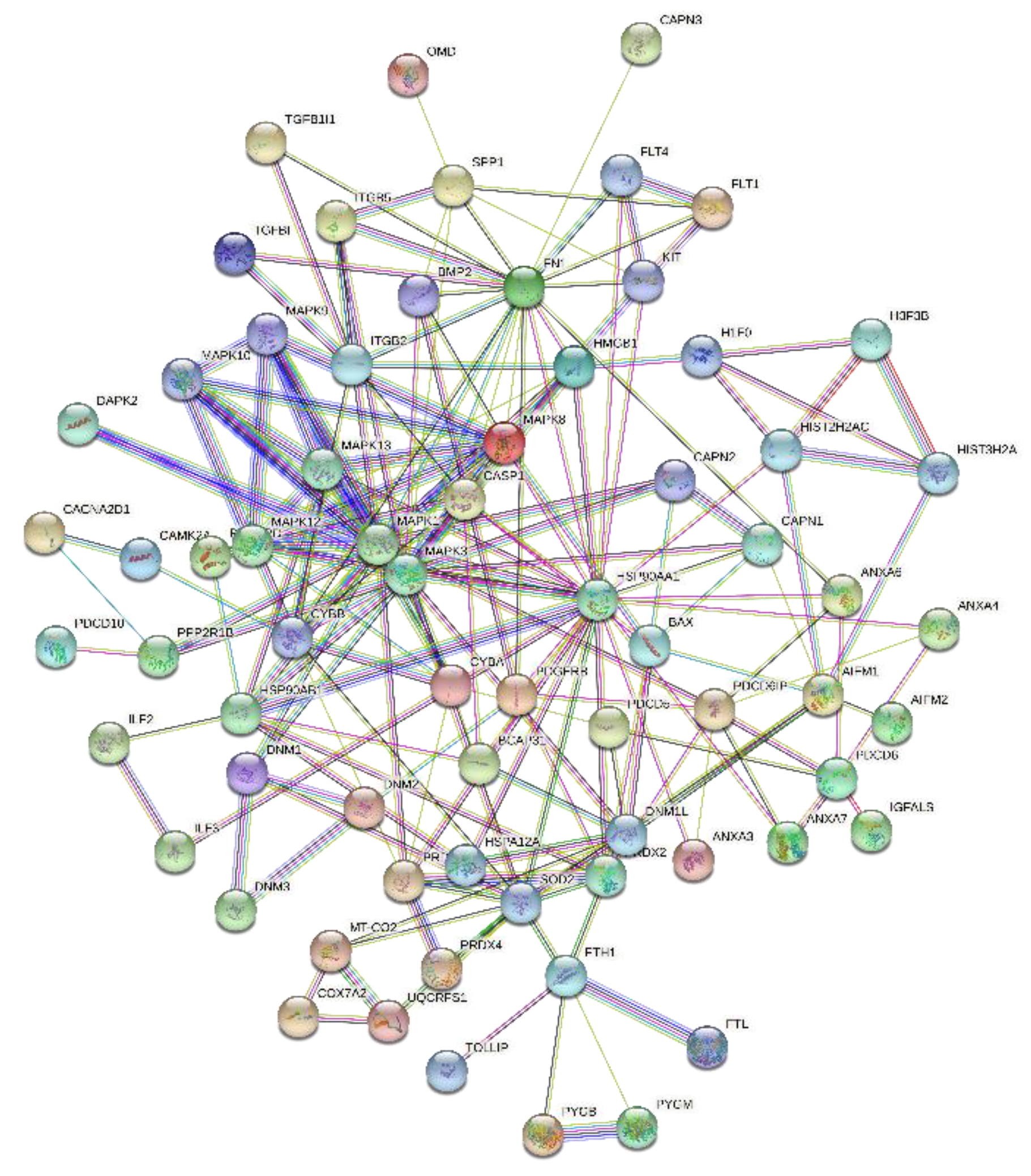
2.4. Protein–Protein Interaction (PPI) Networks
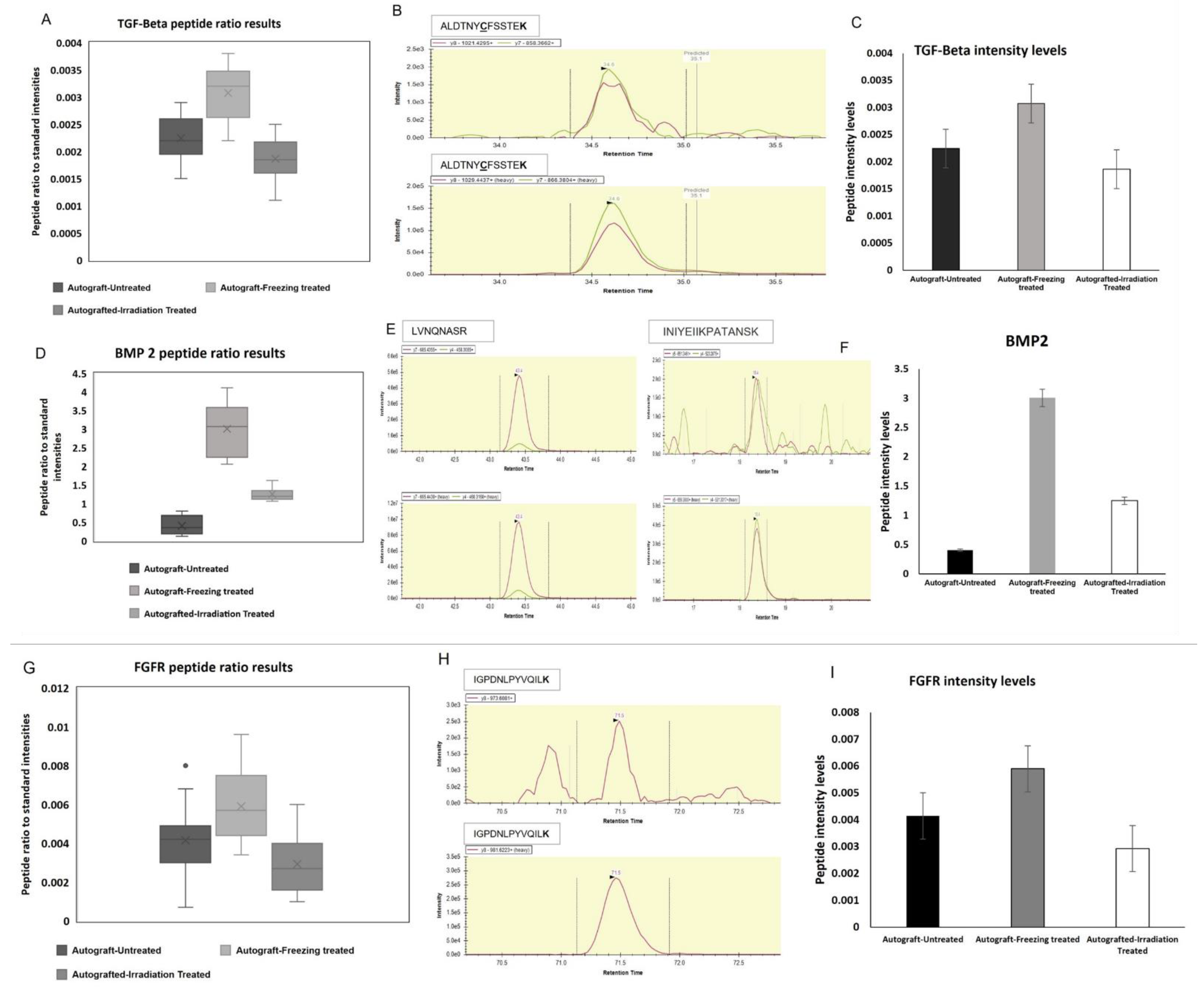
2.5. Bone Growth and Activity Preserving Protein Expressions Validation
2.6. Target Proteins Validation
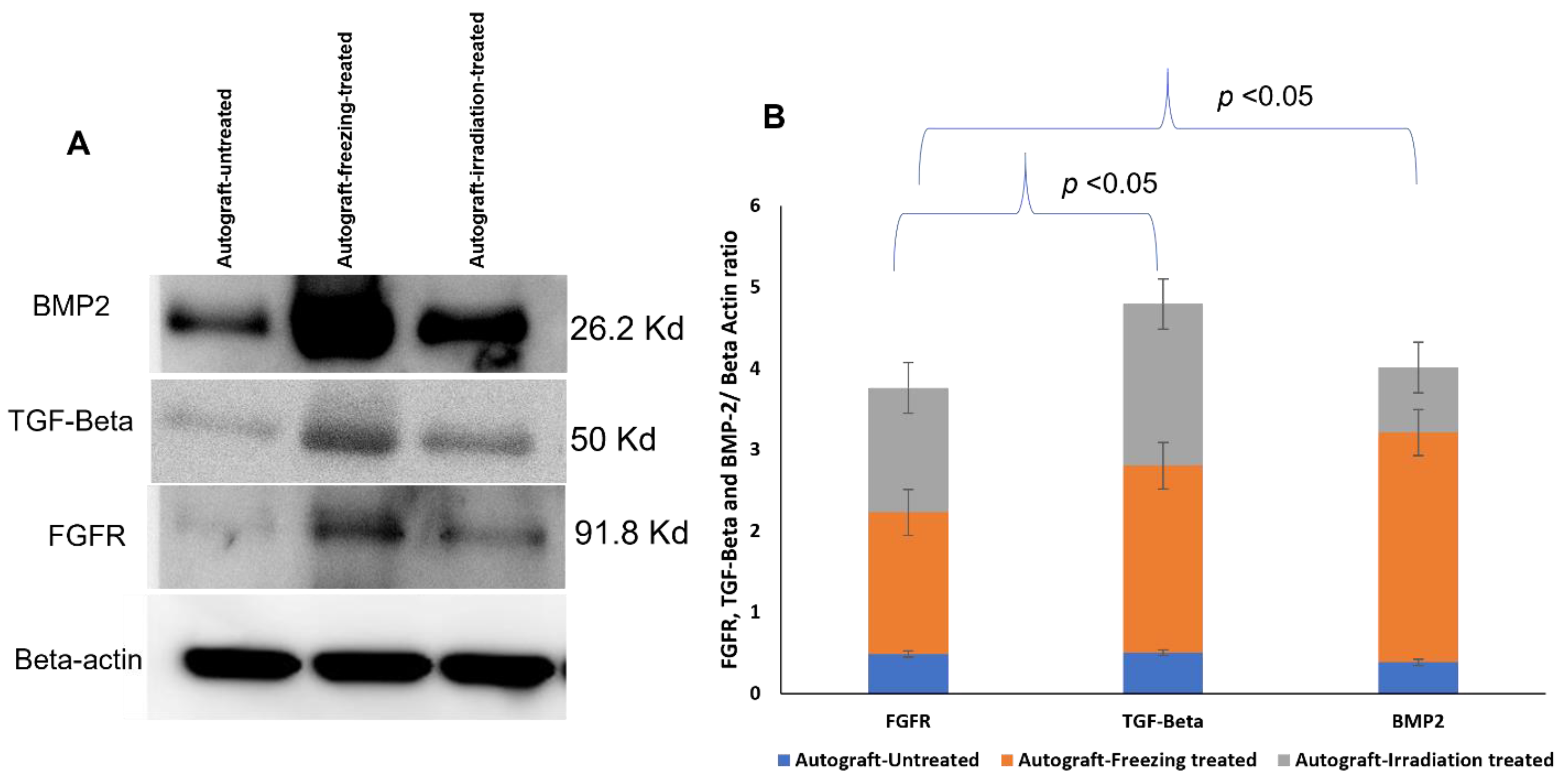
2.7. Western Blotting Analysis
3. Discussion
4. Methods and Materials
4.1. Patients and Clinical Information
4.2. Extraction of Protein from Autograft Untreated and Treated Samples Preparation
4.3. Protein Precipitation and In-Solution Digestion
4.4. Nano UPLC and Mass Spectrometry Conditions
4.5. Target Proteins Validation Using Selection Reaction Monitoring
4.6. Protein Quantification
4.7. Protein Identification
4.8. Bioinformatics Analysis
4.9. Statistical Analysis
4.10. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Harris, M.A.; Hawkins, C.J. Recent and Ongoing Research into Metastatic Osteosarcoma Treatments. Int. J. Mol. Sci. 2022, 23, 3817. [Google Scholar] [CrossRef]
- Sugiyama, T.; Fushimi, K.; Nagano, A.; Tsugita, M.; Nozawa, S.; Iwai, C.; Akiyama, H. The malignant transformation of osteoid osteoma in the cervical spine to high-grade osteosarcoma: A case report and review of literature. Br. J. Neurosurg. 2020, 1–5. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Zhao, N.; Wang, C.; Kamar, S.; Zhou, Y.; He, Z.; Yang, J.; Sun, B.; Shi, X.; et al. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol. Lett. 2018, 16, 6228–6237. [Google Scholar] [CrossRef] [Green Version]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Andrade, J.M.; Mantyh, W.G.; Bloom, A.P.; Ferng, A.S.; Geffre, C.P.; Mantyh, P.W. Bone cancer pain. Ann. N. Y. Acad. Sci. 2010, 1198, 173–181. [Google Scholar] [CrossRef] [Green Version]
- San-Julian, M.; Vazquez-Garcia, B. Biological Reconstruction in Bone Sarcomas: Lessons from Three Decades of Experience. Orthop. Surg. 2016, 8, 111–121. [Google Scholar] [CrossRef]
- Yamamoto, N.; Hayashi, K.; Tsuchiya, H. Progress in biological reconstruction and enhanced bone revitalization for bone defects. J. Orthop. Sci. 2019, 24, 387–392. [Google Scholar] [CrossRef]
- Kwak, K.; Yu, B.; Lewandowski, R.J.; Kim, D.-H. Recent progress in cryoablation cancer therapy and nanoparticles mediated cryoablation. Theranostics 2022, 12, 2175–2204. [Google Scholar] [CrossRef]
- Callstrom, M.R.; Kurup, A.N. Percutaneous ablation for bone and soft tissue metastases—Why cryoablation? Skelet. Radiol. 2009, 38, 835–839. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.L.; Chen, C.M.; Chen, C.F.; Cheng, Y.C.; Lin, Y.K.; Tsai, S.W.; Chen, T.H.; Wu, P.K.; Chen, W.M. Comparable outcomes of recycled autografts and allografts for reconstructions in patients with high-grade osteosarcoma. Int. Orthop. 2021, 45, 2973–2981. [Google Scholar] [CrossRef]
- Kuroda, Y.; Kawai, T.; Goto, K.; Matsuda, S. Clinical application of injectable growth factor for bone regeneration: A systematic review. Inflamm. Regen. 2019, 39, 20. [Google Scholar] [CrossRef] [PubMed]
- Takata, M.; Sugimoto, N.; Yamamoto, N.; Shirai, T.; Hayashi, K.; Nishida, H.; Tanzawa, Y.; Kimura, H.; Miwa, S.; Takeuchi, A.; et al. Activity of bone morphogenetic protein-7 after treatment at various temperatures: Freezing vs. pasteurization vs. allograft. Cryobiology 2011, 63, 235–239. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.M.; Chen, C.F.; Wang, J.Y.; Madda, R.; Tsai, S.W.; Wu, P.K.; Chen, W.M. Bone morphogenetic protein activity preservation with extracorporeal irradiation- and liquid nitrogen freezing-treated recycled autografts for biological reconstruction in malignant bone tumor. Cryobiology 2019, 89, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Miwa, S.; Yamamoto, N.; Hayashi, K.; Takeuchi, A.; Igarashi, K.; Higuchi, T.; Taniguchi, Y.; Araki, Y.; Yonezawa, H.; et al. Pedicle frozen autograft-prosthesis composite reconstructions for malignant bone tumors of the proximal femur. BMC Musculoskelet. Disord. 2020, 21, 81. [Google Scholar] [CrossRef] [Green Version]
- Madda, R.; Chen, C.M.; Chen, C.F.; Wang, J.Y.; Wu, P.K.; Chen, W.M. Effect of Cryoablation Treatment on the Protein Expression Profile of Low-Grade Central Chondrosarcoma Identified by LC-ESI-MS/MS. J. Am. Soc. Mass. Spectrom. 2021, 32, 1469–1489. [Google Scholar] [CrossRef]
- Madda, R.; Chen, C.M.; Wang, J.Y.; Chen, C.F.; Chao, K.Y.; Yang, Y.M.; Wu, H.Y.; Chen, W.M.; Wu, P.K. Proteomic profiling and identification of significant markers from high-grade osteosarcoma after cryotherapy and irradiation. Sci. Rep. 2020, 10, 2105. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Cai, Q.; Wang, J.; Zhang, P.; Wang, X.; Du, X.; Niu, X. Biological reconstruction in the treatment of extremity sarcoma in femur, tibia, and humerus. Medicine 2020, 99, e20715. [Google Scholar] [CrossRef]
- Mistry, H.; Metcalfe, A.; Colquitt, J.; Loveman, E.; Smith, N.A.; Royle, P.; Waugh, N. Autograft or allograft for reconstruction of anterior cruciate ligament: A health economics perspective. Knee Surg. Sports Traumatol. Arthrosc. 2019, 27, 1782–1790. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Ling, L.; Zhang, Q.; Liu, Y.; Guo, X. Evaluation of the Efficacy of Pasteurized Autograft and Intramedullary Vascularized Fibular Transfer for Osteosarcoma of the Femoral Diaphysis. Orthop. Surg. 2019, 11, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Zekry, K.M.; Yamamoto, N.; Hayashi, K.; Takeuchi, A.; Alkhooly, A.Z.A.A.; Abd-Elfattah, A.S.; Elsaid, A.N.S.; Ahmed, A.R.; Tsuchiya, H. Reconstruction of intercalary bone defect after resection of malignant bone tumor. J. Orthop. Surg. 2019, 27, 2309499019832970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Zou, J.; Wang, W.; Yang, A. BMP2 induces hMSC osteogenesis and matrix remodeling. Mol. Med. Rep. 2021, 23. [Google Scholar] [CrossRef]
- Katagiri, T.; Watabe, T. Bone Morphogenetic Proteins. Cold Spring Harb. Perspect Biol. 2016, 8, a021899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lademann, F.; Hofbauer, L.C.; Rauner, M. The Bone Morphogenetic Protein Pathway: The Osteoclastic Perspective. Front. Cell Dev. Biol. 2020, 8, 586031. [Google Scholar] [CrossRef]
- Hutton, D.L.; Moore, E.M.; Gimble, J.M.; Grayson, W.L. Platelet-derived growth factor and spatiotemporal cues induce development of vascularized bone tissue by adipose-derived stem cells. Tissue Eng. Part A 2013, 19, 2076–2086. [Google Scholar] [CrossRef] [Green Version]
- Kasagi, S.; Chen, W. TGF-beta1 on osteoimmunology and the bone component cells. Cell Biosci. 2013, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Thielen, N.G.M.; van der Kraan, P.M.; van Caam, A.P.M. TGFβ/BMP Signaling Pathway in Cartilage Homeostasis. Cells 2019, 8, 969. [Google Scholar] [CrossRef] [Green Version]
- Ogilvie, C.M.; Lu, C.; Marcucio, R.; Lee, M.; Thompson, Z.; Hu, D.; Helms, J.A.; Miclau, T. Vascular endothelial growth factor improves bone repair in a murine nonunion model. Iowa Orthop. J. 2012, 32, 90–94. [Google Scholar]
- Ornitz, D.M.; Marie, P.J. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015, 29, 1463–1486. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Sun, H.; Zhang, J.; Brasier, A.R.; Zhao, Y. Quantitative Assessment of the Effects of Trypsin Digestion Methods on Affinity Purification–Mass Spectrometry-based Protein–Protein Interaction Analysis. J. Proteome Res. 2017, 16, 3068–3082. [Google Scholar] [CrossRef]
- Erve, J.C.L.; DeMaio, W.; Talaat, R.E. Rapid metabolite identification with sub parts-per-million mass accuracy from biological matrices by direct infusion nanoelectrospray ionization after clean-up on a ZipTip and LTQ/Orbitrap mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 3015–3026. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Megger, D.A.; Bracht, T.; Meyer, H.E.; Sitek, B. Label-free quantification in clinical proteomics. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 1581–1590. [Google Scholar] [CrossRef]
- Tran, N.H.; Qiao, R.; Xin, L.; Chen, X.; Liu, C.; Zhang, X.; Shan, B.; Ghodsi, A.; Li, M. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat. Methods 2019, 16, 63–66. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Zhang, J.; Wu, S.; Li, D.; Zhu, Y.; He, F. Improving the sensitivity of MASCOT search results validation by combining new features with Bayesian nonparametric model. Proteomics 2010, 10, 4293–4300. [Google Scholar] [CrossRef]
- Soudy, M.; Anwar, A.M.; Ahmed, E.A.; Osama, A.; Ezzeldin, S.; Mahgoub, S.; Magdeldin, S. UniprotR: Retrieving and visualizing protein sequence and functional information from Universal Protein Resource (UniProt knowledgebase). J. Proteom. 2020, 213, 103613. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Su, Z. EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genom. 2007, 8, 246. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. Panther version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [Green Version]
- Algina, J.; Olejnik, S. Conducting Power Analyses for Anova and Ancova in between-Subjects Designs. Eval. Health Prof. 2003, 26, 288–314. [Google Scholar] [CrossRef]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef]

| Classification | Irradiation-Treated (n = 24) | Freezing-Treated (n = 24) |
|---|---|---|
| Gender | ||
| Male | 12 | 12 |
| Female | 12 | 12 |
| Age (mean) | 40.5 ± 4.2 | 40.5 ± 6.1 |
| Tumor Location | ||
| Distal femur | 4 | 4 |
| Proximal femur | 2 | 2 |
| Proximal tibia | 4 | 4 |
| Proximal humerus | 2 | 2 |
| Tumor Length (mean) | 10.5 ± 6.2 | 11.2 ± 5.8 |
| Follow-up (mean, months) | 55.78 ± 13.6 | 55.96 ± 13.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madda, R.; Chen, C.-M.; Chen, C.-F.; Wang, J.-Y.; Wu, H.-Y.; Wu, P.-K.; Chen, W.-M. Analyzing BMP2, FGFR, and TGF Beta Expressions in High-Grade Osteosarcoma Untreated and Treated Autografts Using Proteomic Analysis. Int. J. Mol. Sci. 2022, 23, 7409. https://doi.org/10.3390/ijms23137409
Madda R, Chen C-M, Chen C-F, Wang J-Y, Wu H-Y, Wu P-K, Chen W-M. Analyzing BMP2, FGFR, and TGF Beta Expressions in High-Grade Osteosarcoma Untreated and Treated Autografts Using Proteomic Analysis. International Journal of Molecular Sciences. 2022; 23(13):7409. https://doi.org/10.3390/ijms23137409
Chicago/Turabian StyleMadda, Rashmi, Chao-Ming Chen, Cheng-Fong Chen, Jir-You Wang, Hsin-Yi Wu, Po-Kuei Wu, and Wei-Ming Chen. 2022. "Analyzing BMP2, FGFR, and TGF Beta Expressions in High-Grade Osteosarcoma Untreated and Treated Autografts Using Proteomic Analysis" International Journal of Molecular Sciences 23, no. 13: 7409. https://doi.org/10.3390/ijms23137409
APA StyleMadda, R., Chen, C.-M., Chen, C.-F., Wang, J.-Y., Wu, H.-Y., Wu, P.-K., & Chen, W.-M. (2022). Analyzing BMP2, FGFR, and TGF Beta Expressions in High-Grade Osteosarcoma Untreated and Treated Autografts Using Proteomic Analysis. International Journal of Molecular Sciences, 23(13), 7409. https://doi.org/10.3390/ijms23137409






