Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress
Abstract
:1. Introduction
2. Result
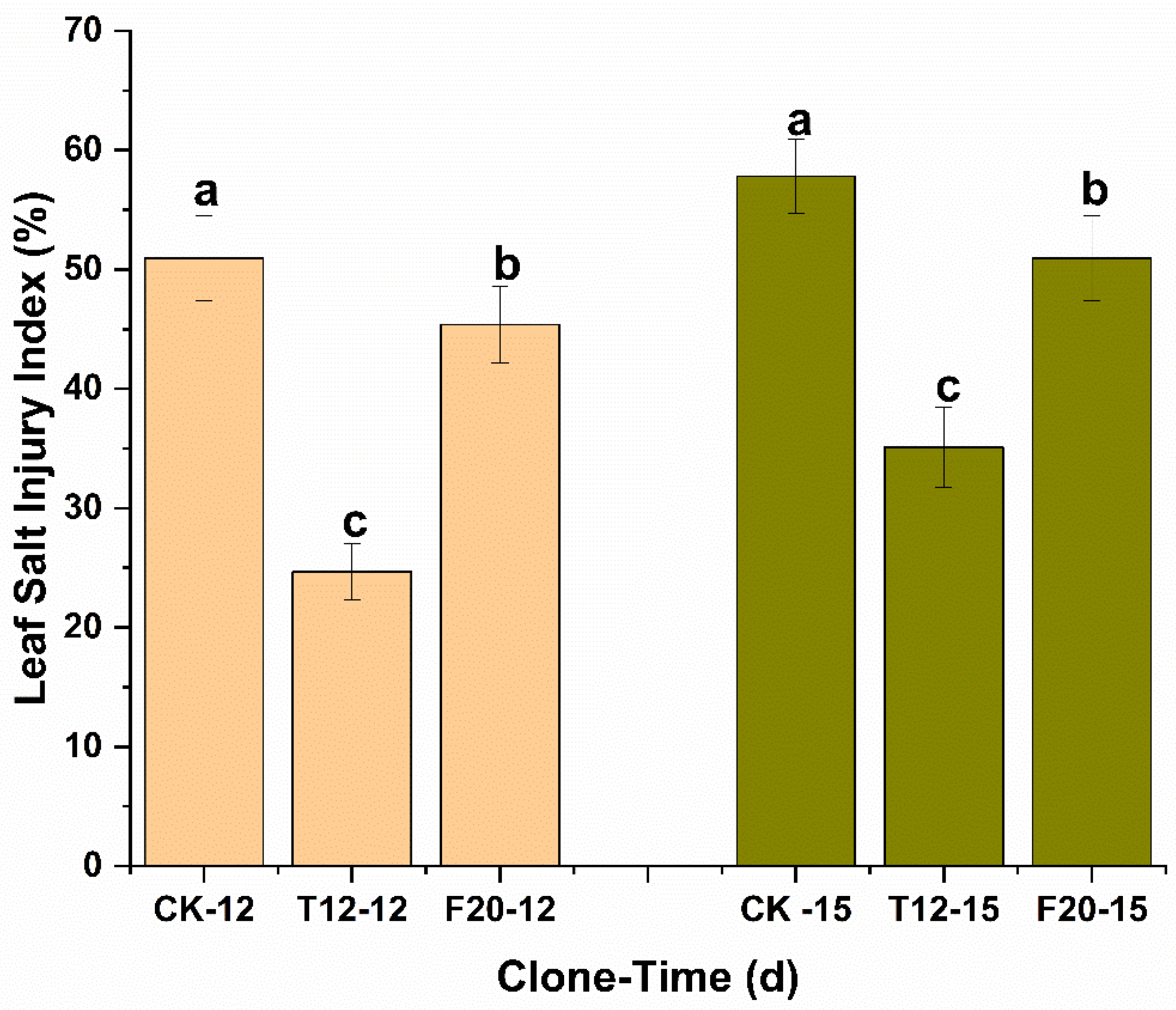
2.1. The Leaf Salt Injury Levels of CK, T12, and F20 under Salt Stress
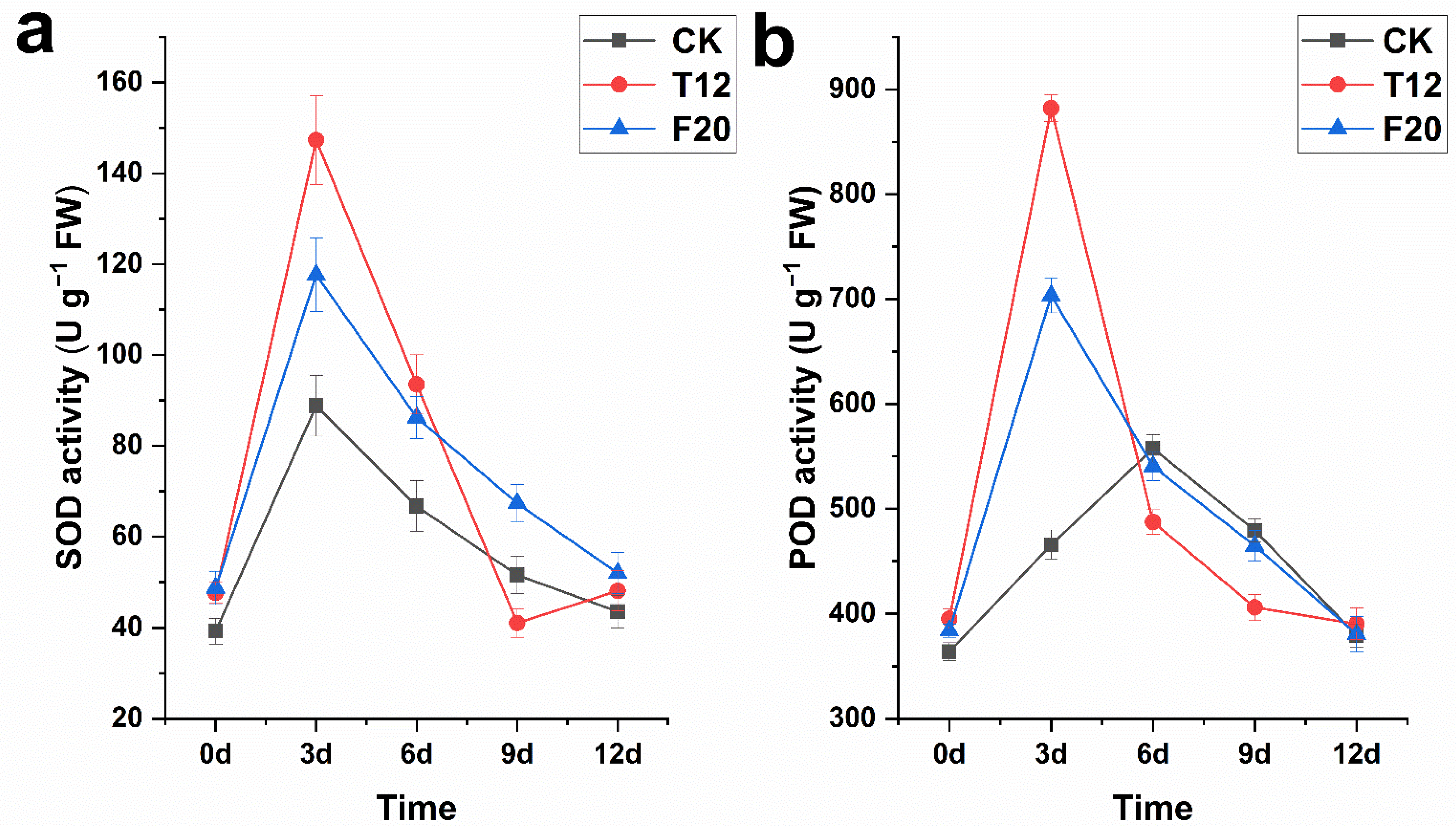
2.2. SOD and POD Activity of CK, T12, and F20 Leaves under Salt Stress
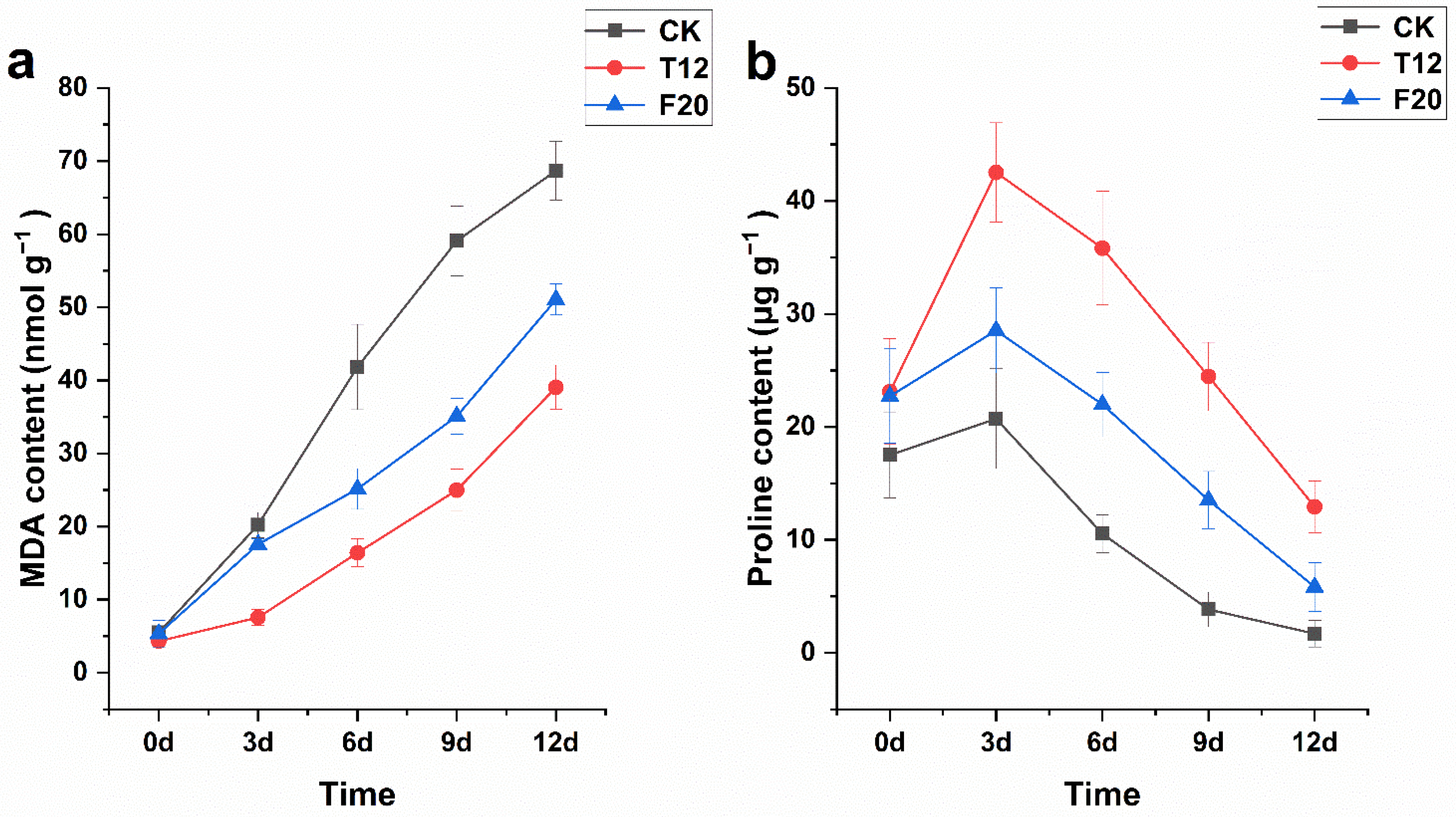
2.3. MDA and Proline Content of CK, T12, and F20 Leaves under Salt Stress
2.4. Relative Electric Conductivity of CK, T12, and F20 Leaves under Salt Stress
2.5. Basic Information of RNA-Seq
2.6. The DEGs among CK, T12, and F20 under Salt Stress Treatment
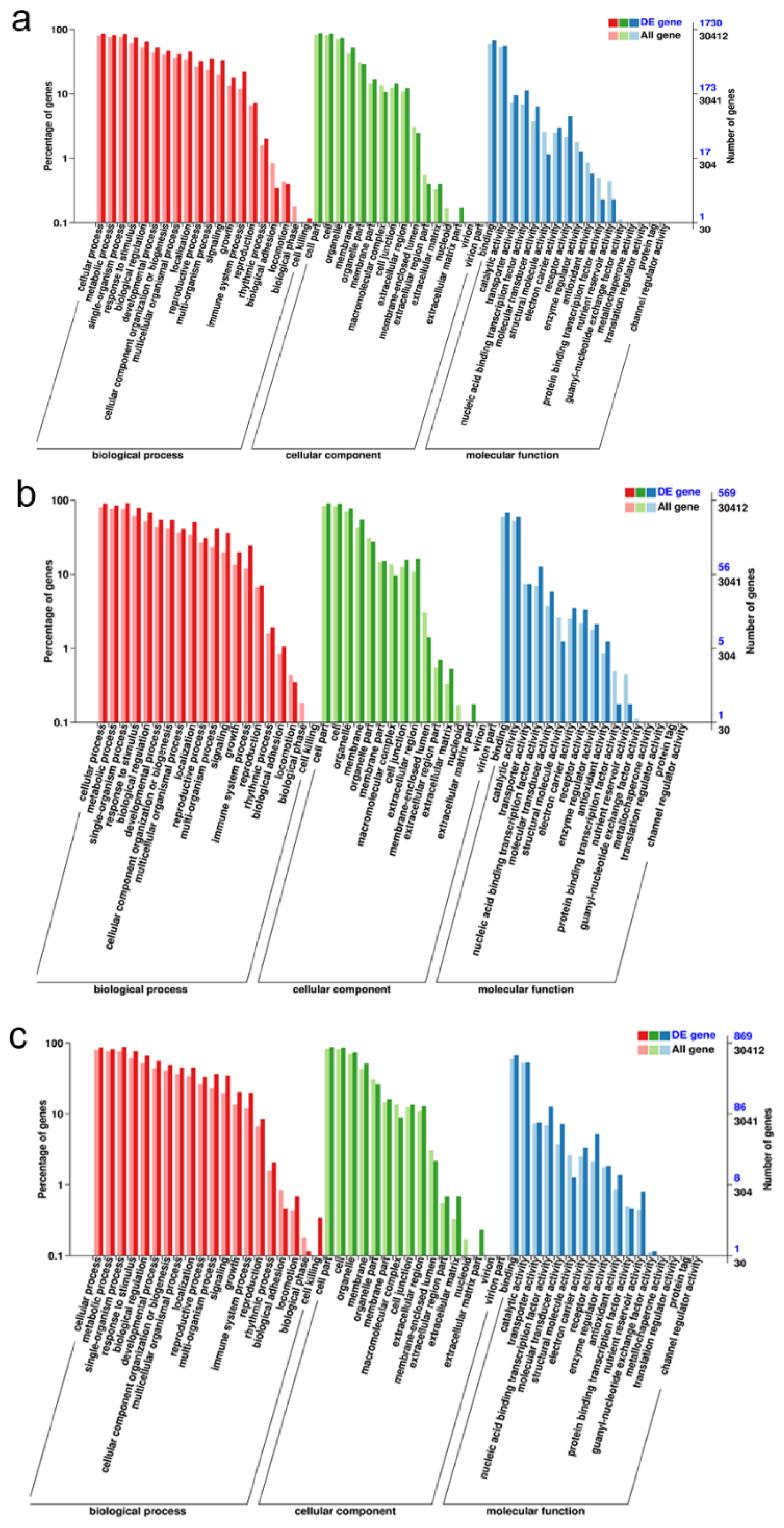
2.7. GO Classification and GO Term Enrichment on the DEGs among CK, T12, and F20 under Salt Stress Treatment
2.8. KEGG Enrichment on the DEGs among CK, T12, and F20 under Salt Stress Treatment
2.9. QRT-PCR Verification
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Salt Stress Treatment
4.2. Leaf Salt Injury Index Survey
4.3. SOD and POD Activity, and MDA and Proline Content Detection
4.4. Relative Electric Conductivity Detection
4.5. RNA-Seq
4.6. QRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; De Vleesschauwer, D.; Sharma, M.K.; Ronald, P.C. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant 2013, 6, 250–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep Learning for Plant Stress Phenotyping: Trends and Future Perspectives. Trends Plant Sci. 2018, 23, 883–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Oh, T.R.; Kim, J.H.; Cho, S.K.; Ryu, M.Y.; Yang, S.W.; Kim, W.T. AtAIRP2 E3 Ligase Affects ABA and High-Salinity Responses by Stimulating Its ATP1/SDIRIP1 Substrate Turnover. Plant Physiol. 2017, 174, 2515–2531. [Google Scholar] [CrossRef] [Green Version]
- Abogadallah, G.M. Antioxidative defense under salt stress. Plant Signal. Behav 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Scholes, D.R.; Paige, K.N. Plasticity in ploidy: A generalized response to stress. Trends Plant Sci. 2015, 20, 165–175. [Google Scholar] [CrossRef]
- Deng, B.; Du, W.; Liu, C.; Sun, W.; Tian, S.; Dong, H. Antioxidant response to drought, cold and nutrient stress in two ploidy levels of tobacco plants: Low resource requirement confers polytolerance in polyploids? Plant Growth Regul. 2012, 66, 37–47. [Google Scholar] [CrossRef]
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Curr. Biol. 2015, 25, R353–R358. [Google Scholar] [CrossRef] [Green Version]
- Tank, D.C.; Eastman, J.M.; Pennell, M.W.; Soltis, P.S.; Soltis, D.E.; Hinchliff, C.E.; Brown, J.W.; Sessa, E.B.; Harmon, L.J. Nested radiations and the pulse of angiosperm diversification: Increased diversification rates often follow whole genome duplications. New Phytol. 2015, 207, 454–467. [Google Scholar] [CrossRef] [Green Version]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.P.; Kiew, R.; Set, O.; Gan, L.H.; Gan, Y.Y. Amplified fragment length polymorphism fingerprinting of 16 banana cultivars (Musa cvs.). Mol. Phylogenet. Evol. 2000, 17, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jin, D.; Wang, Z.; Guo, H.; Zhang, L.; Wang, L.; Li, J.; Paterson, A.H. Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytol. 2015, 205, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Davis, T.M. A New Perspective on Polyploid Fragaria (Strawberry) Genome Composition Based on Large-Scale, Multi-Locus Phylogenetic Analysis. Genome Biol. Evol. 2017, 9, 3433–3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and Applications of a High Throughput Genotyping Tool for Polyploid Crops: Single Nucleotide Polymorphism (SNP) Array. Front. Plant Sci. 2018, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Zhang, B.; Lisch, D.; Ma, J. Patterns and Consequences of Subgenome Differentiation Provide Insights into the Nature of Paleopolyploidy in Plants. Plant Cell 2017, 29, 2974–2994. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, Y.J.; Scheen, A.C.; Marcussen, T.; Pfeil, B.E.; de Sousa, F.; Oxelman, B. Assignment of homoeologs to parental genomes in allopolyploids for species tree inference, with an example from Fumaria (papaveraceae). Syst. Biol. 2015, 64, 448–471. [Google Scholar] [CrossRef] [Green Version]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Van Laere, K.; França, S.C.; Vansteenkiste, H.; Van Huylenbroeck, J.; Steppe, K.; Van Labeke, M. Influence of ploidy level on morphology, growth and drought susceptibility in Spathiphyllum wallisii. Acta Physiol. Plant. 2011, 33, 1149–1156. [Google Scholar] [CrossRef]
- Si, C.L.; Li, S.M.; Liu, Z.; Kim, J.K.; Bae, Y.S. Antioxidant phenolic glycosides from the bark of Populus ussuriensis Kom. Nat. Prod. Res. 2011, 25, 1396–1401. [Google Scholar] [CrossRef]
- Si, C.L.; Kim, J.K.; Bae, Y.S.; Li, S.M. Phenolic compounds in the leaves of Populus ussuriensis and their antioxidant activities. Planta Med. 2009, 75, 1165–1167. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.C.; Malczewski, G.; Saprunoff, M. Assessing the potential of native tree species for carbon sequestration forestry in Northeast China. J. Environ. Manag. 2007, 85, 663–671. [Google Scholar] [CrossRef]
- Xu, J.; Jin, J.; Zhao, H.; Li, K. Drought stress tolerance analysis of Populus ussuriensis clones with different ploidies. J. For. Res. 2019, 30, 1267–1275. [Google Scholar] [CrossRef]
- Lou, L.; Zhao, H.; Qi, C.; Lu, Q.; Wang, B.; Li, K. Autotriploid Induction in Populus ussuriensis Kom. by Colchicine. Plant Physiol. J. 2011, 47, 699–704. [Google Scholar]
- Zhao, H.; Zhao, X.; Li, M.; Jiang, Y.; Xu, J.; Jin, J.; Li, K. Ectopic expression of Limonium bicolor (Bag.) Kuntze DREB (LbDREB) results in enhanced salt stress tolerance of transgenic Populus ussuriensis Kom. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 132, 123–136. [Google Scholar] [CrossRef]
- Anjum, N.A.; Sofo, A.; Scopa, A.; Roychoudhury, A.; Gill, S.S.; Iqbal, M.; Lukatkin, A.S.; Pereira, E.; Duarte, A.C.; Ahmad, I. Lipids and proteins--major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. Int. 2015, 22, 4099–4121. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Yoshida, K.; Kaothien, P.; Matsui, T.; Kawaoka, A.; Shinmyo, A. Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 2003, 60, 665–670. [Google Scholar] [CrossRef]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taulavuori, E.; Hellstrom, E.K.; Taulavuori, K.; Laine, K. Comparison of two methods used to analyse lipid peroxidation from Vaccinium myrtillus (L.) during snow removal, reacclimation and cold acclimation. J. Exp. Bot. 2001, 52, 2375–2380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sochor, J.; Ruttkay-Nedecky, B.; Babula, P.; Adam, V.; Kizek, R. Automation of Methods for Determination of Lipid Peroxidation. In Lipid Peroxidation; IntechOpen: London, UK, 2012. [Google Scholar]
- Sofo, A.; Dichio, B.; Xiloyannis, C.; Masia, A. Lipoxygenase activity and proline accumulation in leaves and roots of olive trees in response to drought stress. Physiol. Plant 2004, 121, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Astorga, G.I.; Alcaraz-Meléndez, L. Salinity effects on protein content, lipid peroxidation, pigments, and proline in Paulownia imperialis (Siebold & Zuccarini) and Paulownia fortunei (Seemann & Hemsley) grown in vitro. Electron. J. Biotechnol. 2011, 13, 13–14. [Google Scholar]
- Ahmad, P.; Jaleel, C.A.; Sharma, S. Antioxidant defense system, lipid peroxidation, proline-metabolizing enzymes, and biochemical activities in two Morus alba genotypes subjected to NaCl stress. Russ. J. Plant Physiol. 2010, 57, 509–517. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Parvin, K.; Bhuiyan, T.F.; Fujita, M. Regulation of ROS Metabolism in Plants under Environmental Stress: A Review of Recent Experimental Evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef]
- Gao, Y.F.; Liu, J.K.; Yang, F.M.; Zhang, G.Y.; Wang, D.; Zhang, L.; Ou, Y.B.; Yao, Y.A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant 2020, 168, 98–117. [Google Scholar] [CrossRef]
- Ju, Y.L.; Yue, X.F.; Min, Z.; Wang, X.H.; Fang, Y.L.; Zhang, J.X. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 98–111. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Zhou, X.; Hou, X.; Yang, G.; Meng, J.; Luan, Y. Tomato MYB49 enhances resistance to Phytophthora infestans and tolerance to water deficit and salt stress. Planta 2018, 248, 1487–1503. [Google Scholar] [CrossRef]
- Li, P.; Chai, Z.; Lin, P.; Huang, C.; Huang, G.; Xu, L.; Deng, Z.; Zhang, M.; Zhang, Y.; Zhao, X. Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane (Saccharum spontaneum L.). BMC Genom. 2020, 21, 685. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Luo, R.; Li, G.; Song, Y.; Zhan, S.; Zhao, K.; Hua, W.; Zhang, Y.; Wu, X.; Yang, C. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics 2019, 9, 4084–4100. [Google Scholar] [CrossRef] [PubMed]
- Spencer, B.G.; Finnie, J.W. The Role of Endoplasmic Reticulum Stress in Cell Survival and Death. J. Comp. Pathol. 2020, 181, 86–91. [Google Scholar] [CrossRef]
- Shalaby, S.; Horwitz, B.A. Plant phenolic compounds and oxidative stress: Integrated signals in fungal-plant interactions. Curr. Genet. 2015, 61, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Wu, H.C.; Jinn, T.L. Coordination of ABA and Chaperone Signaling in Plant Stress Responses. Trends Plant Sci. 2019, 24, 636–651. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Matamoros, M.A.; Becana, M. Molecular responses of legumes to abiotic stress: Post-translational modifications of proteins and redox signaling. J. Exp. Bot. 2021, 72, 5876–5892. [Google Scholar] [CrossRef]
- Rouhier, N.; Santos, C.; Tarrago, L.; Rey, P. Plant methionine sulfoxide reductase A and B multigenic families. Photosynth. Res. 2006, 89, 247–262. [Google Scholar] [CrossRef]
- Sanchez-Pujante, P.J.; Borja-Martinez, M.; Pedreno, M.A.; Almagro, L. Biosynthesis and bioactivity of glucosinolates and their production in plant in vitro cultures. Planta 2017, 246, 19–32. [Google Scholar] [CrossRef]
- Schafer, M.; Brutting, C.; Meza-Canales, I.D.; Grosskinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef] [Green Version]
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 2018, 51, 46. [Google Scholar] [CrossRef] [Green Version]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Horvath, E.; Galle, A.; Szabados, L.; Tari, I.; Csiszar, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Jiang, J.; Li, K.; Liu, G. Populus simonii × Populus nigra WRKY70 is involved in salt stress and leaf blight disease responses. Tree Physiol. 2017, 37, 827–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- He, J.; Li, H.; Luo, J.; Ma, C.; Li, S.; Qu, L.; Gai, Y.; Jiang, X.; Janz, D.; Polle, A.; et al. A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus × canescens. Plant Physiol. 2013, 162, 424–439. [Google Scholar] [CrossRef] [Green Version]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Wang, S.; Chen, S.; Jiang, J.; Liu, G. Phylogenetic and stress-responsive expression analysis of 20 WRKY genes in Populus simonii × Populus nigra. Gene 2015, 565, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Clone | REC-0 d | REC-3 d | REC-6 d | REC-9 d | REC-12 d |
|---|---|---|---|---|---|
| CK | 17.65 ± 1.16 a | 21.09 ± 1.84 a | 25.96 ± 1.35 a | 31.02 ± 3.00 a | 35.64 ± 0.85 a |
| T12 | 11.70 ± 1.37 b | 15.19 ± 1.44 b | 16.45 ± 1.03 c | 19.03 ± 1.40 c | 22.79 ± 1.59 c |
| F20 | 13.87 ± 1.62 b | 16.92 ± 0.19 b | 19.37 ± 1.08 b | 24.36 ± 1.02 b | 28.69 ± 1.42 b |
| Sample Name | Clean Reads | Mapped Clean Reads | %≥ Q30 |
|---|---|---|---|
| T01 | 41,910,366 | 30,375,971 | 92.38% |
| T02 | 41,040,924 | 29,443,067 | 92.26% |
| T03 | 40,111,090 | 28,728,047 | 92.32% |
| T04 | 44,212,562 | 32,116,907 | 91.93% |
| T05 | 45,811,970 | 32,832,194 | 91.54% |
| T06 | 42,112,362 | 30,218,367 | 92.16% |
| T07 | 41,468,618 | 30,189,953 | 91.86% |
| T08 | 45,156,026 | 32,502,831 | 92.58% |
| T09 | 40,742,788 | 28,892,712 | 91.59% |
| T10 | 40,754,178 | 28,732,594 | 91.63% |
| T11 | 45,714,746 | 32,271,613 | 91.74% |
| T12 | 45,579,522 | 32,301,725 | 92.16% |
| T13 | 57,068,718 | 40,927,502 | 92.16% |
| T14 | 45,700,546 | 32,576,326 | 91.78% |
| T15 | 47,126,662 | 33,622,560 | 92.31% |
| T16 | 48,398,450 | 34,772,614 | 92.76% |
| T17 | 59,470,590 | 42,382,740 | 92.82% |
| T18 | 47,650,394 | 34,081,348 | 92.47% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Liu, H.; Jin, J.; Ma, X.; Li, K. Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress. Int. J. Mol. Sci. 2022, 23, 7529. https://doi.org/10.3390/ijms23147529
Zhao H, Liu H, Jin J, Ma X, Li K. Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress. International Journal of Molecular Sciences. 2022; 23(14):7529. https://doi.org/10.3390/ijms23147529
Chicago/Turabian StyleZhao, Hui, Huanzhen Liu, Jiaojiao Jin, Xiaoyu Ma, and Kailong Li. 2022. "Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress" International Journal of Molecular Sciences 23, no. 14: 7529. https://doi.org/10.3390/ijms23147529
APA StyleZhao, H., Liu, H., Jin, J., Ma, X., & Li, K. (2022). Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress. International Journal of Molecular Sciences, 23(14), 7529. https://doi.org/10.3390/ijms23147529





