Transcription Factor AtOFP1 Involved in ABA-Mediated Seed Germination and Root Growth through Modulation of ROS Homeostasis in Arabidopsis
Abstract
:1. Introduction
2. Results
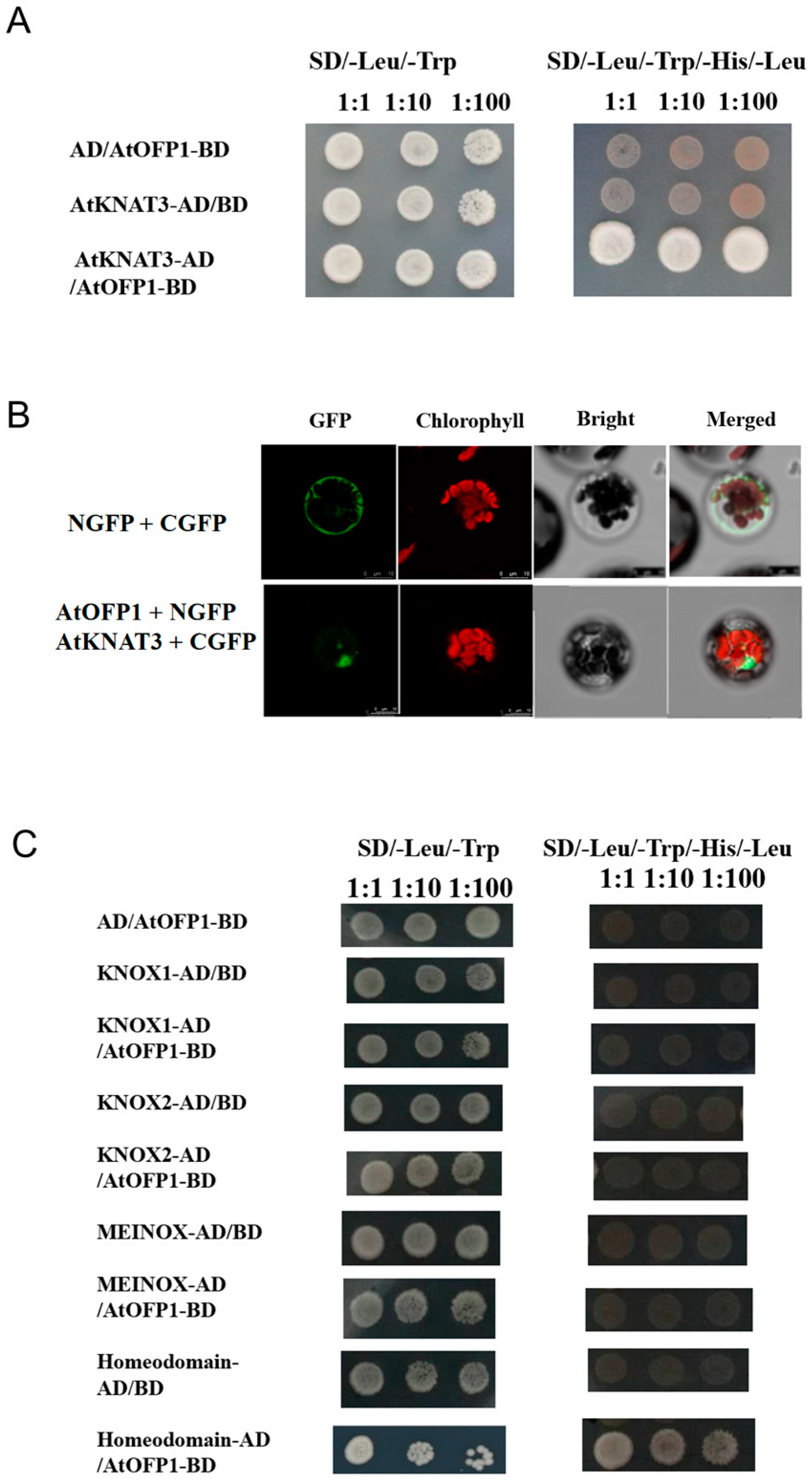
2.1. AtOFP1 Can Physically Interact with AtKNAT3
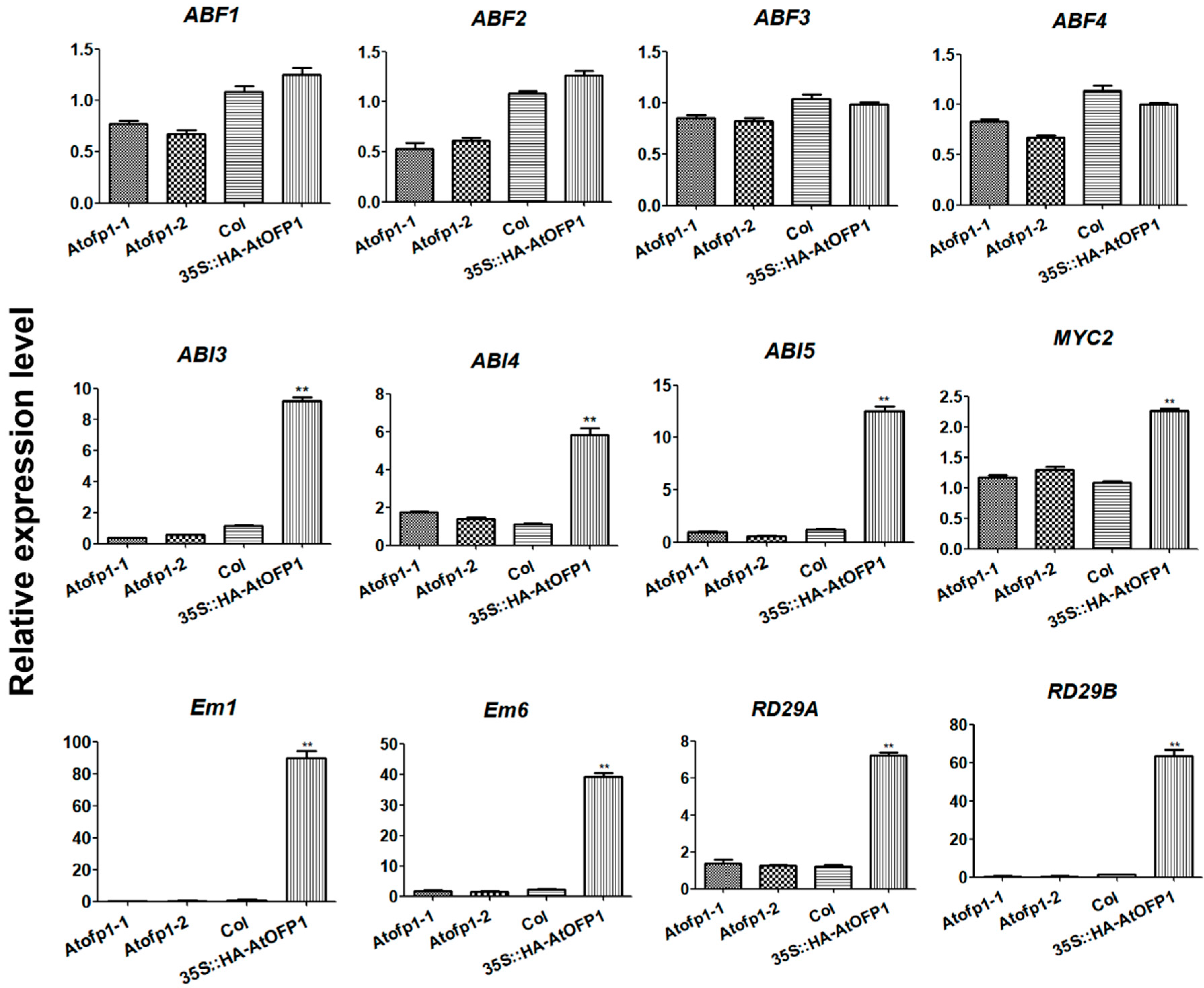
2.2. AtOFP1 Influences Expression Patterns of ABA-Pathway-Related Genes
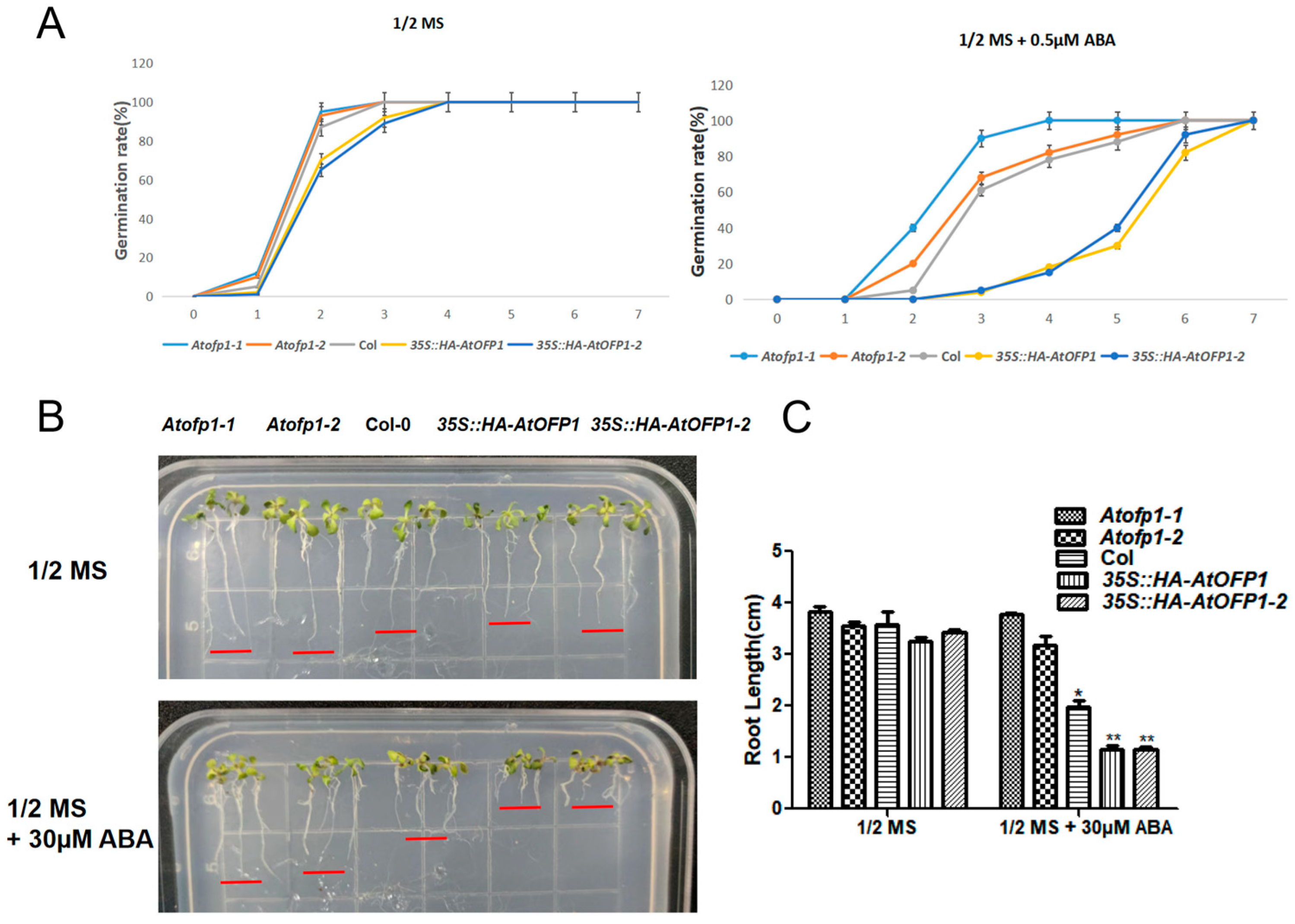
2.3. 35S::HAOFP1 Is Likely to ABA-Hypersensitive during Seed Germination and Primary Root Growth
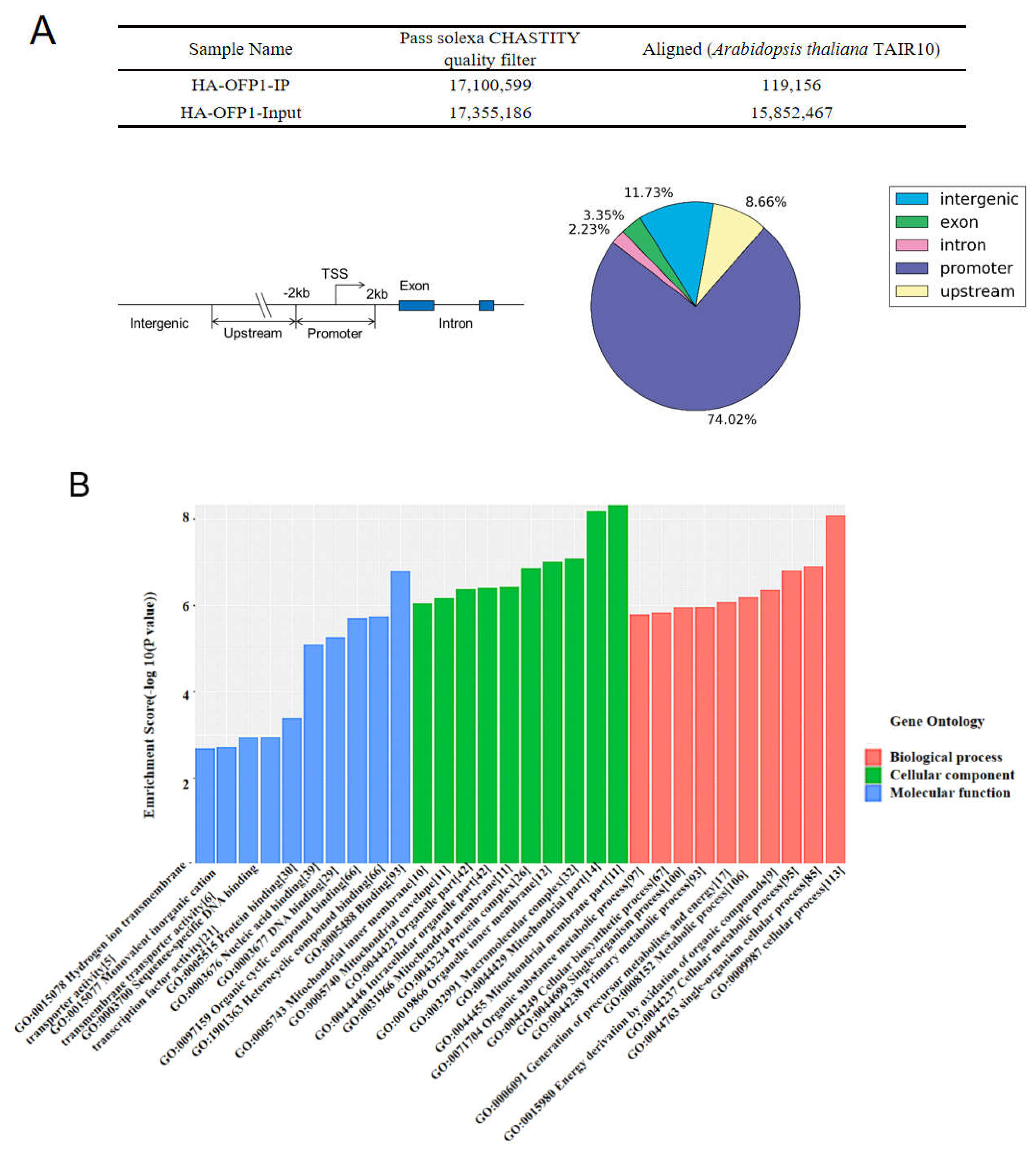
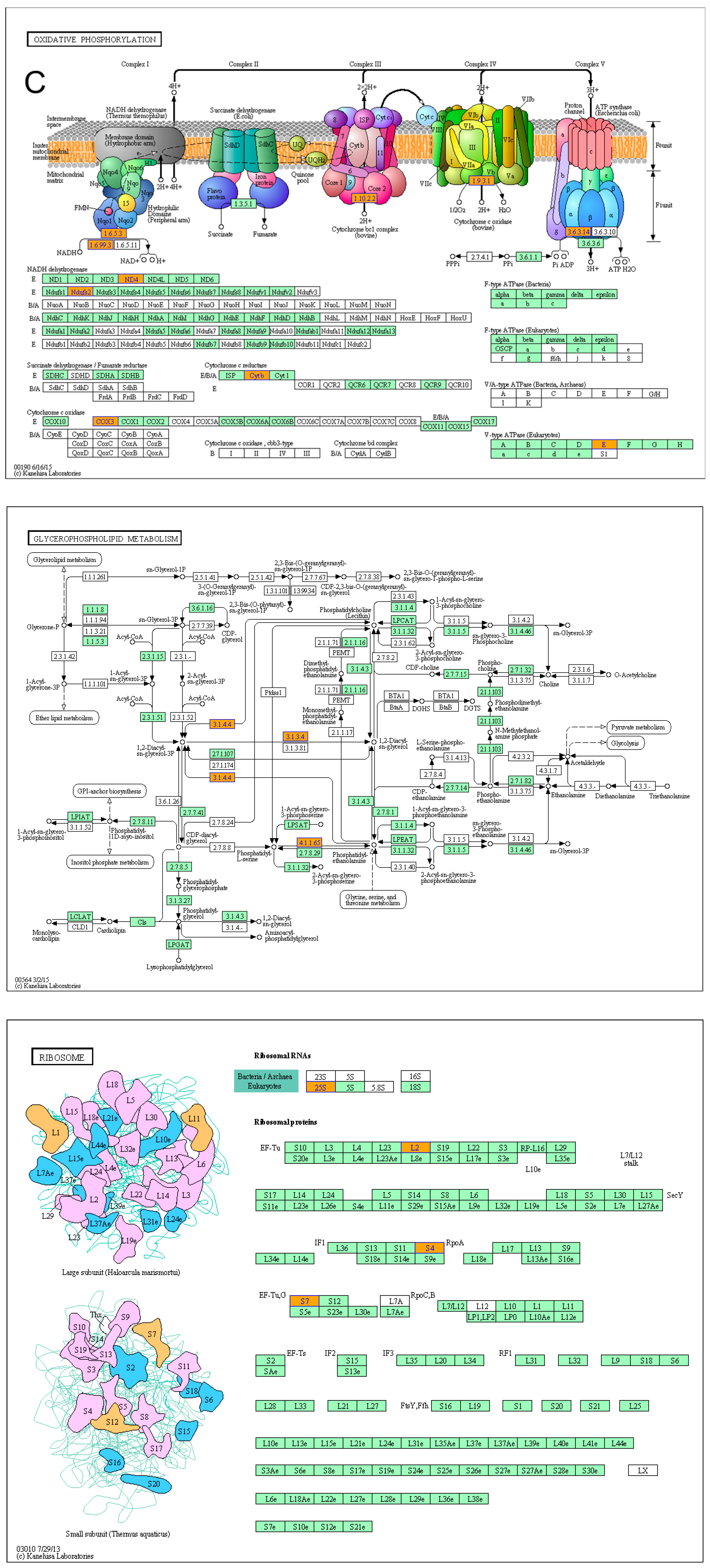
2.4. AtOFP1 Target Genes Obtained from ChIP-SEQ Peaks Were Mainly Mitochondrial-Complex Subunits
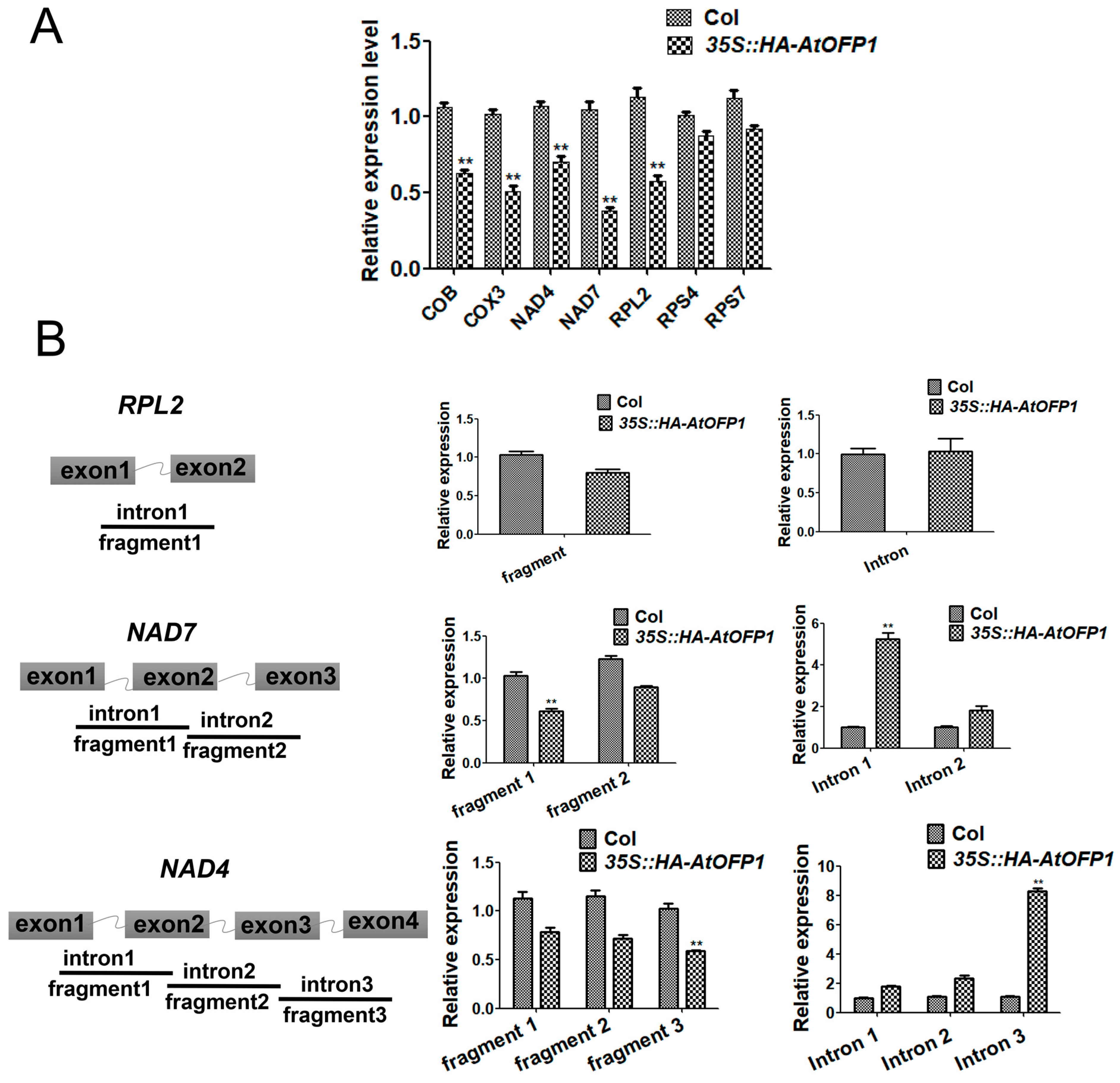
2.5. AtOFP1 Regulates Mitochondrial mRNA Splicing of NAD4 and NAD7
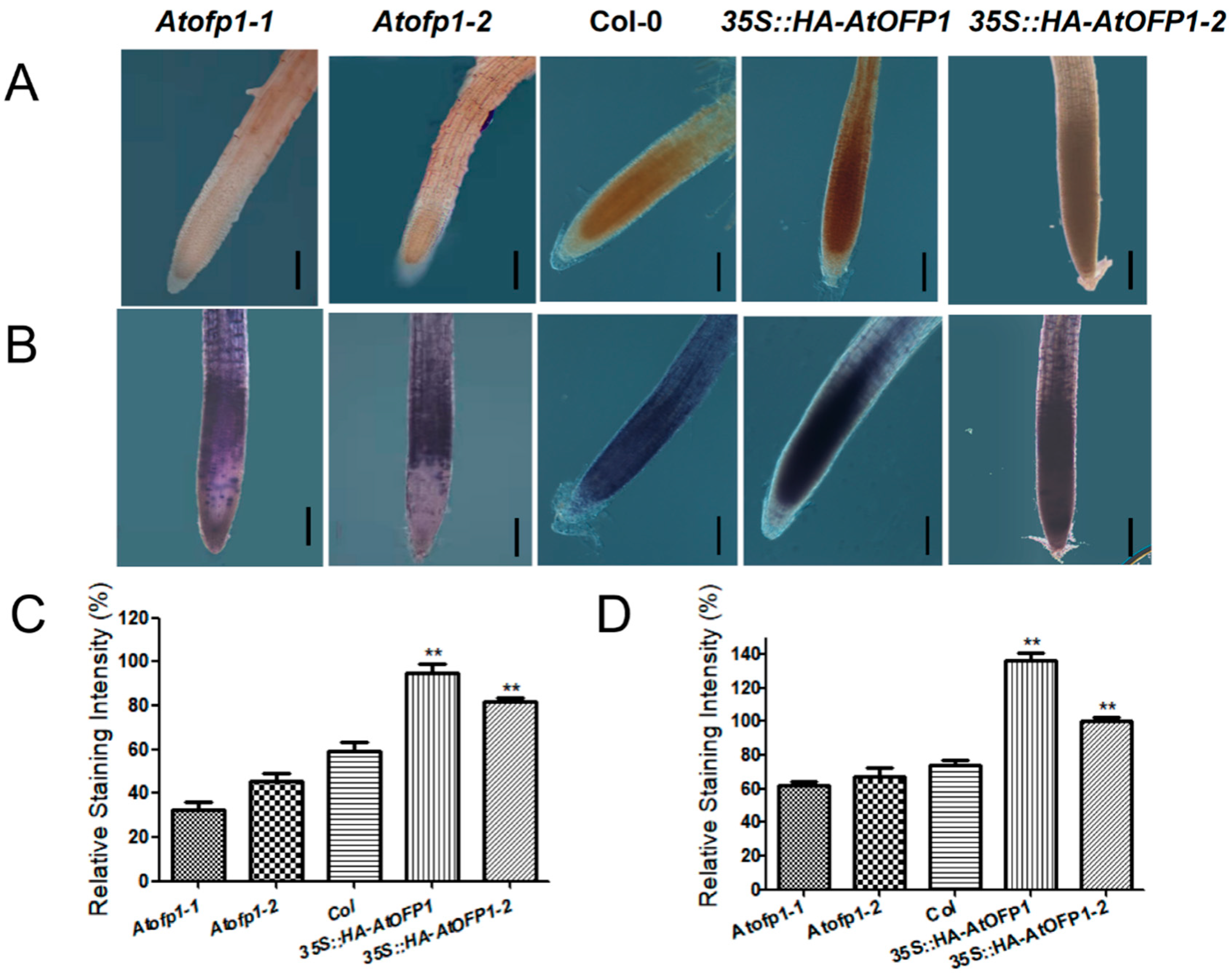
2.6. 35S::HAOFP1 OE Line Accumulates More ROS in Root Tips
2.7. Elevated ROS Levels in 35S::HAOFP1 OE Line Cause ABA-Hypersensitive Phenotype and Are Partially Restored by Additional GSH
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and Gene Expression Profile Analysis
4.3. Plasmid Constrction and Protein-Protein Interaction Assays
4.4. ABA Treatment, Seed Germination and Root Length Measurement
4.5. CHIP-SEQ
4.6. Analysis of Root-Tip ROS Staining
4.7. Measurement of Enzymatic Activity (SOD and POD) and Estimation of Malondialdehyde (MDA)
4.8. Data Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| At | Arabidopsis thaliana |
| OFP | Ovate Family Protein |
| Col-0 | Columbia-0 |
| qRT-PCR | Real-Time Quantitative Reverse Transcription PCR |
| ABA | Abscisic acid |
| ABREs | ABA-responsive elements |
| Em1 | Embryogenesis abundant 1 |
| RD29A | Responsive to desiccation 29A |
| ETC | Electron transfer chain |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DAB | 3,3N-Diaminobenzidine Tertrahydrochloride |
| NBT | Nitrotetrazolium Blue chloride |
| SOD | Superoxide dismutase |
| POD | Peroxidase |
| MDA | Malondialdehyde |
References
- Himmelbach, A.; Yi, Y.; Grill, E. Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 2003, 6, 470–479. [Google Scholar] [CrossRef]
- Yuri, K.; Yusuke, J.; Atsushi, H.; Eiji, N.; Abrams, S.R.; Yuji, K.; Mitsunori, S. Comprehensive Hormone Profiling in Developing Arabidopsis Seeds: Examination of the Site of ABA Biosynthesis, ABA Transport and Hormone Interactions. Plant Cell Physiol. 2010, 51, 1988–2001. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef]
- Hong, L.; Liu, X. AREB/ABF Transcription Factors and Their Involvement in ABA Signal Transduction. Plant Physiol. J. 2011, 47, 211–217. [Google Scholar] [CrossRef]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113. [Google Scholar] [CrossRef] [Green Version]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. ABI4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.P.; Zhao, B.G.; Chao, Q.; Wang, B.C.; Li, X.H. The Maize AP2/EREBP Transcription Factor ZmEREB160 Enhances Drought Tolerance in Arabidopsis. Trop. Plant Biol. 2020, 13, 251–261. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Cai, H.; Guo, M.; Chai, M.; She, Z.; Ye, L.; Cheng, Y.; Wang, B.; Qin, Y. The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean. Int. J. Mol. Sci. 2020, 21, 7778. [Google Scholar] [CrossRef]
- Li, L.; Zhu, T.; Song, Y.; Feng, L.; Ren, M. ABSCISIC ACID INSENSITIVE5 Interacts With RIBOSOMAL S6 KINASE2 to Mediate ABA Responses During Seedling Growth in Arabidopsis. Front. Plant Sci. 2021, 11, 598654. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Aquea, F.; Espinoza, C.; Vega, A.; Cavallini, E.; Dal Santo, S.; Cañón, P.; Rodríguez-Hoces de la Guardia, A.; Serrano, J.; Tornielli, G.B.; et al. Inspection of the Grapevine BURP Superfamily Highlights an Expansion of RD22 Genes with Distinctive Expression Features in Berry Development and ABA-Mediated Stress Responses. PLoS ONE 2014, 9, e110372. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Cha, J.Y.; Kang, S.; Park, B.; Lee, H.J.; Hong, H.; Chun, H.J.; Kim, D.H.; Kim, M.C.; Lee, S.Y.; et al. The Arabidopsis a zinc finger domain protein ARS1 is essential for seed germination and ROS homeostasis in response to ABA and oxidative stress. Front. Plant Sci. 2015, 6, 963. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Vega, J.C.; Cady, B.; Kayanja, G.; Mauriello, A.; Cervantes, N.; Gillespie, A.; Lavia, L.; Trujillo, J.; Alkio, M.; Colón-Carmona, A. Detoxification of polycyclic aromatic hydrocarbons (PAHs) in Arabidopsis thaliana involves a putative flavonol synthase. J. Hazard. Mater. 2017, 321, 268–280. [Google Scholar] [CrossRef] [Green Version]
- Bi, C.; Ma, Y.; Wu, Z.; Yu, Y.T.; Liang, S.; Lu, K.; Wang, X.F. Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol. Biol. 2017, 94, 197–213. [Google Scholar] [CrossRef] [Green Version]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Alamri, S.; Siddiqui, M.H.; Kushwaha, K.; Singh, V.P.; Kushwaha, B.K. Mitigation of arsenate toxicity by indole-3-acetic acid in brinjal roots: Plausible association with endogenous hydrogen peroxide. J. Hazard. Mater. 2020, 405, 124336. [Google Scholar] [CrossRef]
- Klodmann, J.; Sunderhaus, S.; Nimtz, M. Jansch, L.; Braun, H.P. Internal Architecture of Mitochondrial Complex I from Arabidopsis thaliana. Plant Cell 2010, 22, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Binder, S.; Brennicke, A. Gene expression in plant mitochondria: Transcriptional and post–transcriptional control. Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Haili, N.; Arnal, N.; Quadrado, M.; Amiar, S.; Tcherkez, G.; Dahan, J.; Briozzo, P.; Colas des Francs-Small, C.; Vrielynck, N.; Mireau, H. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013, 41, 6650–6663. [Google Scholar] [CrossRef] [Green Version]
- Keren, I.; Tal, L.; Francs-Small, C.C.D.; Araújo, W.L.; Shevtsov, S.; Shaya, F.; Fernie, A.R.; Small, I.; Ostersetzer-Biran, O. nMAT1, a nuclear—Encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 2012, 71, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cui, Y.; Zhang, X.; Yang, Z.; Lai, J.; Song, W.; Liang, J.; Li, X. Maize PPR278 Functions in Mitochondrial RNA Splicing and Editing. Int. J. Mol. Sci. 2022, 23, 3035. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chang, Y.; Guo, J.; Zeng, Q.; Ellis, B.E.; Chen, J.G. Arabidopsis Ovate Family Proteins, a Novel Transcriptional Repressor Family, Control Multiple Aspects of Plant Growth and Development. PLoS ONE 2011, 6, e23896. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ying, C.; Guo, J.; Chen, J.G. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2010, 50, 858–872. [Google Scholar] [CrossRef]
- Li, E.; Wang, S.; Liu, Y.; Chen, J.G.; Douglas, C.J. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2011, 67, 328–341. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Ellis, B. Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators. Front. Plant Sci. 2016, 7, 417. [Google Scholar] [CrossRef] [Green Version]
- Hackbusch, J.; Richter, K.; Müller, J.; Hackbusch, J.; Richter, K.; Muller, J.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, X.; Ju, H.; Chen, J.; Chang, Y. OVATE FAMILY PROTEIN1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 470, 492–497. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, L.; Zhang, X.; Zhang, S.; Xie, D.; Liang, C.; Huang, W.; Fan, L.; Fang, Y.; Chang, Y. OFP1 Interaction with ATH1 Regulates Stem Growth, Flowering Time and Flower Basal Boundary Formation in Arabidopsis. Genes 2018, 9, 399. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Liu, X.; He, C.; Xu, B.; Li, Y.; Hu, Y. Functional divergence and adaptive selection of KNOX gene family in plants. Open Life Sci. 2020, 15, 346–363. [Google Scholar] [CrossRef]
- Kim, D.; Cho, Y.H.; Ryu, H.; Kim, Y.; Kim, T.H.; Hwang, I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 2013, 75, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Jothi, R.; Cuddapah, S.; Barski, A.; Cui, K.; Zhao, K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008, 36, 5221–5231. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein-Protein Interactions in Vivo. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Song, A.; Zhang, X.; Li, S.; Chen, F. The core regulatory networks and hub genes regulating flower development in Chrysanthemum morifolium. Plant Mol. Biol. 2020, 103, 669–688. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, J.; Yu, Q.; Liu, R.; Sun, Y. AtOFPs regulate cell elongation by modulating microtubule orientation via direct interaction with TONNEAU2. Plant Sci. 2020, 292, 110405. [Google Scholar] [CrossRef]
- Martin, R.C.; Vining, K.; Dombrowski, J.E. Genome-wide (ChIP-seq) identification of target genes regulated by BdbZIP10 during paraquat-induced oxidative stress. BMC Plant Biol. 2018, 18, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Zhang, Y.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Cao, S.K.; Sayyed, A.; Xu, C.; Sun, F.; Wang, X.; Tan, B.C. The Mitochondrial Pentatricopeptide Repeat Protein PPR18 Is Required for the cis-Splicing of nad4 Intron 1 and Essential to Seed Development in Maize. Int. J. Mol. Sci. 2020, 21, 4047. [Google Scholar] [CrossRef]
- Hsieh, W.Y.; Liao, J.C.; Chang, C.Y.; Harrison, T.; Boucher, C.; Hsieh, M.H. The SLOW GROWTH3 Pentatricopeptide Repeat Protein Is Required for the Splicing of Mitochondrial NADH Dehydrogenase Subunit7 Intron 2 in Arabidopsis. Plant Physiol. 2015, 168, 490–501. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhang, J.; He, J.; Qin, Y.; Hua, D.; Duan, Y.; Chen, Z.; Gong, Z. ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 2014, 10, e1004791. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Zhang, F.; Li, H.; Qiu, B.; Yang, Z. Antioxidant defense system in lettuces tissues upon various As species exposure. J. Hazard. Mater. 2020, 399, 123003. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhang, D.; Zhou, X.; Zhou, G.; Zong, W.; Chen, L.; Chang, Y.; Wu, X. Transcription Factor AtOFP1 Involved in ABA-Mediated Seed Germination and Root Growth through Modulation of ROS Homeostasis in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 7427. https://doi.org/10.3390/ijms23137427
Wang H, Zhang D, Zhou X, Zhou G, Zong W, Chen L, Chang Y, Wu X. Transcription Factor AtOFP1 Involved in ABA-Mediated Seed Germination and Root Growth through Modulation of ROS Homeostasis in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(13):7427. https://doi.org/10.3390/ijms23137427
Chicago/Turabian StyleWang, Hemeng, Dongrui Zhang, Xi’nan Zhou, Ganghua Zhou, Wenbo Zong, Lingling Chen, Ying Chang, and Xiaoxia Wu. 2022. "Transcription Factor AtOFP1 Involved in ABA-Mediated Seed Germination and Root Growth through Modulation of ROS Homeostasis in Arabidopsis" International Journal of Molecular Sciences 23, no. 13: 7427. https://doi.org/10.3390/ijms23137427






