Cloning and Characterization of a Thermostable Endolysin of Bacteriophage TP-84 as a Potential Disinfectant and Biofilm-Removing Biological Agent
Abstract
:1. Introduction
2. Results
2.1. Bioinformatic Analysis of TP84_28 Endolysin Gene
2.2. Biosynthesis and Isolation of Native and Recombinant TP84_28 Endolysins
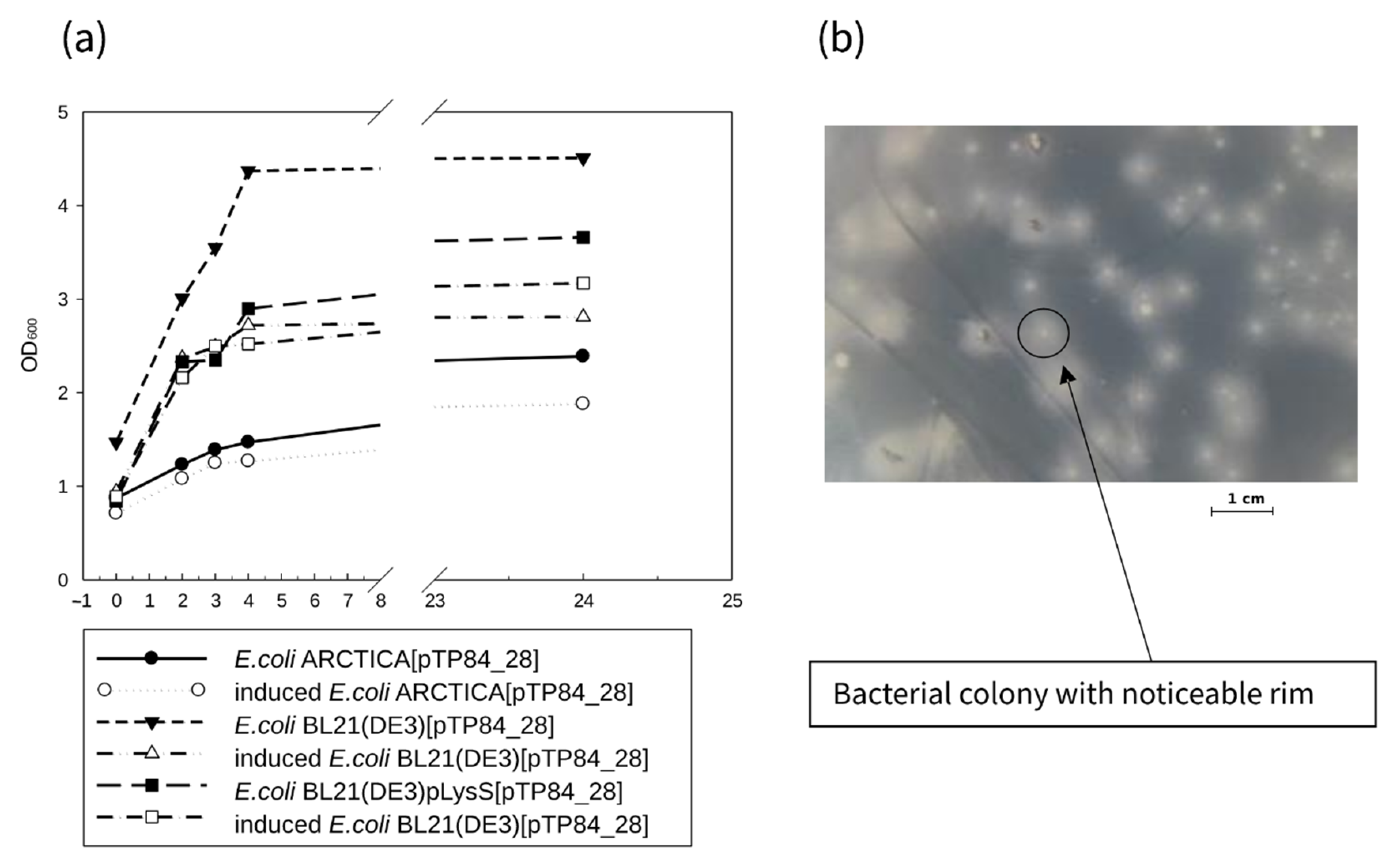
2.2.1. Cloning, Overexpression, and Purification of Recombinant TP84_28 Endolysin
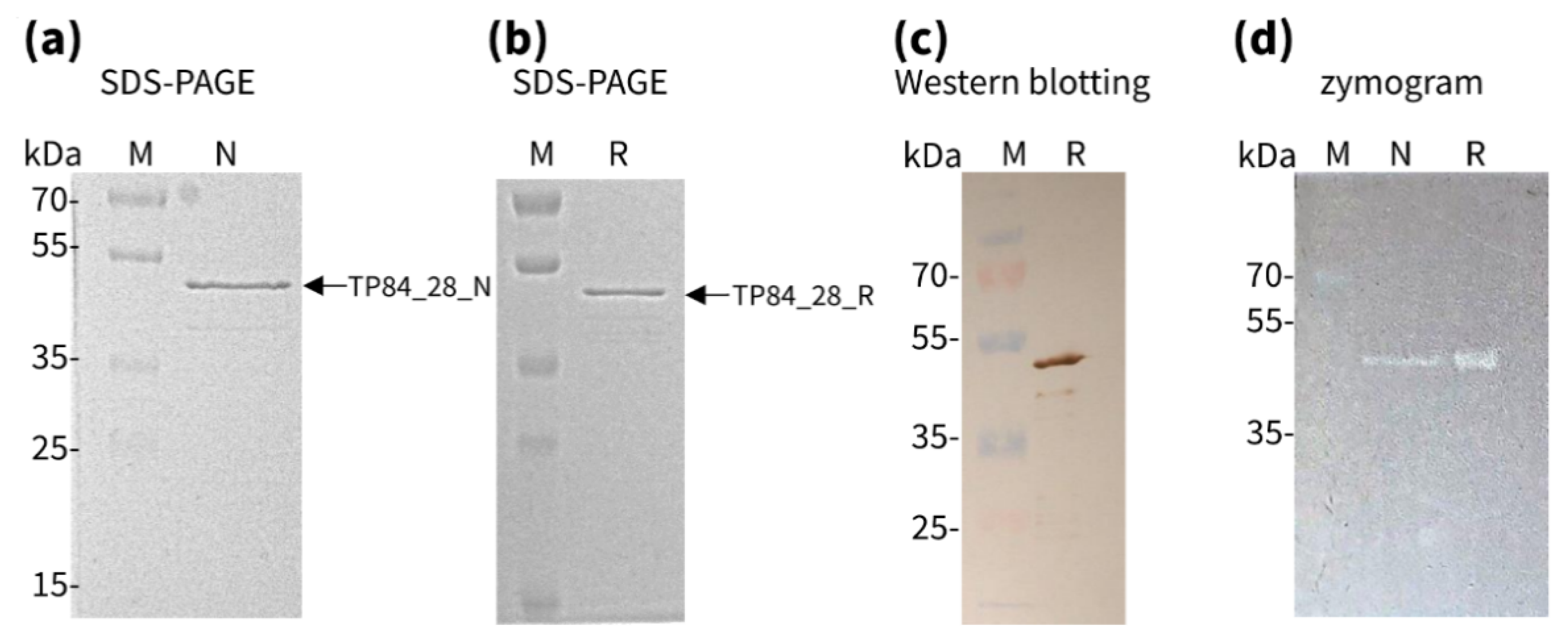
2.2.2. Isolation and Purification of Native TP84_28 Endolysin
2.3. Properties of TP84_28 Endolysin
2.3.1. Activity of Recombinant versus Native TP84_28 Endolysin
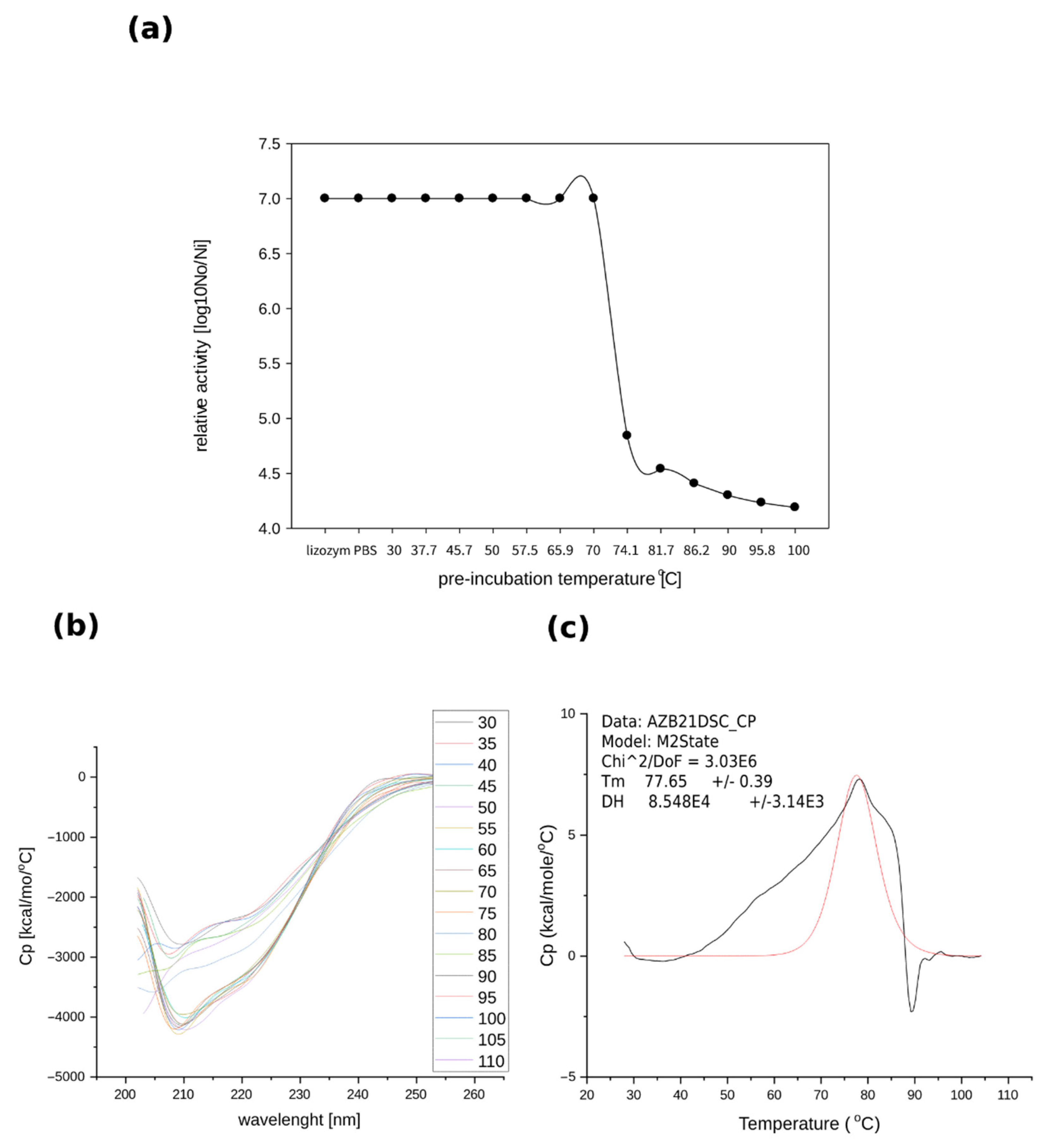
2.3.2. Thermostability of Recombinant TP84_28 Endolysin
2.3.3. pH Effect on TP84_28 Endolysin Activity
2.4. Lytic Spectrum of TP84_28 Endolysin on Bacterial Substrates
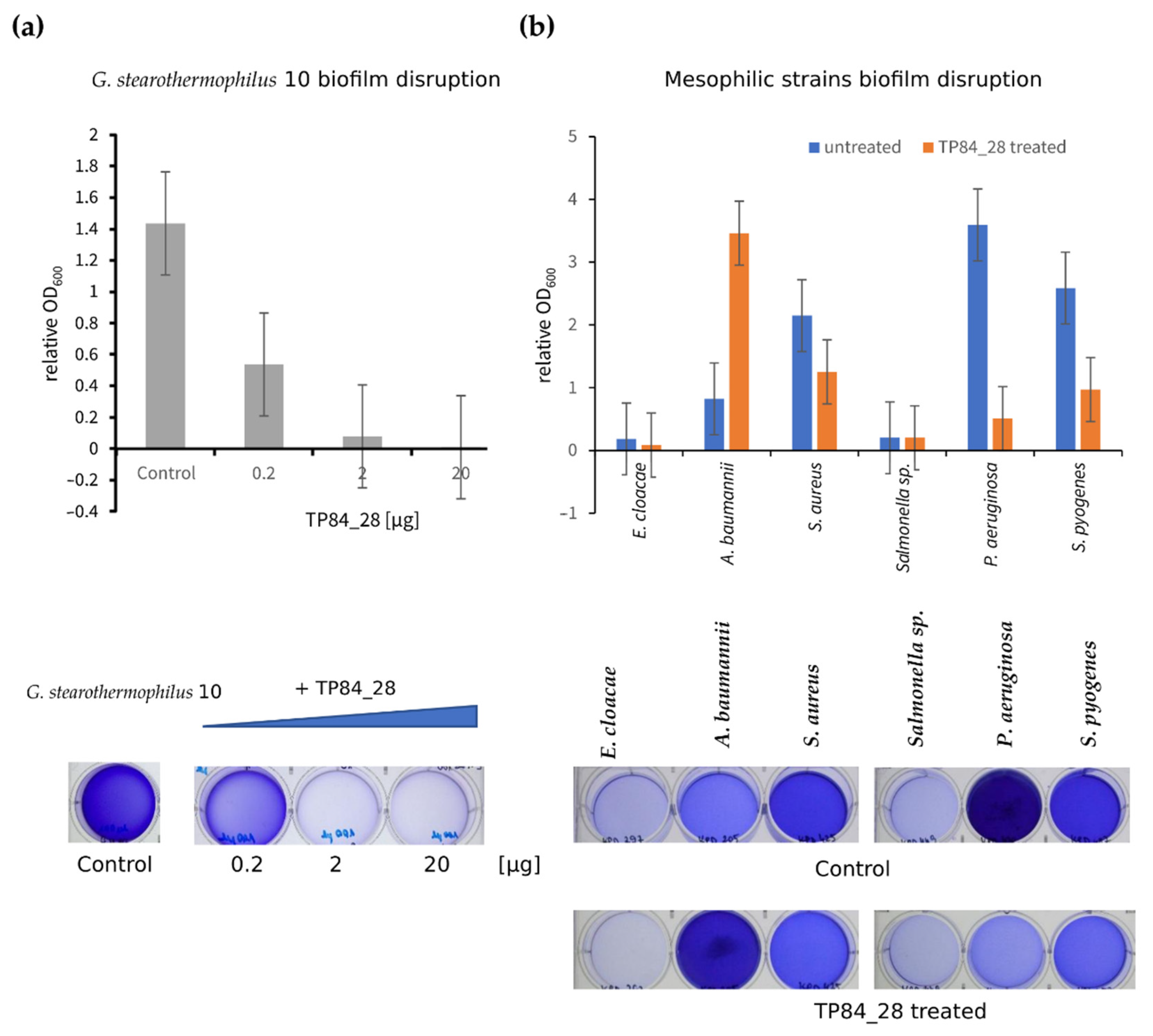
2.5. Disruption of G. stearothermophilus Strain 10 and Mesophilic Bacterial Biofilms
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmid, Media, and Reagents
4.2. Recombinant TP84_28 Endolysin Production
4.2.1. Cloning of TP-84 Endolysin Gene into pET21d_His Vector
4.2.2. Recombinant TP84_28 Endolysin Purification
4.3. Native TP84_28 Endolysin Production
4.3.1. Bacterial Culture and Bacteriophage TP84-Mediated Cell Lysis
4.3.2. Protein Purification
4.4. Western Blotting Analysis
4.5. LC–MS–MS/MS Analysis
4.6. Analysis of Protein Activity
4.6.1. Diffusion Test
4.6.2. Zymogram Analysis
4.6.3. Turbidity-Reduction Assay
4.7. Antibacterial Activity
4.8. CD Spectroscopy—Thermostability
4.9. DSC Analysis
4.10. pH Effect Evaluation Using TRA
4.11. Biofilm Survey
4.12. Biofilm Disruption
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- SchmelcherSchmelcher, M.; Donovan, D.; Loessner, M. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skowron, P.M.; Kropinski, A.M.; Zebrowska, J.; Janus, L.; Szemiako, K.; Czajkowska, E.; Maciejewska, N.; Skowron, M.; Łoś, J.; Łoś, M. Sequence, genome organization, annotation and proteomics of the thermophilic, 47.7-kb Geobacillus stearothermophilus bacteriophage TP-84 and its classification in the new Tp84virus genus. PLoS ONE 2018, 13, e0195449. [Google Scholar] [CrossRef]
- Epstein, I.; Campbell, L. Production and Purification of the Thermophilic Bacteriophage TP-84. Microbiology 1975, 29, 219–223. [Google Scholar] [CrossRef]
- Łubkowska, B.; Czajkowska, E.; Stodolna, A.; Zylicz-Stachula, A.; Skoron, P. Rapid polyethyleneimine processing of a bacteriophage-expressed proteins from cell lysates: A novel thermostable TP-84 glycosylase-depolymerase as a potential antimicrobial agent. Microb. Cell Fact. 2022, submitted.
- De Vos, P.; Garrity, G.M.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.; Schleifer, K.-H.; Whitman, W.B. Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Volume 3: The Firmicutes; Springer: New York, NY, USA, 2009; p. 182. [Google Scholar]
- Guttman, B.; Raya, P.; Kutter, E. Basic Phage Biology. In Bacteriophages: Biology and Application; Kutter, E.B., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 29–66. [Google Scholar]
- Fischetti, V.A. Using phage Lytic Enzymes to Control Pathogenic Bacteria. BMC Oral Health 2006, 6 (Suppl. 1), S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldentey, J.; Hänninen, A.L.; Bamford, D.H. Gene XV of bacteriophage PRD1 encodes a lytic enzyme with muramidase activity. Eur. J. Biochem. 1994, 225, 341–346. [Google Scholar] [CrossRef]
- Rydman, P.S.; Bamford, D.H. Identification and mutational analysis of bacteriophage PRD1 holin protein P35. J. Bacteriol. 2003, 185, 3795–3803. [Google Scholar] [CrossRef] [Green Version]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef]
- Swift, S.M.; Reid, K.P.; Donovan, D.M.; Ramsay, T.G. Thermophile Lytic Enzyme Fusion Proteins that Target Clostridium perfringens. Antibiotics 2019, 8, 214. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar] [CrossRef] [Green Version]
- Dewey, J.S.; Savva, C.G.; White, R.L.; Vitha, S.; Holzenburg, A.; Young, R. Micron-scale holes terminate the phage infection cycle. Proc. Natl. Acad. Sci. USA 2010, 107, 2219–2223. [Google Scholar] [CrossRef] [Green Version]
- Borysowski, J.; Weber-Dąbrowska, B.; Górski, A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Hermoso, J.A.; Garcia, J.L.; Garcia, P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007, 10, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Mesnage, S.; Chau, F.; Dubost, L.; Arthur, M. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J. Biol. Chem. 2008, 283, 19845–19853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lood, R.; Raz, A.; Molina, H.; Euler, C.W.; Fischetti, V.A. A highly active and negatively charged Streptococcus pyogenes lysin with a rare d-alanyl-l-alanine endopeptidase activity protects mice against streptococcal bacteremia. Antimicrob. Agents Chemother. 2014, 58, 3073–3084. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.; Thiagarajan, V.; Walmagh, M.; Sillankorva, S.; Lavigne, R.; Neves-Petersen, M.T.; Kluskens, L.D.; Azeredo, J. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against Gram-negative pathogens in presence of weak acids. PLoS ONE 2014, 9, e108376. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Mao, J.; Xie, J. Bacteriophage Polysaccharide Depolymerases and Biomedical Applications. BioDrugs 2013, 28, 265–274. [Google Scholar] [CrossRef]
- Maszewska, A. Phage associated polysaccharide depolymerases—Characteristics and application. Postepy Hig. Med. Dosw. 2015, 69, 690–702. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Mirski, T.; Mizak, L.; Nakonieczna, A.; Gryko, R. Bacteriophages, phage endolysins and antimicrobial peptides—The possibilities for their common use to combat infections and in the design of new drugs. Ann. Agric. Environ. Med. 2019, 26, 203–209. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zebrowska, J.; Witkowska, M.; Struck, A.; Laszuk, P.E.; Raczuk, E.; Ponikowska, M.; Skowron, P.M.; Zylicz-Stachula, A. Antimicrobial Potential of the Genera Geobacillus and Parageobacillus, as Well as Endolysins Biosynthesized by Their Bacteriophages. Antibiotics 2022, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.U.; Wang, W.; Sun, Q.; Shah, J.A.; Li, C.; Sun, Y.; Li, Y.; Zhang, B.; Chen, W.; Wang, S. Endolysin, a Promising Solution against Antimicrobial Resistance. Antibiotics 2021, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y. Bacteriophage-Derived Endolysins Applied as Potent Biocontrol Agents to Enhance Food Safety. Microorganisms 2020, 8, 724. [Google Scholar] [CrossRef] [PubMed]
- Andre, S.; Vallaeys, T.; Planchon, S. Spore-forming bacteria responsible for food spoilage. Res. Microbiol. 2017, 168, e379–e387. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Loessner, M.J. Bacteriophage endolysins: Applications for food safety. Curr. Opin. Biotechnol. 2016, 37, 76–87. [Google Scholar] [CrossRef]
- National Library of Medicine; National Centre for Biotechnology Information. BLAST: Basic Local Aligment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 22 April 2022).
- Plotka, M.; Kaczorowska, A.K.; Stefanska, A.; Morzywolek, A.; Fridjonsson, O.H.; Dunin-Horkawicz, S.; Kozlowski, L.; Hreggvidsson, G.O.; Kristjansson, J.K.; Dabrowski, S.; et al. Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl. Environ. Microbiol. 2014, 80, 886–895. [Google Scholar] [CrossRef] [Green Version]
- Schuch, R.; Khan, B.K.; Raz, A.; Rotolo, J.A.; Wittekind, M. Bacteriophage Lysin CF-301, a Potent Antistaphylococcal Biofilm Agent. Am. Soc. Microbiol. Antimicrob. Agents Chemother. 2017, 61, e02666-16. [Google Scholar] [CrossRef] [Green Version]
- Donovan, D.; Dong, S.; Garrett, W.; Rousseau, G.M.; Moineau, S.; Pritchard, D. Peptidoglycan Hydrolase Fusions Maintain Their Parental Specificities. Appl. Environ. Microbiol. 2006, 72, 2988–2996. [Google Scholar] [CrossRef] [Green Version]
- Skowron, P.M.; Krawczun, N.; Żebrowska, J.; Krefft, D.; Żołnierkiewicz, O.; Bielawa, M.; Jeżewska-Frąckowiak, J.; Janus, Ł.; Witkowska, M.; Palczewska, M.; et al. Data regarding a new, vector-enzymatic DNA fragment amplification-expression technology for the construction of artificial, concatemeric DNA, RNA and proteins, as well as biological effects of selected polypeptides obtained using this method. Data Brief. 2019, 28, 105069. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Donovan, D.M.; Martínez, B.; Rodríguez, A.; García, P. Peptidoglycan Hydrolytic Activity of Bacteriophage Lytic Proteins in Zymogram Analysis. In Bacteriophages: Methods in Molecular Biology; Clokie, M., Kropinski, A., Lavigne, R., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1898, pp. 107–115. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Szadkowska, M.; Kaczorowska, A.K. A novel cryptic clostridial peptide that kills bacteria by cell membrane permeabilization mechanism. Microbiol. Spectr. 2022; rewieved. [Google Scholar]


| Barrier | Enzyme | Enzyme Action | References | |
|---|---|---|---|---|
| Phospholipid membrane | Holins (small proteins accumulate in the cytoplasmic membrane without impairing the cell during the expression of late bacteriophage genes) | Permeabilization of the cell membrane (allows endolysin to transfer from the cytoplasm to the cell wall) and preliminary degradation of the peptidoglycan layer | [7,8,9,10,11,12,13] | |
| Peptidoglycan wall | Endolysins (peptidoglycan hydrolases and depolymerases) divide into four groups and are expressed in the final stage of bacteriophage infection | Glycosidases | Hydrolyzation of the β-1,4-glycosidic linkage of peptidoglycan behind the N-acetylglucosamine (GlcNAc) terminal and hydrolyzation of the glycosidic bonds at the end of N-acetylmuramyl (MurNAc) | [12,14,15,16,17,18] |
| Amido- hydrolases | Digestion of the amide bonds between MurNAc and the peptide residue L-alanine of bacterial peptidoglycan (commonly found in bacteriophage endolysins) | |||
| Endo- peptidases | Hydrolyzation of peptide bonds in the peptides’ core directly attached to N-acetylmuramine acid or between peptides forming cross bridges and connecting the peptides | |||
| Lytic trans- glicosylases | Digestion of the β-1,4-glycosidic bond between residues MurNAc acid and GlcNAc acid with a different mechanism than glycosidases | |||
| Polysaccharide layer | Polysaccharide depolymerases (present in a bacteriophage tail or diffused to the medium) | Glycanases | Degradation of the bacterial macromolecular carbohydrates in the capsule or structural polysaccharides (these help the bacteriophage in every stage of the lytic cycle) | [19,20] |
| Polysaccharide lyases | ||||
| Species | Strain | Growth Temperature | Gram Staining | Activity of TP84_28 | |
|---|---|---|---|---|---|
| Turbidity Assay | Spot Assay | ||||
| G. stearothermophilus | strain 10 | 55 °C/T | P | ++ | ++ |
| Bacillus stearothermophilus | 55 °C/T | P | + | ++ | |
| Geobacillus | ICI | 60 °C/T | P | +++ | +++ |
| Bacillus cereus | 37 °C/M | P | +/− | + | |
| Bacillus subtilis | 37 °C/M | P | + | + | |
| Escherichia coli | DH11S/ATCC | 37 °C/M | N | +/− | + |
| Thermus thermophilus | HB27/ATCC | 65 °C/T | N | +/− | + |
| Thermus aquaticus | YT/ATCC | 65 °C/T | N | +/− | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Żebrowska, J.; Żołnierkiewicz, O.; Ponikowska, M.; Puchalski, M.; Krawczun, N.; Makowska, J.; Skowron, P. Cloning and Characterization of a Thermostable Endolysin of Bacteriophage TP-84 as a Potential Disinfectant and Biofilm-Removing Biological Agent. Int. J. Mol. Sci. 2022, 23, 7612. https://doi.org/10.3390/ijms23147612
Żebrowska J, Żołnierkiewicz O, Ponikowska M, Puchalski M, Krawczun N, Makowska J, Skowron P. Cloning and Characterization of a Thermostable Endolysin of Bacteriophage TP-84 as a Potential Disinfectant and Biofilm-Removing Biological Agent. International Journal of Molecular Sciences. 2022; 23(14):7612. https://doi.org/10.3390/ijms23147612
Chicago/Turabian StyleŻebrowska, Joanna, Olga Żołnierkiewicz, Małgorzata Ponikowska, Michał Puchalski, Natalia Krawczun, Joanna Makowska, and Piotr Skowron. 2022. "Cloning and Characterization of a Thermostable Endolysin of Bacteriophage TP-84 as a Potential Disinfectant and Biofilm-Removing Biological Agent" International Journal of Molecular Sciences 23, no. 14: 7612. https://doi.org/10.3390/ijms23147612
APA StyleŻebrowska, J., Żołnierkiewicz, O., Ponikowska, M., Puchalski, M., Krawczun, N., Makowska, J., & Skowron, P. (2022). Cloning and Characterization of a Thermostable Endolysin of Bacteriophage TP-84 as a Potential Disinfectant and Biofilm-Removing Biological Agent. International Journal of Molecular Sciences, 23(14), 7612. https://doi.org/10.3390/ijms23147612






