Evolutionary and Expression Analysis of MOV10 and MOV10L1 Reveals Their Origin, Duplication and Divergence
Abstract
:1. Introduction
2. Results
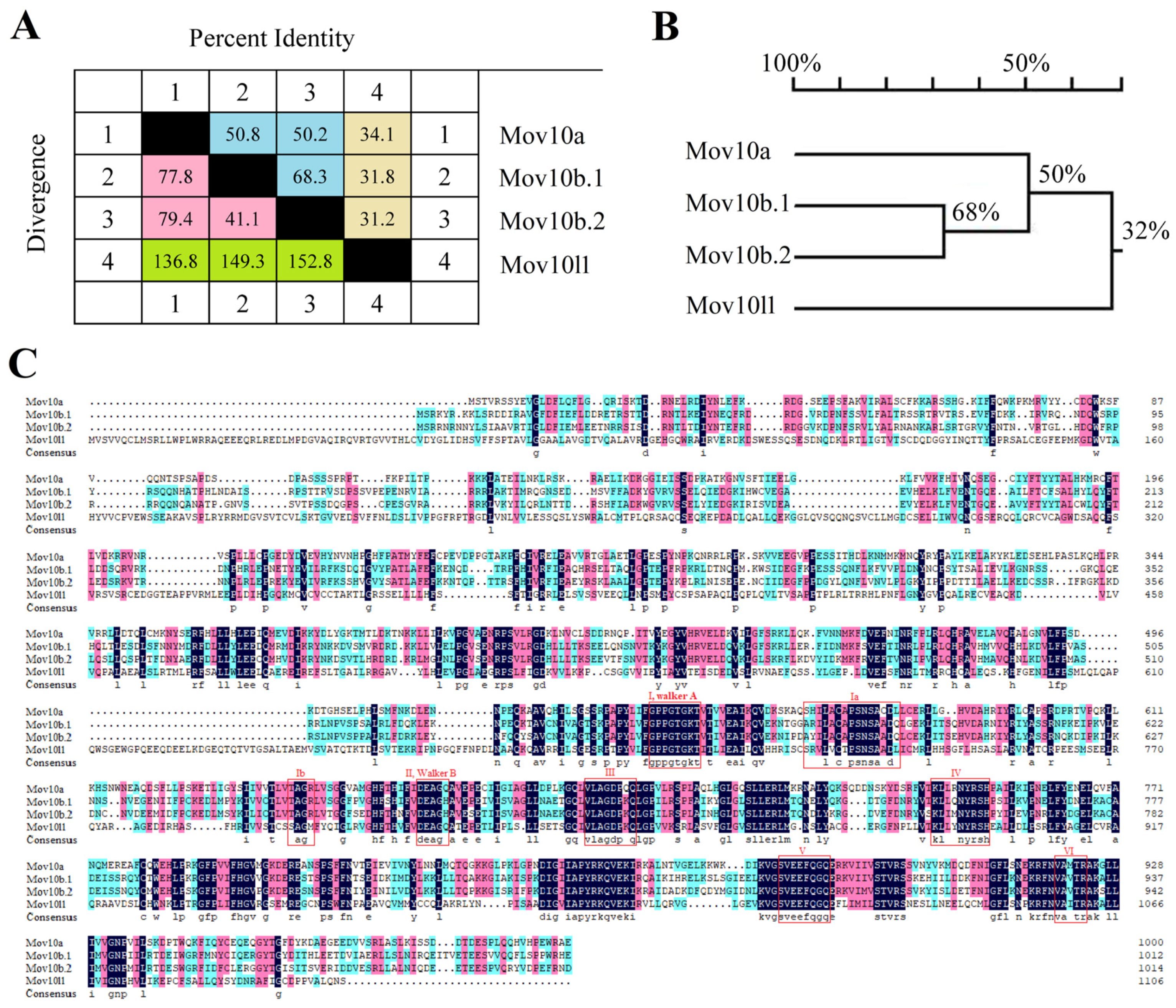
2.1. Three mov10 Genes and mov10l1 in Zebrafish
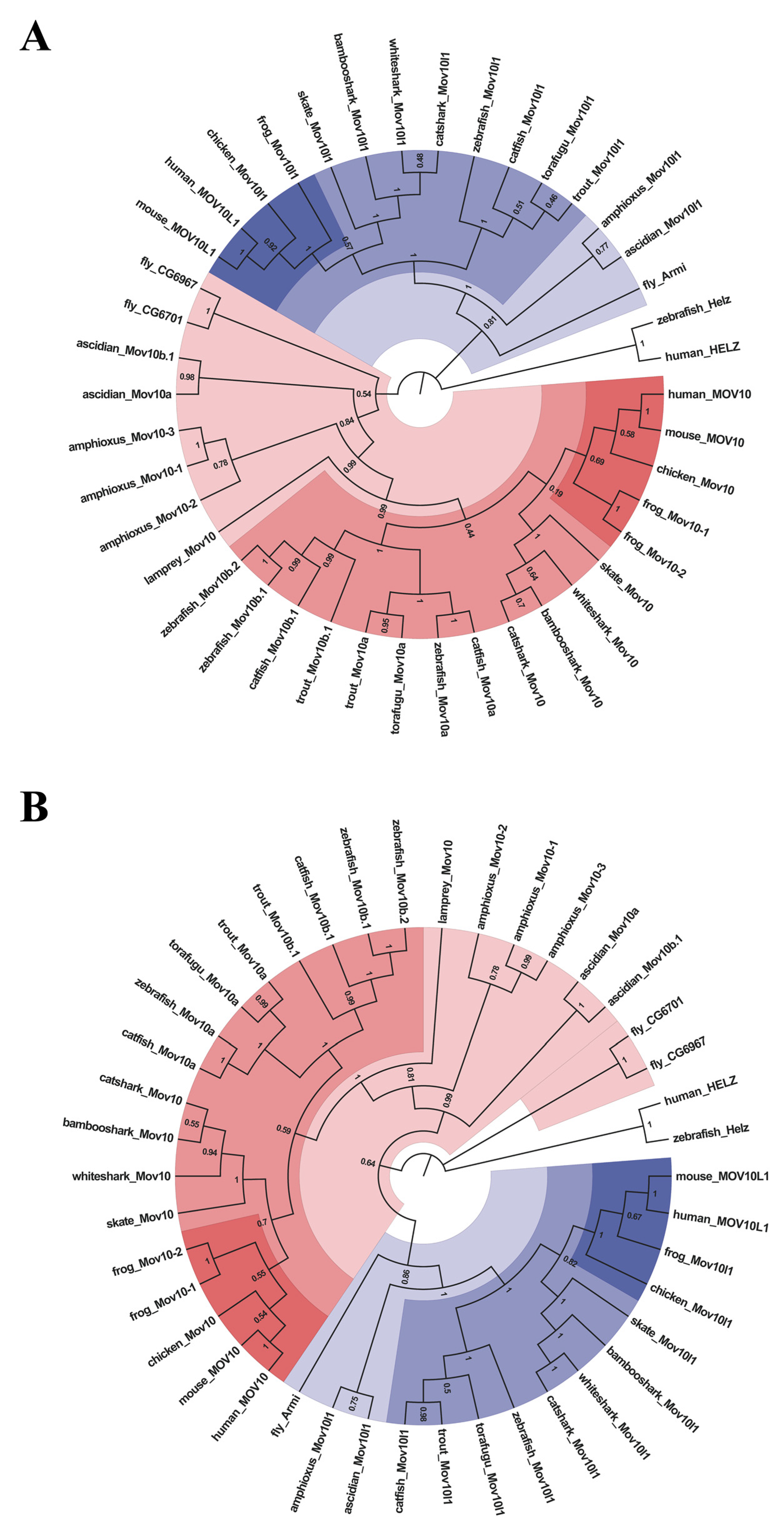
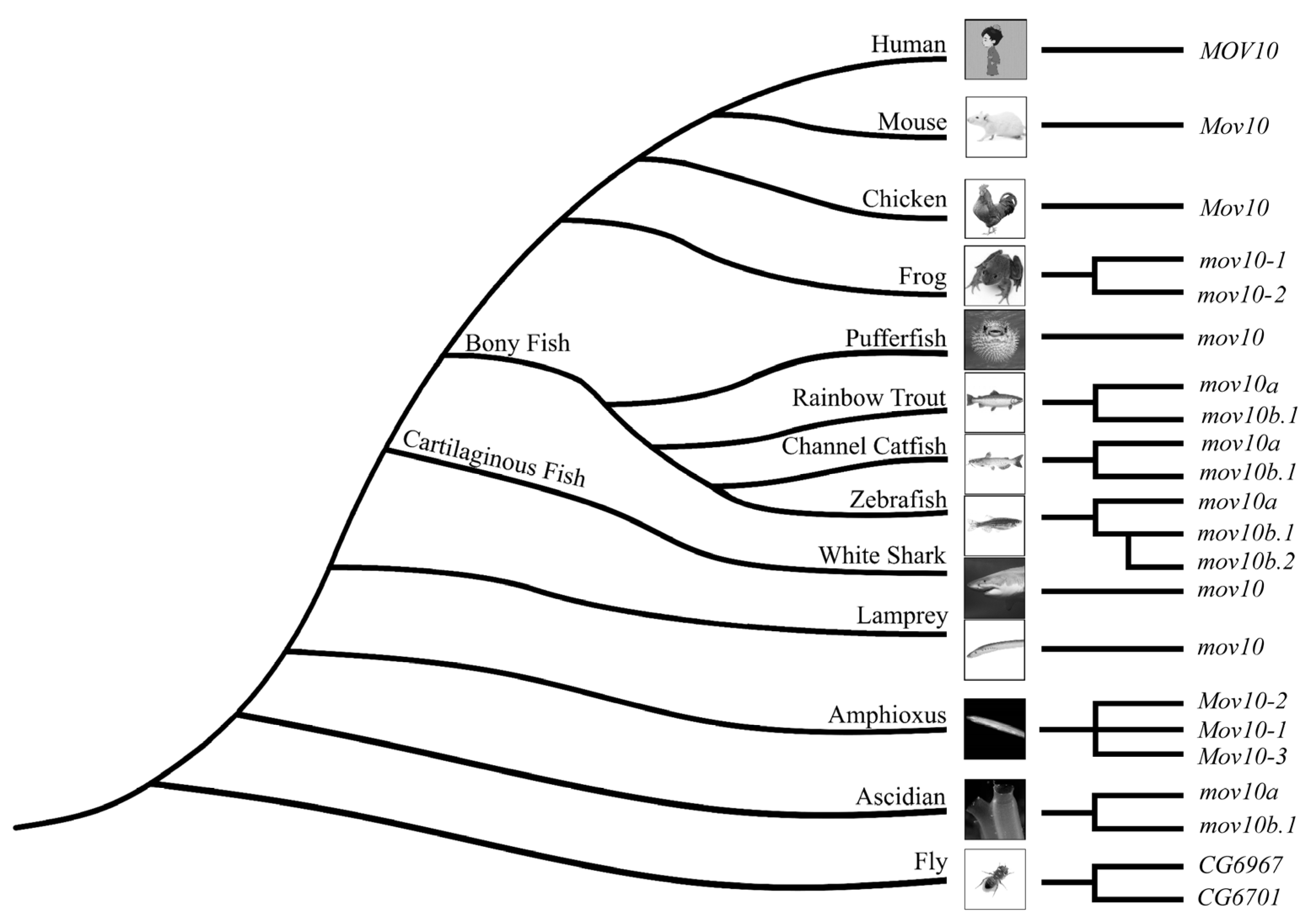
2.2. Lineage-Specific Gene Duplication of MOV10 in Multiple Species
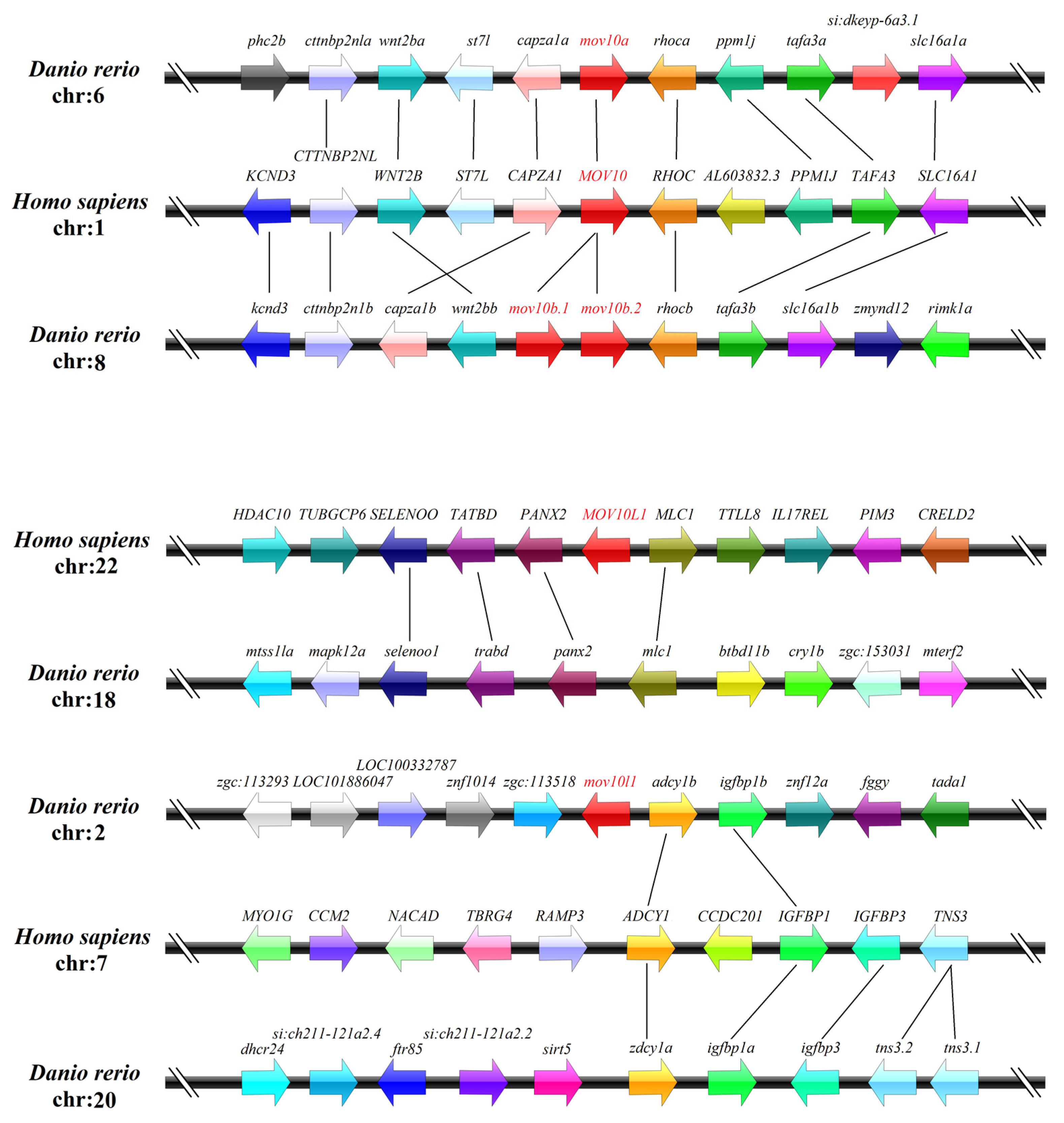
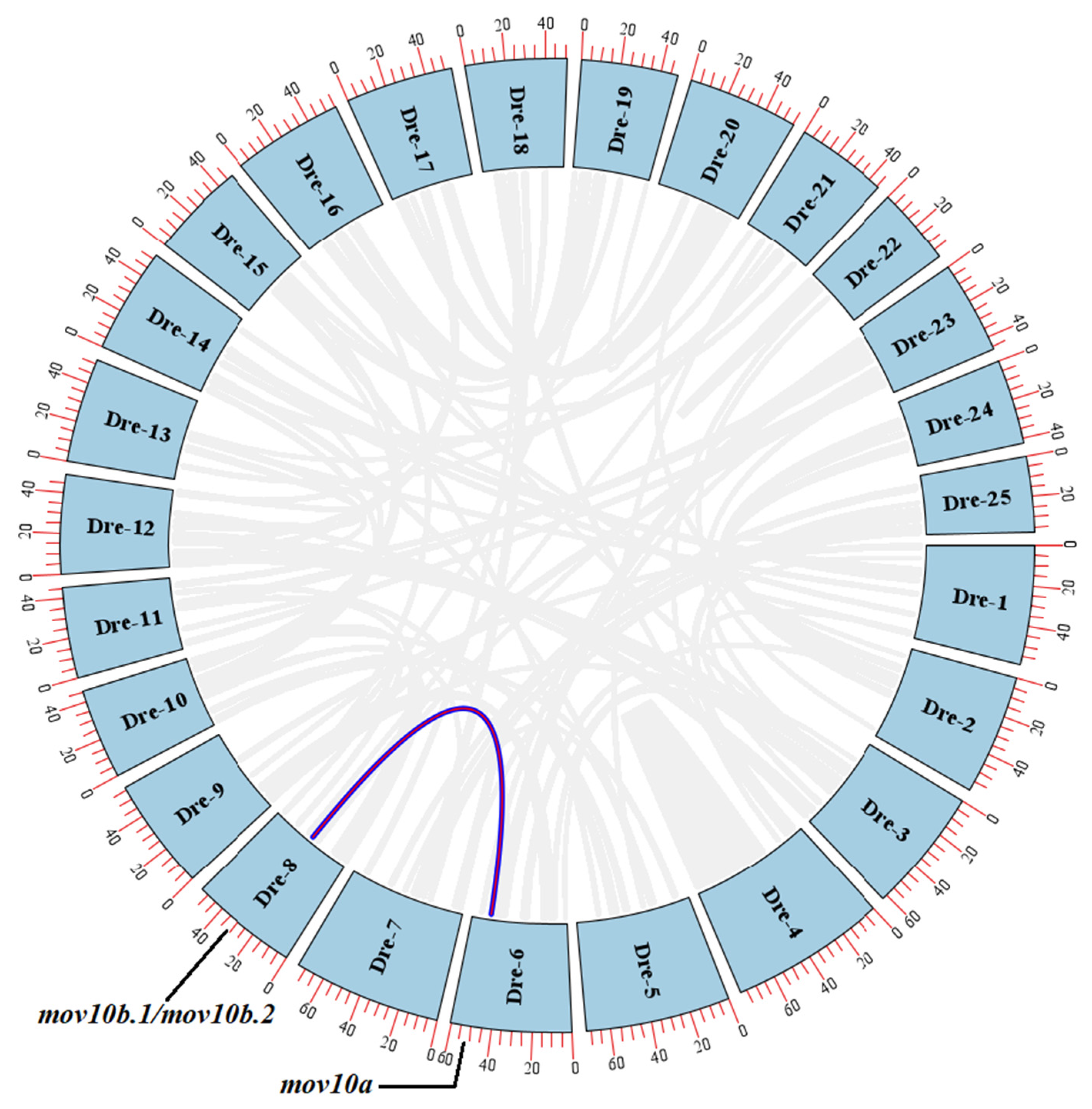
2.3. Genomic Structure and Synteny of MOV10 Genes
2.4. Gene Duplication and Selective Pressure Analysis
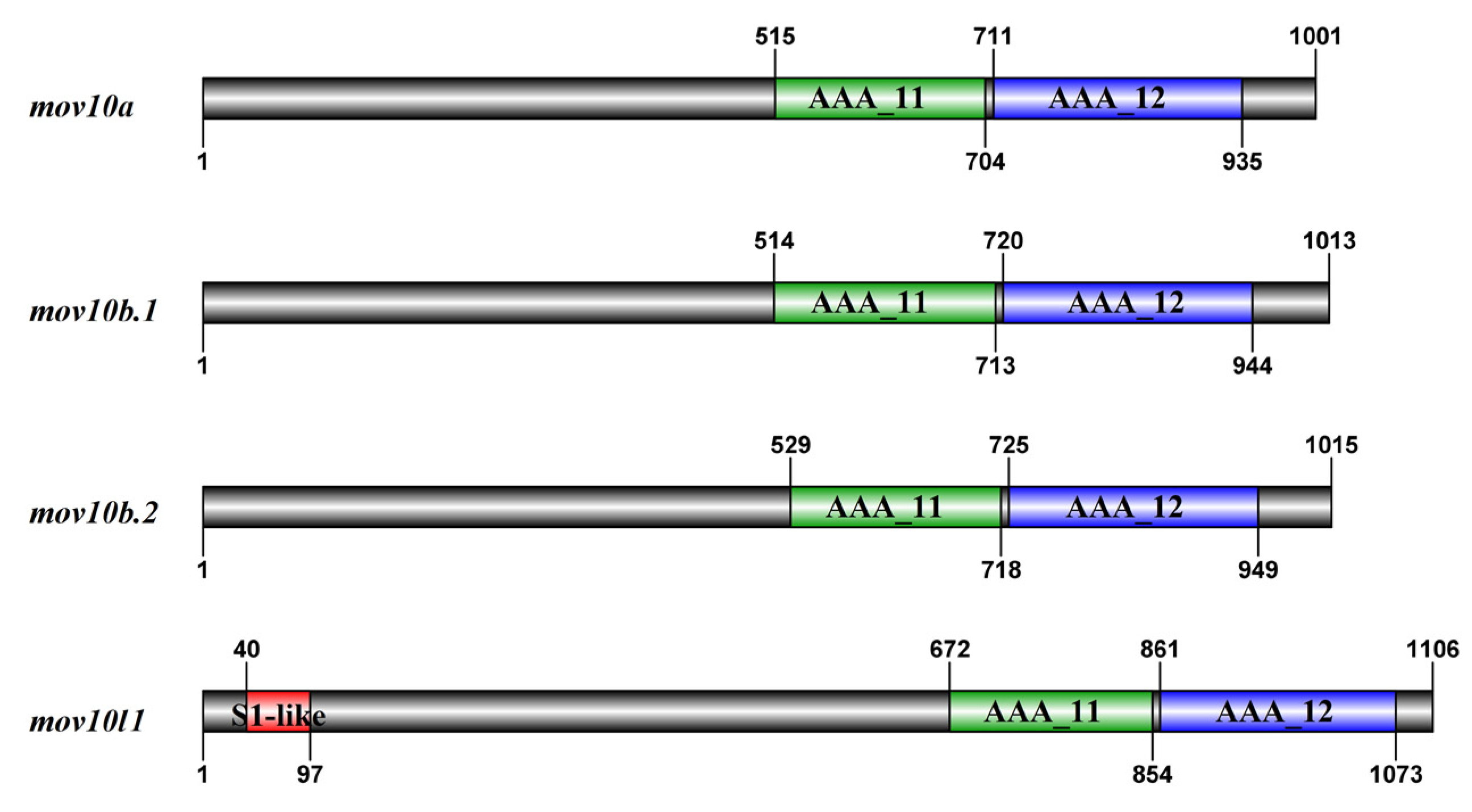
2.5. Domain and Motif Distribution Analysis
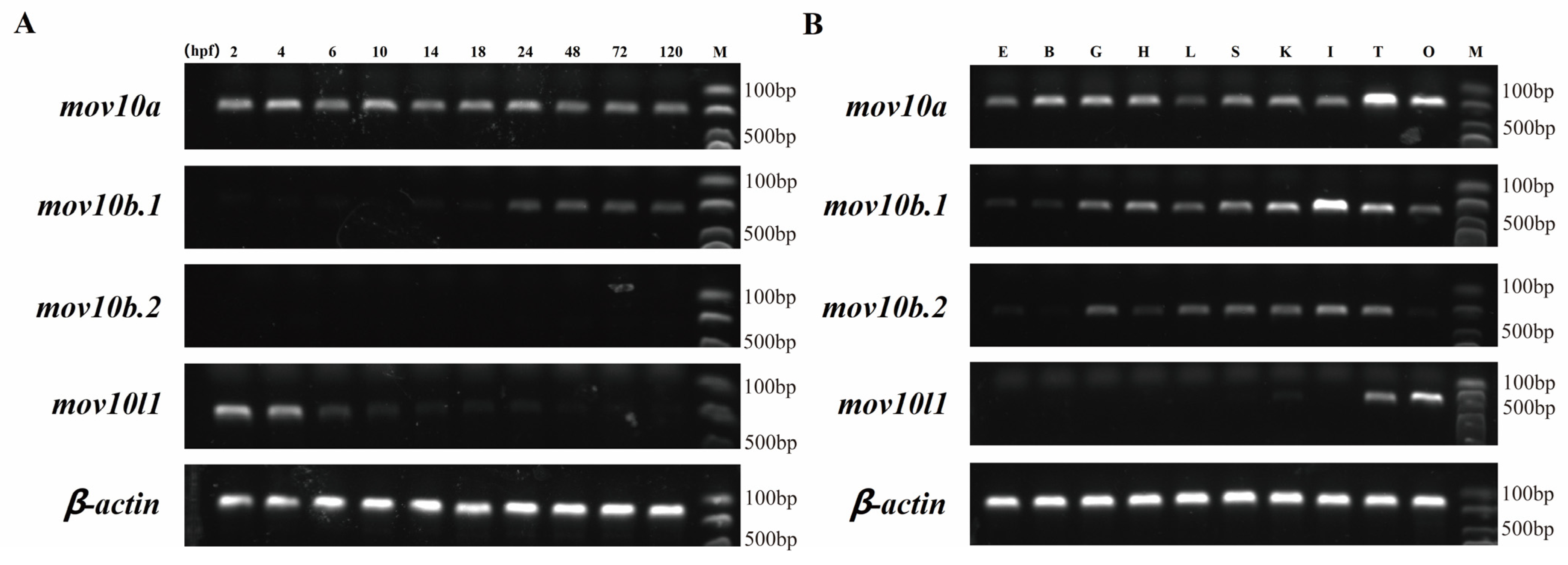
2.6. Expression Pattern of mov10s and mov10l1 in Zebrafish
2.7. Expression Profiles of mov10s and mov10l1 upon Virus Challenge
3. Discussion
4. Materials and Methods
4.1. Zebrafish Strain
4.2. Sequence Retrieval and Phylogenetics Analysis
4.3. Synteny Analysis and Selective Pressure Analysis
4.4. Virus Amplification, Adult Zebrafish Injection and Expression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, K.; Tian, S.; Tan, H.; Wang, C.; Wang, H.; Wang, M.; Wang, Y.; Chen, Z.; Wang, Y.; Yue, Q.; et al. Biological and Rna Regulatory Function of Mov10 in Mammalian Germ Cells. BMC Biol. 2019, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Jahner, D.; Nobis, P.; Simon, I.; Lohler, J.; Harbers, K.; Grotkopp, D. Chromosomal Position and Activation of Retroviral Genomes Inserted into the Germ Line of Mice. Cell 1981, 24, 519–529. [Google Scholar] [CrossRef]
- Wang, P.J.; McCarrey, J.R.; Yang, F.; Page, D.C. An Abundance of X-Linked Genes Expressed in Spermatogonia. Nat. Genet. 2001, 27, 422–426. [Google Scholar] [CrossRef]
- Hanet, A.; Rasch, F.; Weber, R.; Ruscica, V.; Fauser, M.; Raisch, T.; Kuzuoglu-Ozturk, D.; Chang, C.T.; Bhandari, D.; Igreja, C.; et al. Helz Directly Interacts with Ccr4-Not and Causes Decay of Bound Mrnas. Life Sci. Alliance 2019, 2, e201900405. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, S.C.; Jia, X.; Ge, Y.; Ling, T.; Nie, M.; Lan, X.H.; Chen, S.W.; Xu, A.L. Mitochondria-Localised Znfx1 Functions as a Dsrna Sensor to Initiate Antiviral Responses through Mavs. Nat. Cell Biol. 2019, 21, 1346. [Google Scholar] [CrossRef]
- Fairman-Williams, M.E.; Guenther, U.P.; Jankowsky, E. Sf1 and Sf2 Helicases: Family Matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, A.; Shilikbay, T.; Skariah, G.; Ceman, S. Unwinding the Roles of Rna Helicase Mov10. Wiley Interdiscip. Rev. RNA 2022, 13, e1682. [Google Scholar] [CrossRef]
- Skariah, G.; Perry, K.J.; Drnevich, J.; Henry, J.J.; Ceman, S. RNA Helicase Mov10 Is Essential for Gastrulation and Central Nervous System Development. Dev. Dyn. 2018, 247, 660–671. [Google Scholar] [CrossRef]
- Takemura, M.; Bowden, N.; Lu, Y.S.; Nakato, E.; O’Connor, M.B.; Nakato, H. Drosophila Mov10 Regulates the Termination of Midgut Regeneration. Genetics 2021, 218, iyab031. [Google Scholar] [CrossRef]
- Skariah, G.; Seimetz, J.; Norsworthy, M.; Lannom, M.C.; Kenny, P.J.; Elrakhawy, M.; Forsthoefel, C.; Drnevich, J.; Kalsotra, A.; Ceman, S. Mov10 Suppresses Retroelements and Regulates Neuronal Development and Function in the Developing Brain. BMC Biol. 2017, 15, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Identification of Novel Argonaute-Associated Proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenny, P.J.; Zhou, H.; Kim, M.; Skariah, G.; Khetani, R.S.; Drnevich, J.; Arcila, M.L.; Kosik, K.S.; Ceman, S. Mov10 and Fmrp Regulate Ago2 Association with Microrna Recognition Elements. Cell Rep. 2014, 9, 1729–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. Mov10 Is a 5’ to 3’ Rna Helicase Contributing to Upf1 Mrna Target Degradation by Translocation Along 3’ Utrs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Zhang, J.; Zhang, Y.; Geng, G.; Liang, J.; Li, Y.; Chen, J.; Liu, C.; Zhang, H. Rna Helicase Mov10 Functions as a Co-Factor of Hiv-1 Rev to Facilitate Rev/Rre-Dependent Nuclear Export of Viral Mrnas. Virology 2015, 486, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Hu, S.; Xu, F.; Mei, S.; Liu, X.; Yin, L.; Zhao, F.; Zhao, X.; Sun, H.; Xiong, Z.; et al. Mov10 Sequesters the Rnp of Influenza a Virus in the Cytoplasm and Is Antagonized by Viral Ns1 Protein. Biochem. J. 2019, 476, 467–481. [Google Scholar] [CrossRef]
- Liu, D.; Ndongwe, T.P.; Puray-Chavez, M.; Casey, M.C.; Izumi, T.; Pathak, V.K.; Tedbury, P.R.; Sarafianos, S.G. Effect of P-Body Component Mov10 on Hcv Virus Production and Infectivity. FASEB J. 2020, 34, 9433–9449. [Google Scholar] [CrossRef]
- Liu, T.; Sun, Q.; Liu, Y.; Cen, S.; Zhang, Q. The Mov10 Helicase Restricts Hepatitis B Virus Replication by Inhibiting Viral Reverse Transcription. J. Biol. Chem. 2019, 294, 19804–19813. [Google Scholar] [CrossRef]
- Song, Z.W.; Ma, Y.X.; Fu, B.Q.; Teng, X.; Chen, S.J.; Xu, W.Z.; Gu, H.X.; Participants Antibiotic Resistance Surveillance Program in Portugal. Altered Mrna Levels of Mov10, A3g, and Ifn-Alpha in Patients with Chronic Hepatitis B. J. Microbiol. 2014, 52, 510–514. [Google Scholar] [CrossRef]
- Su, F.; Liu, X.; Jiang, Y. Roles of Mov10 in Animal Rna Virus Infection. Front. Vet. Sci. 2020, 7, 569737. [Google Scholar] [CrossRef]
- Frost, R.J.; Hamra, F.K.; Richardson, J.A.; Qi, X.; Bassel-Duby, R.; Olson, E.N. Mov10l1 Is Necessary for Protection of Spermatocytes against Retrotransposons by Piwi-Interacting Rnas. Proc. Natl. Acad. Sci. USA 2010, 107, 11847–11852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, R.R.; Homolka, D.; Chen, K.M.; Sachidanandam, R.; Fauvarque, M.O.; Pillai, R.S. Recruitment of Armitage and Yb to a Transcript Triggers Its Phased Processing into Primary Pirnas in Drosophila Ovaries. PLoS Genet. 2017, 13, e1006956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vourekas, A.; Zheng, K.; Fu, Q.; Maragkakis, M.; Alexiou, P.; Ma, J.; Pillai, R.S.; Mourelatos, Z.; Wang, P.J. The RNA Helicase Mov10l1 Binds Pirna Precursors to Initiate Pirna Processing. Genes Dev. 2015, 29, 617–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, K.; Wang, P.J. Blockade of Pachytene Pirna Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity. PloS Genet. 2012, 8, e1003038. [Google Scholar] [CrossRef] [Green Version]
- Zheng, K.; Xiol, J.; Reuter, M.; Eckardt, S.; Leu, N.A.; McLaughlin, K.J.; Stark, A.; Sachidanandam, R.; Pillai, R.S.; Wang, P.J. Mouse Mov10l1 Associates with Piwi Proteins and Is an Essential Component of the Piwi-Interacting Rna (Pirna) Pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 11841–11846. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Pandey, R.R.; Leu, N.A.; Pillai, R.S.; Wang, P.J. Mutations in the Mov10l1 Atp Hydrolysis Motif Cause Pirna Biogenesis Failure and Male Sterility in Mice. Biol. Reprod. 2016, 95, 103. [Google Scholar] [CrossRef]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The Spotted Gar Genome Illuminates Vertebrate Evolution and Facilitates Human-Teleost Comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Dehal, P.; Boore, J.L. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef] [Green Version]
- Holland, P.W.; Garcia-Fernandez, J.; Williams, N.A.; Sidow, A. Gene Duplications and the Origins of Vertebrate Development. Dev. Suppl. 1994, 125–133. Available online: https://journals.biologists.com/dev/article/1994/Supplement/125/49474/Gene-duplications-and-the-origins-of-vertebrate (accessed on 5 December 2021). [CrossRef]
- Gu, Z.; Steinmetz, L.M.; Gu, X.; Scharfe, C.; Davis, R.W.; Li, W.H. Role of Duplicate Genes in Genetic Robustness against Null Mutations. Nature 2003, 421, 63–66. [Google Scholar] [CrossRef]
- Zhu, C.; Yin, Z.; Ren, J.; McCormick, R.J.; Ford, S.P.; Guo, W. Rbm20 Is an Essential Factor for Thyroid Hormone-Regulated Titin Isoform Transition. J. Mol. Cell Biol. 2015, 7, 88–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodier, J.L.; Cheung, L.E.; Kazazian, H.H., Jr. Mov10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells. PLoS Genet. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, J.; Jia, R.; Cheng, V.; Xu, X.; Qiao, W.; Guo, F.; Liang, C.; Cen, S. The Mov10 Helicase Inhibits Line-1 Mobility. J. Biol. Chem. 2013, 288, 21148–21160. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Hwang, S.Y.; Ahn, K. Interplay between Rnaseh2 and Mov10 Controls Line-1 Retrotransposition. Nucleic. Acids Res. 2018, 46, 1912–1926. [Google Scholar] [CrossRef]
- El Messaoudi-Aubert, S.; Nicholls, J.; Maertens, G.N.; Brookes, S.; Bernstein, E.; Peters, G. Role for the Mov10 Rna Helicase in Polycomb-Mediated Repression of the Ink4a Tumor Suppressor. Nat. Struct. Mol. Biol. 2010, 17, 862–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, D.T.; Wang, W.; Tipping, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The Rna-Binding Atpase, Armitage, Couples Pirna Amplification in Nuage to Phased Pirna Production on Mitochondria. Mol. Cell 2019, 74, 982–995.e6. [Google Scholar] [CrossRef] [PubMed]
- Ishizu, H.; Kinoshita, T.; Hirakata, S.; Komatsuzaki, C.; Siomi, M.C. Distinct and Collaborative Functions of Yb and Armitage in Transposon-Targeting Pirna Biogenesis. Cell Rep. 2019, 27, 1822–1835.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizu, H.; Nagao, A.; Siomi, H. Gatekeepers for Piwi-Pirna Complexes to Enter the Nucleus. Curr. Opin. Genet. Dev. 2011, 21, 484–490. [Google Scholar] [CrossRef]
- Furtak, V.; Mulky, A.; Rawlings, S.A.; Kozhaya, L.; Lee, K.; Kewalramani, V.N.; Unutmaz, D. Perturbation of the P-Body Component Mov10 Inhibits Hiv-1 Infectivity. PLoS ONE 2010, 5, e9081. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Q.; Wang, Z.; Feng, S.; Li, H.; Ji, D.; Zhang, S. Evolutionary and Expression Analyses Show Co-Option of Khdrbs Genes for Origin of Vertebrate Brain. Front. Genet. 2017, 8, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
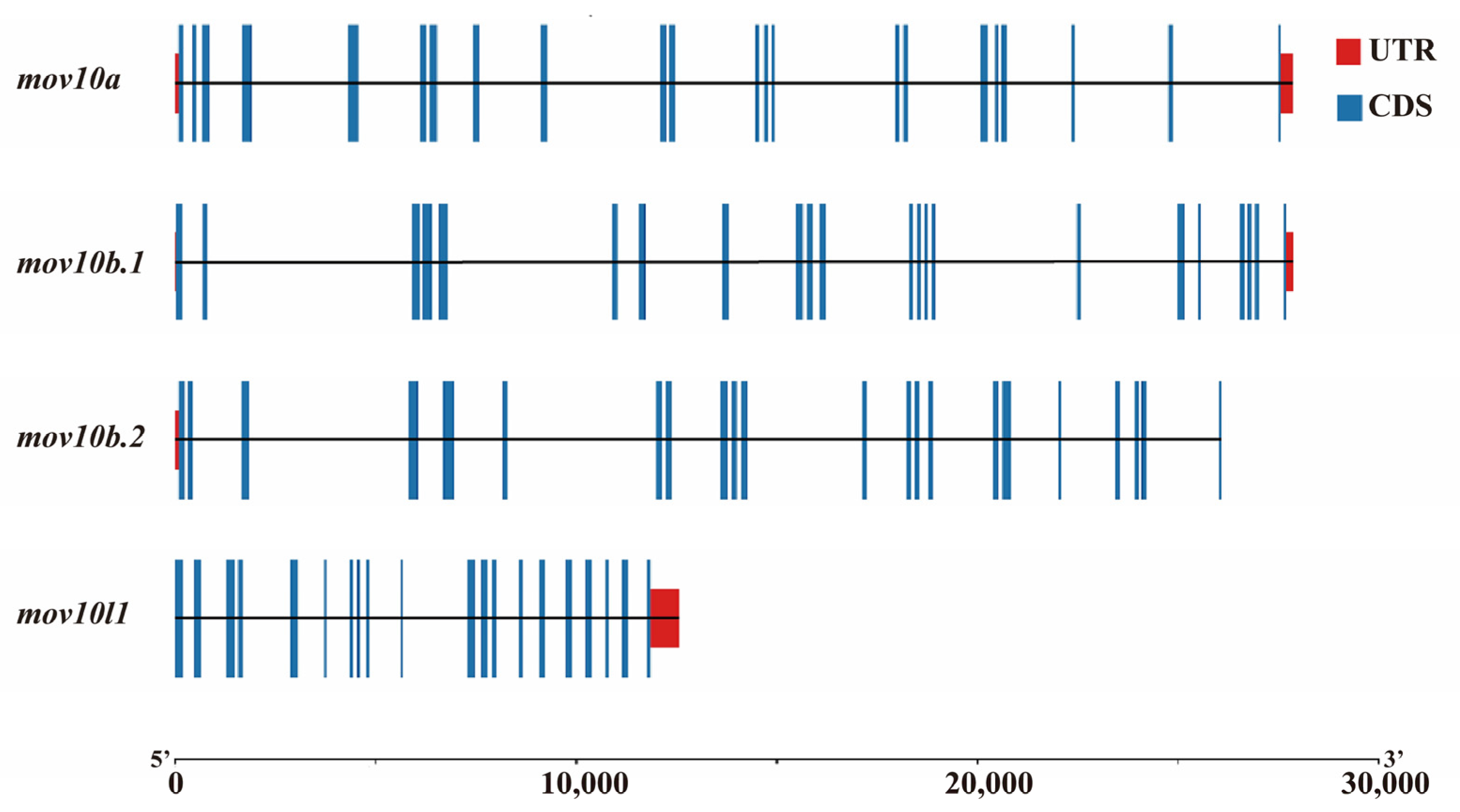


| Gene | Chromosome | No. Exon | Coding Exon | CDS (bp) | Amino Acids (aa) | Molecular Weight (kDa) | pI | Localization |
|---|---|---|---|---|---|---|---|---|
| mov10a | 6 | 22 | 22 | 3006 | 1001 | 113.92 | 9.02 | C, N, P |
| mov10b.1 | 8 | 22 | 22 | 3042 | 1013 | 116.41 | 8.87 | C, N, M |
| mov10b.2 | 8 | 22 | 22 | 3048 | 1015 | 116.51 | 8.53 | C, N, M |
| mov10l1 | 8 | 21 | 20 | 3321 | 1106 | 122.61 | 5.89 | C, N, M, E |
| Class | Species | MOV10 | MOV10L1 |
|---|---|---|---|
| Mammalia | human | 1 | 1 |
| mouse | 1 | 1 | |
| Aves | chicken | 1 | 1 |
| Amphibia | frog | 2 (1, 2) | 1 |
| Osteichthyes | torafugu | 1 | 1 |
| catfish | 2 (a, b.1) | 1 | |
| trout | 2 (a, b.1) | 1 | |
| zebrafish | 3 (a, b.1, b.2) | 1 | |
| Chondrichthyes 1 | shark | 1 | 1 |
| Cyclostomata | lamprey | 1 | 1 |
| Leptocardii | amphioxus | 3 (1, 2, 3) | 1 |
| Ascidiacea | ascidian | 2 (a, b.1) | 1 |
| Insecta | fly | 2 * | 1 |
| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Dre-mov10a | Dre-mov10b.1 | 0.429010153 | 2.306444827 | 0.186004949 |
| Dre-mov10a | Dre-mov10b.2 | 0.459220325 | 2.228487154 | 0.206068195 |
| Dre-mov10b.1 | Dre-mov10b.2 | 0.215764568 | 0.641967028 | 0.336099143 |
| Dre-mov10a | Dre-mov10l1 | 0.73581865 | 2.163735682 | 0.34006864 |
| Dre-mov10b.1 | Dre-mov10l1 | 0.782334787 | NaN | NaN |
| Dre-mov10b.2 | Dre-mov10l1 | 0.779131356 | 3.864842108 | 0.201594615 |
| Dre-mov10a | Hsa-MOV10 | 0.52853637 | 2.393945269 | 0.220780473 |
| Dre-mov10b.1 | Hsa-MOV10 | 0.558510073 | 2.654557187 | 0.2103967 |
| Dre-mov10b.2 | Hsa-MOV10 | 0.572195229 | NaN | NaN |
| Dre-mov10l1 | Hsa-MOV10L1 | 0.440656796 | 3.495808363 | 0.126052904 |
| Dre-mov10a | Ipu-mov10a | 0.20299231 | 1.431621356 | 0.141791898 |
| Dre-mov10b.1 | Ipu-mov10b.1 | 0.309622035 | 1.920636668 | 0.16120802 |
| Dre-mov10b.2 | Ipu-mov10b.1 | 0.300891219 | 2.060577589 | 0.146022756 |
| Dre-mov10l1 | Ipu-mov10l1 | 0.216627513 | 1.314532038 | 0.164794396 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Zhang, X.; Li, X.; Yin, X.; Teng, L.; Ji, G.; Li, H. Evolutionary and Expression Analysis of MOV10 and MOV10L1 Reveals Their Origin, Duplication and Divergence. Int. J. Mol. Sci. 2022, 23, 7523. https://doi.org/10.3390/ijms23147523
Yang S, Zhang X, Li X, Yin X, Teng L, Ji G, Li H. Evolutionary and Expression Analysis of MOV10 and MOV10L1 Reveals Their Origin, Duplication and Divergence. International Journal of Molecular Sciences. 2022; 23(14):7523. https://doi.org/10.3390/ijms23147523
Chicago/Turabian StyleYang, Shuaiqi, Xiangmin Zhang, Xianpeng Li, Xiu Yin, Lei Teng, Guangdong Ji, and Hongyan Li. 2022. "Evolutionary and Expression Analysis of MOV10 and MOV10L1 Reveals Their Origin, Duplication and Divergence" International Journal of Molecular Sciences 23, no. 14: 7523. https://doi.org/10.3390/ijms23147523
APA StyleYang, S., Zhang, X., Li, X., Yin, X., Teng, L., Ji, G., & Li, H. (2022). Evolutionary and Expression Analysis of MOV10 and MOV10L1 Reveals Their Origin, Duplication and Divergence. International Journal of Molecular Sciences, 23(14), 7523. https://doi.org/10.3390/ijms23147523






